How the Microbiota May Affect Celiac Disease and What We Can Do
Abstract
1. Introduction
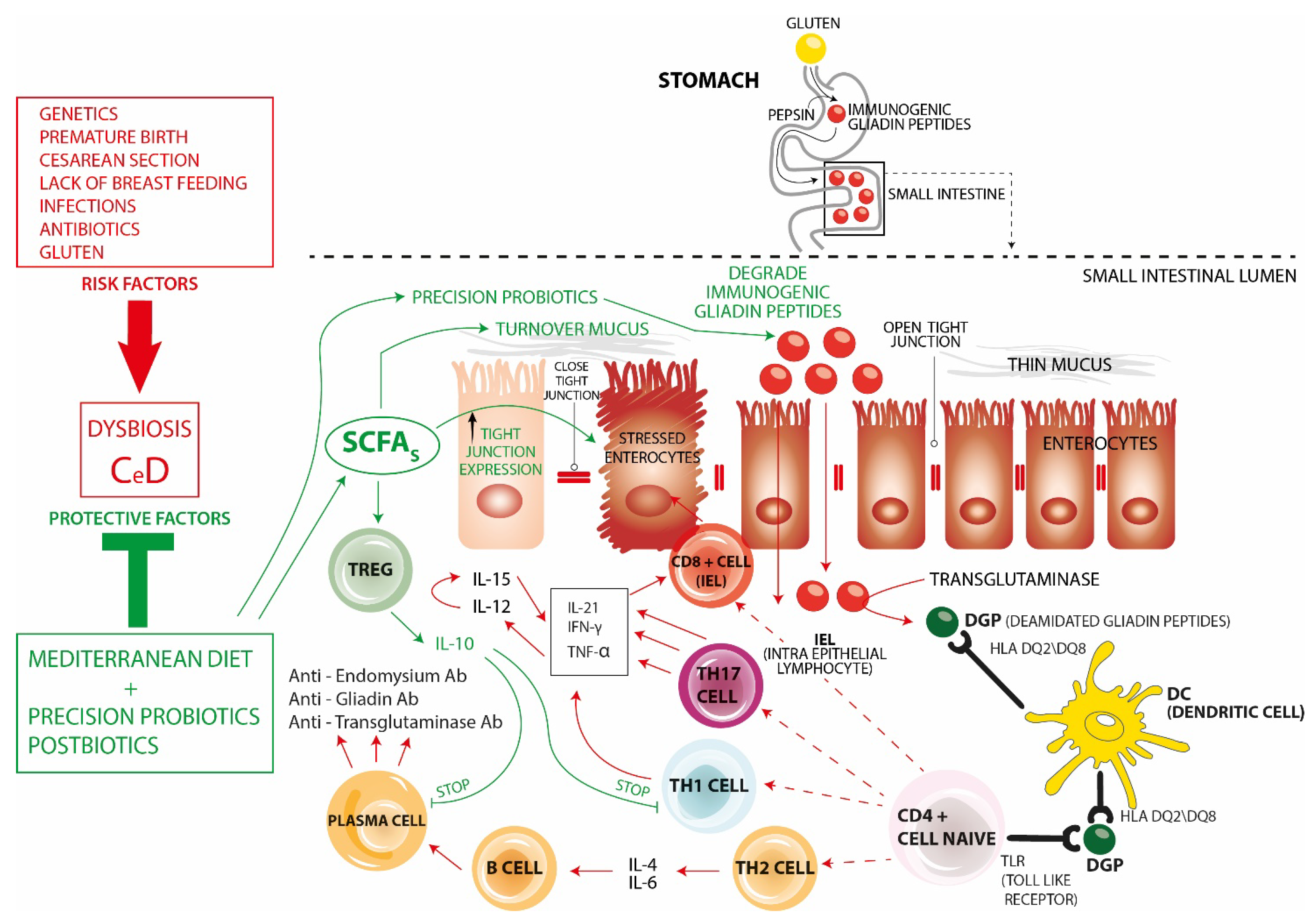
2. Gluten and the Pathophysiology of Celiac Disease
- (1)
- CD8+ cells, which are cytotoxic cells becoming intraepithelial lymphocytes (IEL) that participate in enterocyte apoptosis.
- (2)
- Th17 inflammatory cells that, like Th1 cells, produce IL-21, IFN-γ, and TNF a.
- (3)
- Th1 inflammatory cells producing high levels of IFN-γ, IL-21, and TNF-a.TNF-a stimulates the production of IL-12 and IL-15 which synergistically promote the increase in gluten-mediated IFN-γ production thus further pushing infiltration of IEL promoting enterocyte apoptosis, crypt hyperplasia, and villous atrophy,
- (4)
- Th2 cells. Through the production of IL-4 and IL-6, they stimulate the progression of B-cells into plasma cells and the production of specific autoantibodies against endomysium, gliadin, and transglutaminase, thus participating in intestinal damage.
- (1)
- The use of a Mediterranean-type diet during the first 2 years of life has been shown to have a protective effect in preventing the development of CeD. Such a diet is rich in fiber and phytochemicals capable of stimulating the intestinal growth of eubiotic commensal bacteria that produce adequate proportions of short-chain fatty acids (SCFAs). SCFAs act by counteracting intestinal permeability (increasing mucus turnover and increasing the expression of tight junctions) and also by modulating the immune system. In fact, they promote homeostasis favoring Tregs that produce IL-10 and counteract both Th1 cells and the production of autoantibodies.
- (2)
- “Precision” probiotics are capable of several effects all blunting the inflammatory changes seen in CeD. In fact, they fight pathogenic and inflammatory species, restore eubiotic species producing SCFAs, produce peptidases capable of degrading immunogenic gliadin peptides, promote immune homeostasis through the enhancement of Tregs, modulate the permeability of the intestinal barrier, and produce Aryl receptor (AhR) ligands correlated with increased IL-22, intestinal stem cell proliferation, and restoration of intestinal mucosal damage.
- (3)
- Postbiotics—though so far less investigated in this regard—have the potential to improve gut barrier function by increasing tight junction expression and preventing the inflammatory effects of gliadin.
3. How the Gut Microbiota Is Made and Its Relationship with CeD
4. Early Environmental Factors, Microbiota and CeD
5. Gut Microbiota and the Pathogenesis of CeD
6. The Role of Oral Microbiota
7. Gluten-Free Diet and Gut Microbiota
8. Targeted Microbiota Therapy for CeD
9. Lactobacilli
10. Bifidobacteria
11. Combining Lactobacilli and Bifidobacteria
12. Postbiotics
13. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Rossi, R.E.; Dispinzieri, G.; Elvevi, A.; Massironi, S. Interaction between Gut Microbiota and Celiac Disease: From Pathogenesis to Treatment. Cells 2023, 12, 823. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Liu, X.; Hadley, D.; Hagopian, W.; Liu, E.; Chen, W.M.; Onengut-Gumuscu, S.; Simell, V.; Rewers, M.; Ziegler, A.G.; et al. Identification of Non-HLA Genes Associated with Celiac Disease and Country-Specific Differences in a Large, International Pediatric Cohort. PLoS ONE 2016, 11, e0152476. [Google Scholar] [CrossRef] [PubMed]
- Losurdo, G.; Principