Impaired Fat Absorption from Intestinal Tract in High-Fat Diet Fed Male Mice Deficient in Proglucagon-Derived Peptides
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Mice and Diet
2.2. Plasma Biochemical Analyses
2.3. Oral Fat Tolerance Test (OFTT) and Intraperitoneal Fat Tolerance Test (IPFTT)
2.4. Isolation of RNA and Quantitative PCR Analysis of Various Tissues
2.5. Measurement of Food Consumption
2.6. Measurement of Hepatic Triglyceride, FFA, Total Cholesterol and Glycogen Content
2.7. Immunoblotting Analysis
2.8. Histology
2.9. Measurement of Fecal Triglyceride, FFA, and Total Cholesterol
2.10. DNA Samples
2.11. 16S rRNA Next-Generation Sequencing
2.12. Quantitative PCR Analysis of Gut Microbiomes
2.13. Statistical Analysis
3. Results
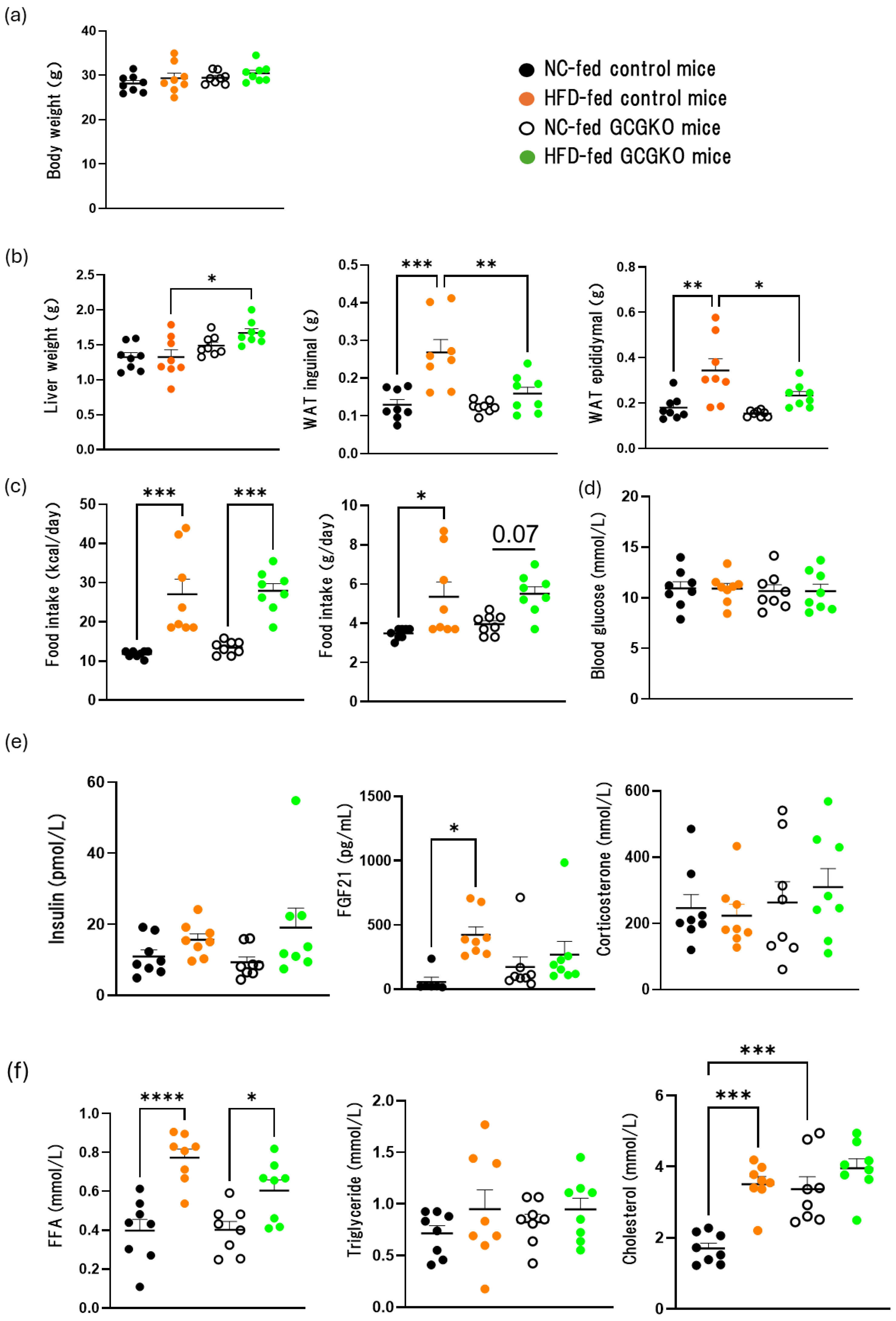
3.1. The Effects of HFD Feeding on the Weight of Various Tissues and Hormones in the Presence and Absence of Proglucagon-Derived Peptides (PGDPs)
3.2. Fat Accumulation in the Liver Was Decreased in HFD-Fed GCGKO Mice Compared to That in HFD-Fed Control Mice
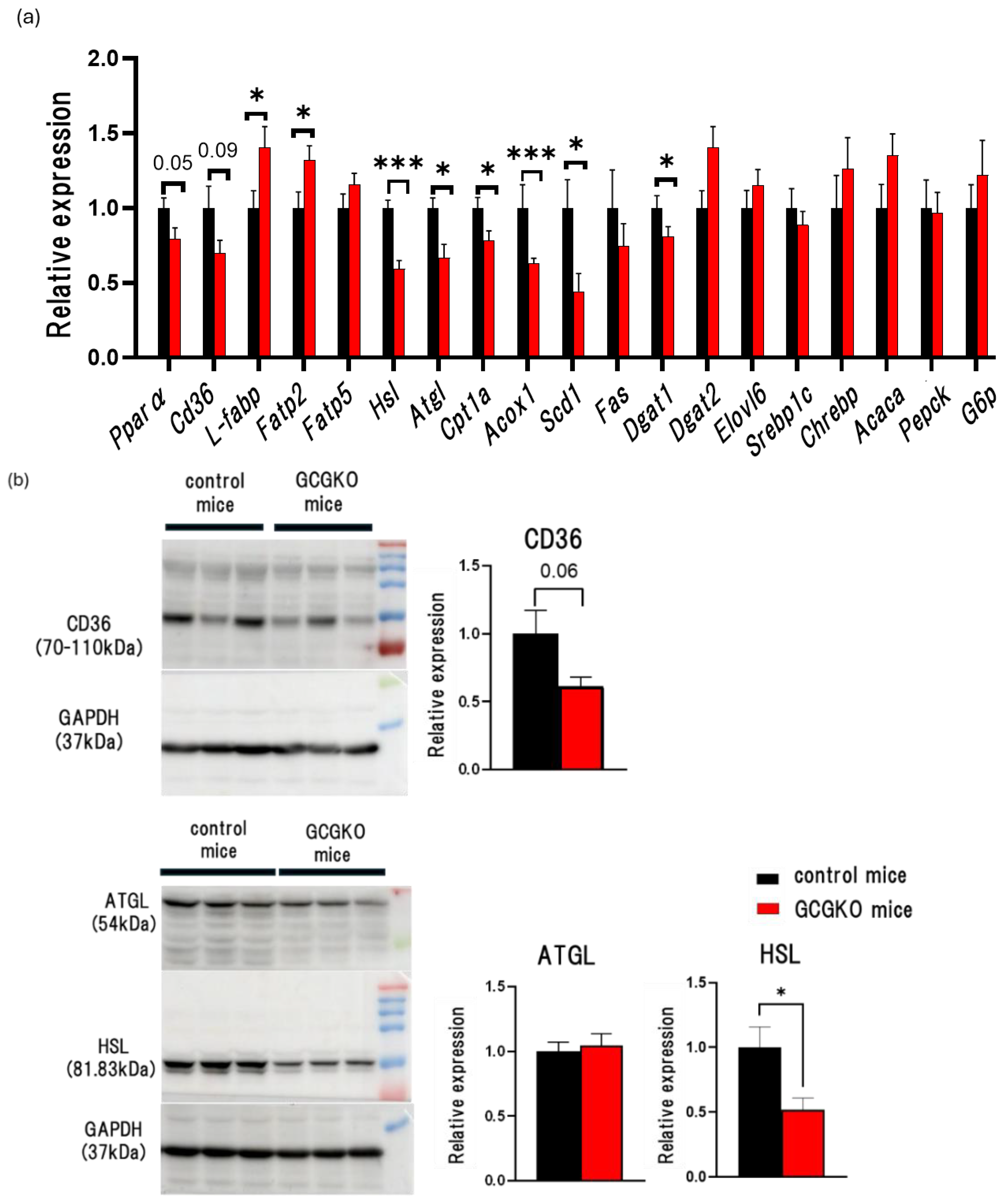
3.3. Expression Levels of Genes Involved in β-Oxidation in the Liver of HFD-Fed GCGKO Mice and HFD-Fed Control Mice
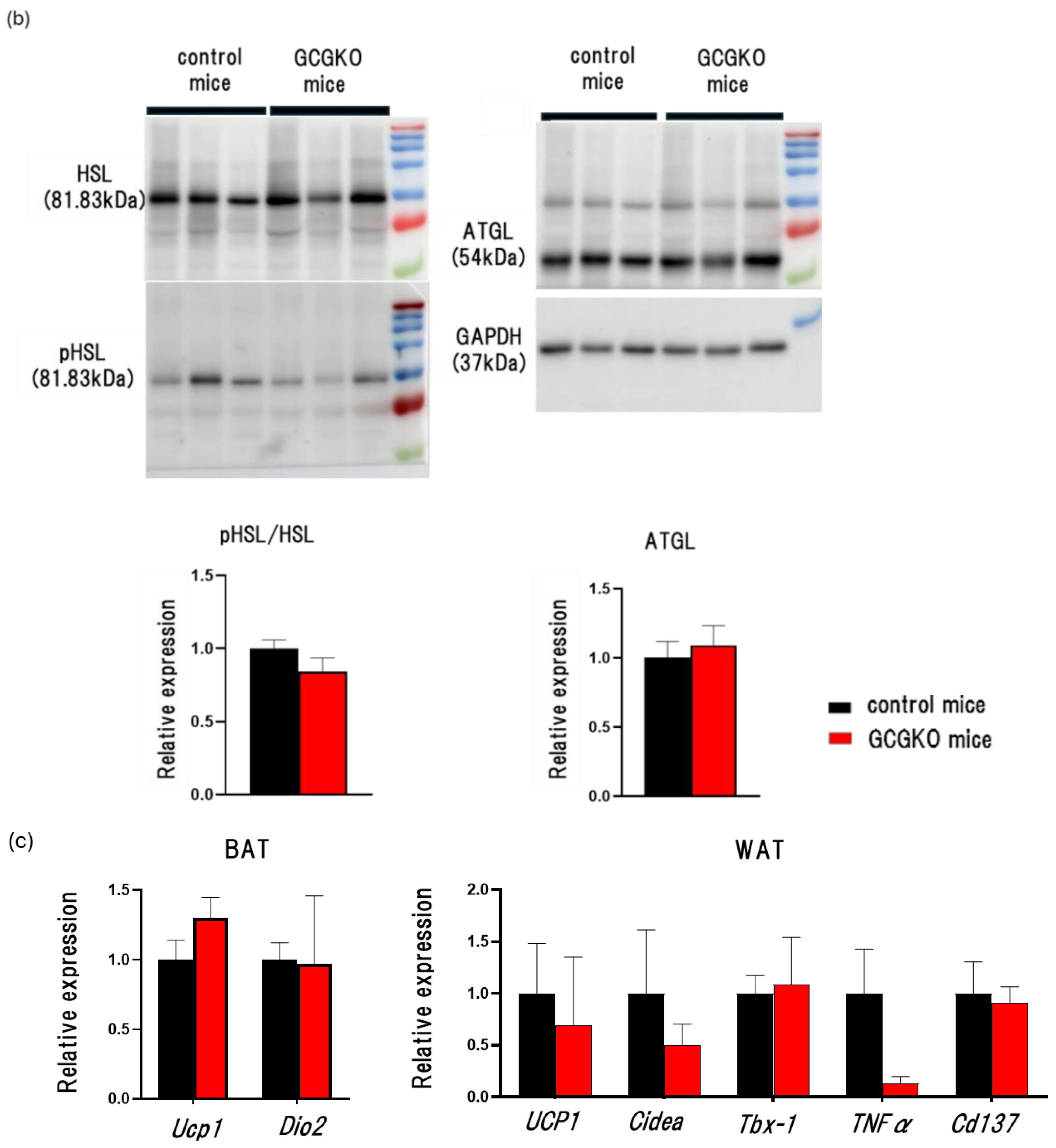
3.4. The Effects of HFD Feeding and Proglucagon-Derived Peptides (PGDPs) on Adipose Tissue
3.5. Fat Absorption from the Intestinal Tract Was Decreased in HFD-Fed GCGKO Mice Compared to That in HFD-Fed Control Mice
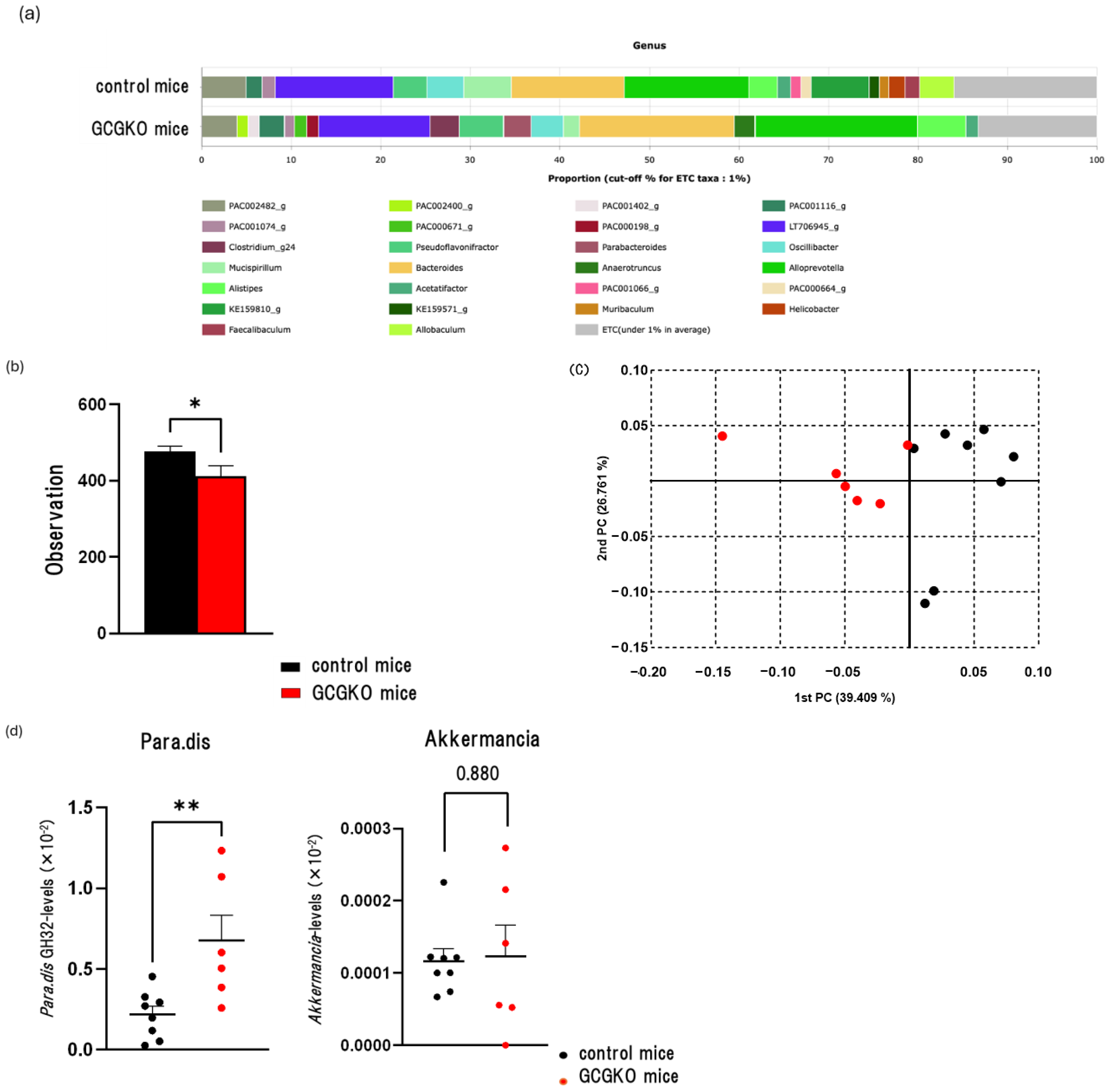
3.6. The Gut Microbiomes Differed between HFD-Fed Control Mice and HFD-Fed GCGKO Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kieffer, T.J.; Habener, J.F. The glucagon-like peptides. Endocr. Rev. 1999, 20, 876–913. [Google Scholar] [CrossRef] [PubMed]
- Penhos, J.C.; Wu, C.H.; Daunas, J.A.; Levine, R. Effect of glucagon on lipid and glycogen content of the perfused rat liver. Biochim. Et Biophys. Acta BBA Lipids Lipid Metab. 1967, 144, 678–680. [Google Scholar] [CrossRef]
- Chiasson, J.L.; Liljenquist, J.E.; Sinclair-Smith, B.C.; Lacy, W.W. Gluconeogenesis from alanine in normal postabsortive man.Intrahepatic stimulatory effect of glucagon. Diabetes 1975, 24, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Boden, G.; Rezvani, I.; Owen, O.E. Effects of glucagon on plasma amino acids. J. Clin. Investig. 1984, 73, 785–793. [Google Scholar] [CrossRef]
- Janah, L.; Kjeldsen, S.; Galsgaard, K.D.; Winther-Sørensen, M.; Stojanovska, E.; Pedersen, J.; Knop, F.K.; Holst, J.J.; Wewer Albrechtsen, N.J. Glucagon Receptor Signaling and Glucagon Resistance. Int. J. Mol. Sci. 2019, 20, 3314. [Google Scholar] [CrossRef]
- Pégorier, J.P.; Garcia-Garcia, M.V.; Prip-Buus, C.; Duée, P.H.; Kohl, C.; Girard, J. Induction of ketogenesis and fatty acid oxidation by glucagon and cyclic AMP in induced hepatocytes from rabbit fetuses. Biochem. J. 1989, 264, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.; Steinberg, D. Effect of hormones on lipolysis and esterification of free fatty acid during incubation of adipose tissue in vitro. J. Lipid Res. 1963, 4, 193–199. [Google Scholar] [CrossRef]
- Rodbell, M.; Jones, A.B. Metabolism of isolated fat cells. 3. The similar inhibitory action of phospholipase C (Clostridium perfringens alpha toxin) and of insulin on lipolysis stimulated by lipolytic hormones and theophylline. J. Biol. Chem. 1966, 241, 140–142. [Google Scholar] [CrossRef]
- Pozza, G.; Pappalettera, A.; Melogli, O.; Viberti, G.; Ghidoni, A. Lipolytic effect of intra-arterial injection of glucagon in man. Horm. Metab. Res. 1971, 3, 291–292. [Google Scholar] [CrossRef]
- Eaton, R.P. Hypolipemic action of glucagon in experimental endogenous lipemia in the rat. J. Lipid Res. 1973, 14, 312–318. [Google Scholar] [CrossRef]
- Aubry, F.; Marcel, Y.L.; Davignon, J. Effects of glucagon on plasma lipids in different types of primary hyperlipoproteinemia. Metabolism 1974, 23, 225–238. [Google Scholar] [CrossRef]
- Guettet, C.; Mathe, D.; Riottot, M.; Lutton, C. Effect of chronic glucagon administration on cholesterol and bile acid metabolism. Biochim. Et Biophys. Acta BBA Lipids Lipid Metab. 1988, 963, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.F.; Salter, A.M.; Fears, R.; Brindley, D.N. Glucagon, cyclin AMP and adrenaline stimulate the degradation of low-density lipoprotein by culture rat hepatocytes. Biochem. J. 1989, 262, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Nason, S.; Holleman, C.; Pepin, M.; Wilson, L.; Berryhill, T.F.; Wende, A.R.; Steele, C.; Young, M.E.; Barnes, S.; et al. Glucagon Receptor Signaling Regulates Energy Metabolism via Hepatic Farnesoid X Receptor and Fibroblast Growth Factor 21. Diabetes 2018, 67, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Nason, S.R.; Antipenko, J.; Presedo, N.; Cunningham, S.E.; Pierre, T.H.; Kim, T.; Paul, J.R.; Holleman, C.; Young, M.E.; Gamble, K.L.; et al. Glucagon receptor signaling regulates weight loss via central KLB receptor complexes. JCI Insight 2021, 6, e141323. [Google Scholar] [CrossRef] [PubMed]
- Sloop, K.W.; Cao, J.X.; Siesky, A.M.; Zhang, H.Y.; Bodenmiller, D.M.; Cox, A.L.; Jacobs, S.J.; Moyers, J.S.; Owens, R.A.; Showalter, A.D.; et al. Hepatic and glucagon-like peptide-1-mediated reversal of diabetes by glucagon receptor anti-sense oligonucleotide inhibitors. J. Clin. Investig. 2004, 113, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.X.; Quittner-Strom, E.B.; Lee, Y.; Johnson, J.A.; Martin, S.A.; Yu, X.; Li, J.; Lu, J.; Cai, Z.; Chen, S.; et al. Glucagon Receptor Antagonism Improves Glucose Metabolism and Cardiac Function by Promoting AMP-Mediated Protein Kinase in Diabetic Mice. Cell Rep. 2018, 22, 1760–1773. [Google Scholar] [CrossRef] [PubMed]
- Conarello, S.L.; Jiang, G.; Mu, J.; Li, Z.; Woods, J.; Zycband, E.; Ronan, J.; Liu, F.; Roy, R.S.; Zhu, L.; et al. Glucagon receptor knockout mice are resistant to diet-induced obesity and streptozotocin-mediated beta cell loss and hyperglycaemia. Diabetologia 2007, 50, 142–150. [Google Scholar] [CrossRef]
- Iida, A.; Seino, Y.; Fukami, A.; Maekawa, R.; Yabe, D.; Shimizu, S.; Kinoshita, K.; Takagi, Y.; Izumoto, T.; Ogata, H.; et al. Endogenous GIP ameliorates impairment of insulin secretion in proglucagon-deficient mice under moderate beta cell damage induced by streptozotocin. Diabetologia 2016, 59, 1533–1541. [Google Scholar] [CrossRef]
- Han, S.; Akiyama, T.E.; Previs, S.F.; Herath, K.; Roddy, T.P.; Jensen, K.K.; Guan, H.P.; Murphy, B.A.; McNamara, L.A.; Shen, X.; et al. Effects of small interfering RNA-mediated hepatic glucagon receptor inhibition on lipid metabolism in db/db mice. J. Lipid Res. 2013, 54, 2615–2622. [Google Scholar] [CrossRef]
- Guan, H.P.; Yang, X.; Lu, K.; Wang, S.P.; Castro-Prez, J.M.; Previs, S.; Wright, M.; Shah, V.; Herath, K.; Xie, D.; et al. Glucagon receptor antagonism induces increased cholesterol absorption. J. Lipid Res. 2015, 56, 2183–2195. [Google Scholar] [CrossRef]
- Ali, S.; Lamont, B.J.; Charron, M.J.; Drucker, D.J. Dual elimination of the glucagon and GLP-1 receptors in mice reveals plasticity in the incretin axis. J. Clin. Investig. 2011, 121, 1917–1929. [Google Scholar] [CrossRef] [PubMed]
- Drucker, D.J.; Habener, J.F.; Holst, J.J. Discovery, characterization, and clinical development of the glucagon-like peptides. J. Clin. Investig. 2017, 127, 4217–4227. [Google Scholar] [CrossRef] [PubMed]
- Hein, G.J.; Baker, C.; Hsieh, J.; Farr, S.; Adeli, K. GLP-1 and GLP-2 as yin and yang of intestinal lipoprotein production: Evidence for predominance of GLP-2-stimulated postprandial lipemia in normal and insulin-resistant states. Diabetes 2013, 62, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Longuet, C.; Baker, C.L.; Qin, B.; Federico, L.M.; Drucker, D.J.; Adeli, K. The glucagon-like peptide 1 receptor is essential for postprandial lipoprotein synthesis and secretion in hamsters and mice. Diabetologia 2010, 53, 552–561. [Google Scholar] [CrossRef]
- Hsieh, J.; Longuet, C.; Maida, A.; Bahrami, J.; Xu, E.; Baker, C.L.; Brubaker, P.L.; Drucker, D.J.; Adeli, K. Glucagon-like peptide-2 increases intestinal lipid absorption and chylomicron production via CD36. Gastroenterology 2009, 137, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Takagi, Y.; Kinoshita, K.; Ozaki, N.; Seino, Y.; Murata, Y.; Oshida, Y.; Hayashi, Y. Mice Deficient in Proglucagon-Derived Peptides Exhibit Glucose Intolerance on a High-Fat Diet but Are Resistant to Obesity. PLoS ONE 2015, 10, e0138322. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Seino, Y.; Hidaka, S.; Maekawa, R.; Takano, Y.; Yamamoto, M.; Hori, M.; Yokota, K.; Masuda, A.; Himeno, T.; et al. High Protein Diet Feeding Aggravates Hyperaminoacidemia in Mice Deficient in Proglucagon-Derived Peptides. Nutrients 2022, 14, 975. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Yamamoto, M.; Mizoguchi, H.; Watanabe, C.; Ito, R.; Yamamoto, S.; Sun, X.Y.; Murata, Y. Mice deficient for glucagon gene-derived peptides display normoglycemia and hyperplasia of islet α-cells but not of intestinal L-cells. Mol. Endocrinol. 2009, 23, 1990–1999. [Google Scholar] [CrossRef]
- Watanabe, C.; Seino, Y.; Miyahira, H.; Yamamoto, M.; Fukami, A.; Ozaki, N.; Takagishi, Y.; Sato, J.; Fukuwatari, T.; Shibata, K.; et al. Remodeling of hepatic metabolism and hyperaminoacidemia in mice deficient in proglucagon-derived peptides. Diabetes 2012, 61, 74–84. [Google Scholar] [CrossRef]
- Varin, E.M.; Hanson, A.A.; Beaudry, J.L.; Nguyen, M.A.; Cao, X.; Baggio, L.L.; Mulvihill, E.E.; Drucker, D.J. Hematopoietic cell- versus enterocyte-derived dipeptidyl peptidase-4 differentially regulates triglyceride excursion in mice. JCI Insight 2020, 5, e140418. [Google Scholar] [CrossRef]
- Lei, X.L.; Wong, G.W. C1q/TNF-related protein 2 (CTRP2) deletion promotes adipose tissue lipolysis and hepatic triglyceride secretion. J. Biol. Chem. 2019, 294, 15638–15649. [Google Scholar] [CrossRef]
- Masuda, A.; Seino, Y.; Murase, M.; Hidaka, S.; Shibata, M.; Takayanagi, T.; Sugimura, Y.; Hayashi, Y.; Suzuki, A. Short-term high-starch, low-protein diet induces reversible increase in β-cell mass independent of body weight gain in mice. Nutrients 2019, 11, 1045. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Fujii, T.; Yamakawa, S.; Yamada, C.; Fujiki, K.; Kondo, N.; Funasaka, K.; Hirooka, Y.; Tochio, T. Combined oral intake of short and long fructans alters the gut microbiota in food allergy model mice and contributes to food allergy pre-vention. BMC Microbiol. 2023, 23, 266. [Google Scholar] [CrossRef] [PubMed]
- Furet, J.P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Doré, J.; Corthier, G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef]
- Hul, M.V.; Cani, P.D. The gut microbiota in obesity and weight management: Microbes as friends or foe? Nat. Rev. Endocrinol. 2023, 19, 258–271. [Google Scholar]
- Soeda, S.; Sasako, T.; Enooku, K.; Kubota, N.; Kobayashi, N.; Matsumoto, Y.; Awazawa, M.; Bouchi, R.; Toda, G.; Yamada, T.; et al. Gut insulin action protects from hepatocarcinogenesis in diabetic mice comorbid with nonalcoholic steatohepatitis. Nat. Commun. 2023, 14, 6584. [Google Scholar] [CrossRef]
- Kato, S.; Sato, T.; Fujita, H.; Kawatani, M.; Yamada, Y. Effects of GLP-1 receptor agonist on changes in the gut bacterium and the underlying mechanisms. Sci. Rep. 2021, 11, 9167. [Google Scholar] [CrossRef]
- Madsen, M.S.A.; Holm, J.B.; Pallejà, A.; Wismann, P.; Fabricius, K.; Rigbolt, K.; Mikkelsen, M.; Sommer, M.; Jelsing, J.; Nielsen, H.B.; et al. Metabolic and gut microbiome changes following GLP-1 or dual GLP-1/GLP-2 receptor agonist treatment in di-et-induced obese mice. Sci. Rep. 2019, 9, 15582. [Google Scholar] [CrossRef]
- Ferré, P.; Foufelle, F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes. Metab. 2010, 2, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Koonen, D.P.Y.; Jacobs, R.L.; Febbraio, M.; Young, M.E.; Soltys, C.M.; Ong, H.; Vance, D.E.; Dyck, J.R.B. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 2007, 56, 2863–2871. [Google Scholar] [CrossRef]
- Ge, F.; Zhou, S.; Hu, C.; Lobdell IV, H.; Berk, P.D. Insulin- and leptin-regulated fatty acid uptake plays a key causal role in hepatic steatosis in mice with intact leptin signaling but not in ob/ob or db/db mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G855–G866. [Google Scholar] [CrossRef]
- Wilson, C.G.; Tran, J.L.; Erion, D.M.; Vera, N.B.; Febbraio, M.; Weiss, E.J. Hepatocyte-Specific Disruption of CD36 Attenuates Fatty Liver and Improves Insulin Sensitivity in HFD-Fed Mice. Endocrinology 2016, 157, 570–585. [Google Scholar] [CrossRef]
- Geisler, C.E.; Renquist, B.J. Hepatic lipid accumulation: Cause and consequence of dysregulated glucoregulatory hormones. J. Endocrinol. 2017, 234, R1–R21. [Google Scholar] [CrossRef]
- Montagner, A.; Polizzi, A.; Fouché, E.; Ducheix, S.; Lippi, Y.; Lasserre, F.; Barquissau, V.; Régnier, M.; Lukowicz, C.; Benhamed, F.; et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 2016, 65, 1202–1214. [Google Scholar] [CrossRef]
- Stec, D.E.; Gordon, D.M.; Hipp, J.A.; Hong, S.; Mitchell, Z.L.; Franco, N.R.; Robison, J.W.; Anderson, C.D.; Stec, D.F.; Hinds, T.D., Jr. Loss of hepatic PPARα promotes inflammation and serum hyperlipidemia in diet-induced obesity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 317, R733–R745. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; MacDougall, M.L.; McDowell, M.T.; Xi, L.; Wei, R.; Zavadoski, W.J.; Molloy, M.P.; Baker, J.D.; Kuhn, M.; Cabrera, O.; et al. Polyomic profiling reveals significant hepatic metabolic alterations in glucagon-receptor (GCGR) knockout mice: Im-plications on anti-glucagon therapies for diabetes. BMC Genom. 2011, 12, 281. [Google Scholar] [CrossRef]
- Galsgaard, K.D.; Elmelund, E.; Johansen, C.D.; Bomholt, A.B.; Kizilkaya, H.S.; Ceutz, F.; Hunt, J.E.; Kissow, H.; Winther-Sørensen, M.; Sørensen, C.M.; et al. Glucagon receptor antagonism impairs and glucagon receptor agonism enhances triglycerides metabolism in mice. Mol. Metab. 2022, 66, 101639. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.J.; Zhang, D.; Guerra, M.T.; Brill, A.L.; Goedeke, L.; Nasiri, A.R.; Rabin-Court, A.; Wang, Y.; Peng, L.; Dufour, S.; et al. Glucagon stimulates gluconeogenesis by INSP3R1-mediated hepatic lipolysis. Nature 2020, 579, 279–283. [Google Scholar] [CrossRef]
- Steneberg, P.; Sykaras, A.G.; Backlund, F.; Straseviciene, J.; Söderström, I.; Edlund, H. Hyperinsulinemia Enhances Hepatic Expression of the Fatty Acid Transporter Cd36 and Provokes Hepatosteatosis and Hepatic Insulin Resistance. J. Biol. Chem. 2015, 290, 19034–19043. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Seino, Y.; Hidaka, S.; Nakatani, M.; Hitachi, K.; Murao, N.; Maeda, Y.; Fujisawa, H.; Shibata, M.; Takayanagi, T.; et al. Blockade of glucagon increases muscle mass and alters fiber type composition in mice deficient in proglucagon-derived peptides. J. Diabetes Investig. 2023, 14, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, A.; Marx, T.; Beaudry, J.L.; Stern, J.H. Glucagon receptor signaling at white adipose tissue does not regulate lipolysis. Am. J. Physiol. Endocrinol. Metab. 2022, 323, E389–E401. [Google Scholar] [CrossRef] [PubMed]
- Uno, K.; Yamada, T.; Ishigaki, Y.; Imai, J.; Hasegawa, Y.; Sawada, S.; Kaneko, K.; Ono, H.; Asano, T.; Oka, Y.; et al. A hepatic amino acid/mTOR/S6K-dependent signalling pathway modulates systemic lipid metabolism via neuronal signals. Nat. Commun. 2015, 6, 7940. [Google Scholar] [CrossRef] [PubMed]
- Izumida, Y.; Yahagi, N.; Takeuchi, Y.; Nishi, M.; Shikama, A.; Takarada, A.; Masuda, Y.; Kubota, M.; Matsuzaka, T.; Nakagawa, Y.; et al. Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization. Nat. Commun. 2013, 4, 2316. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.; Sendo, M.; Dezaki, K.; Hira, T.; Sato, T.; Nakata, M.; Goswami, C.; Aoki, R.; Arai, T.; Kumari, P.; et al. GLP-1 release and vagal afferent activation mediate the beneficial metabolic and chronotherapeutic effects of D-allulose. Nat. Commun. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Yusta, B.; Baggio, L.L.; Varin, E.M.; Matthews, D.; Drucker, D.J. Loss of Glp2r signaling activates hepatic stellate cells and exacerbates diet-induced steatohepatitis in mice. JCI Insight 2020, 5, e136907. [Google Scholar] [CrossRef] [PubMed]
- Taher, J.; Baker, C.; Alvares, D.; Ijaz, L.; Hussain, M.; Adeli, K. GLP-2 Dysregulates Hepatic Lipoprotein Metabolism, Inducing Fatty Liver and VLDL Overproduction in Male Hamsters and Mice. Endocrinology 2018, 159, 3340–3350. [Google Scholar] [CrossRef]
- Baldassano, S.; Rappa, F.; Amato, A.; Cappello, F.; Mulè, F. GLP-2 as Beneficial Factor in the Glucose Homeostasis in Mice Fed a High Fat Diet. J. Cell Physiol. 2015, 230, 3029–3036. [Google Scholar] [CrossRef]
- Stojanović, O.; Altirriba, J.; Rigo, D.; Spiljar, M.; Evrard, E.; Roska, B.; Fabbiano, S.; Zamboni, N.; Maechler, P.; Rohner-Jeanrenaud, F.; et al. Dietary excess regulates absorption and surface of gut epithelium through intestinal PPARα. Nat. Commun. 2021, 12, 7031. [Google Scholar] [CrossRef]
- Longuet, C.; Sinclair, E.M.; Maida, A.; Baggio, L.L.; Maziarz, M.; Charron, M.J.; Drucker, D.J. The glucagon receptor is required for the adaptive metabolic response to fasting. Cell Metab. 2008, 8, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Fujisaka, S.; Morinaga, Y.; Watanabe, S.; Allah, N.; Hatta, H.; Kado, T.; Nishimura, A.; Bilal, M.; Aslam, M.R.; et al. Isoxanthohumol improves obesity and glucose metabolism via inhibiting intestinal lipid absorption with a bloom of Akkermansia muciniphila in mice. Mol. Metab. 2023, 77, 101797. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Qin, X.; Jia, H.; Chen, S.; Sun, W.; Wang, X. The association between gut microbiota composition and BMI in Chinese male college students, as analysed by next-generation sequencing. Br. J. Nutr. 2019, 122, 986–995. [Google Scholar] [CrossRef] [PubMed]




| CATYGGCTGTTATCCGCAGC | |
| CP042830.1 | TTGAACGACCGGTTCAGCTATGGAATCGGCATCGGCTGTTATCCGCAGCTTGGGGATTATCCTTTCCTGCCCCTTATCCAGTTTAAATGG |
| CP048438.1 | TTGAACGACCGGTTCAGCTATGGAATCGGCATTGGCTGTTATCCGCAGCTTGGGGATTATCCTTTCCTGCCCCTTATCCAGTTTAAATGG |
| CP084201.1 | TTGAACGACCGGTTCAGCTATGGAATCGGCATTGGCTGTTATCCGCAGCTTGGGGATTATCCTTTCCTGCCCCTTATCCAGTTTAAATGG |
| CP010553.1 | TTGAACGACCGGTTCAGCTATGGAATCGGCATCGGCTGTTATCCGCAGCTTGGGGATTATCCTTTCCTGCCCCTTATCCAGTTTAAATGG |
| CP025823.1 | TTGAACGACCGGTTCAGCTATGGAATCGGCATCGGCTGTTATCCGCAGCTTGGGGATTATCCTTTCCTGCCCCTTATCCAGTTTAAATGG |
| AGGTGAGCGATGGGTTGAAG | |
| CP042830.1 | GAAGCTTCCCGCAATTGGACGCTTCAGCTGGAAGGGGCCCGCCTTTCCTATATCAACAAGGTGAGCGATGGGTTGAAGTGGGGACCCTTC |
| CP048438.1 | GAAGCTTCCCGCAATTGGACGCTTCAGCTGGAAGGGGCCCGCCTTTCCTATATCAACAAGGTGAGCGATGGGTTGAAGTGGGGACCCTTC |
| CP084201.1 | GAAGCTTCCCGCAATTGGACGCTTCAGCTGGAAGGGGCCCGCCTTTCCTATATCAACAAGGTGAGCGATGGGTTGAAGTGGGGACCCTTC |
| CP010553.1 | GAAGCTTCCCGCAATTGGACGCTTCAGCTGGAAGGGGCCCGCCTTTCCTATATCAACAAGGTGAGCGATGGGTTGAAGTGGGGACCCTTC |
| CP025823.1 | GAAGCTTCCCGCAATTGGACGCTTCAGCTGGAAGGGGCCCGCCTTTCCTATATCAACAAGGTGAGCGATGGGTTGAAGTGGGGACCCTTC |
| Taxon Name | LDA Effect Size | p-Value | Taxonomic Relative Abundance | |
|---|---|---|---|---|
| Ctrl | GCGKO | |||
| Anaerotruncus | 4.0296 | 0.001 | 0.26952 | 2.31236 |
| Muribaculum | 3.51248 | 0.001 | 1.06293 | 0.33961 |
| Lactobacillus | 3.02518 | 0.001 | 0.21994 | 0.03216 |
| ASTB_g | 3.20119 | 0.006 | 0.40544 | 0.04149 |
| Parabacteroides | 4.12931 | 0.006 | 0.56143 | 3.07072 |
| Odoribacter | 3.16979 | 0.014 | 0.34725 | 0.07691 |
| PAC001066_g | 3.5805 | 0.014 | 1.14625 | 0.44854 |
| Faecalibaculum | 3.78907 | 0.019 | 1.62686 | 0.25278 |
| KE159600_g | 3.25465 | 0.020 | 0.42597 | 0.08407 |
| PAC000664_g | 3.64829 | 0.020 | 1.15243 | 0.29079 |
| Helicobacter | 3.81307 | 0.038 | 1.81157 | 0.40755 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishida, K.; Ueno, S.; Seino, Y.; Hidaka, S.; Murao, N.; Asano, Y.; Fujisawa, H.; Shibata, M.; Takayanagi, T.; Ohbayashi, K.; et al. Impaired Fat Absorption from Intestinal Tract in High-Fat Diet Fed Male Mice Deficient in Proglucagon-Derived Peptides. Nutrients 2024, 16, 2270. https://doi.org/10.3390/nu16142270
Nishida K, Ueno S, Seino Y, Hidaka S, Murao N, Asano Y, Fujisawa H, Shibata M, Takayanagi T, Ohbayashi K, et al. Impaired Fat Absorption from Intestinal Tract in High-Fat Diet Fed Male Mice Deficient in Proglucagon-Derived Peptides. Nutrients. 2024; 16(14):2270. https://doi.org/10.3390/nu16142270
Chicago/Turabian StyleNishida, Koki, Shinji Ueno, Yusuke Seino, Shihomi Hidaka, Naoya Murao, Yuki Asano, Haruki Fujisawa, Megumi Shibata, Takeshi Takayanagi, Kento Ohbayashi, and et al. 2024. "Impaired Fat Absorption from Intestinal Tract in High-Fat Diet Fed Male Mice Deficient in Proglucagon-Derived Peptides" Nutrients 16, no. 14: 2270. https://doi.org/10.3390/nu16142270
APA StyleNishida, K., Ueno, S., Seino, Y., Hidaka, S., Murao, N., Asano, Y., Fujisawa, H., Shibata, M., Takayanagi, T., Ohbayashi, K., Iwasaki, Y., Iizuka, K., Okuda, S., Tanaka, M., Fujii, T., Tochio, T., Yabe, D., Yamada, Y., Sugimura, Y., ... Suzuki, A. (2024). Impaired Fat Absorption from Intestinal Tract in High-Fat Diet Fed Male Mice Deficient in Proglucagon-Derived Peptides. Nutrients, 16(14), 2270. https://doi.org/10.3390/nu16142270








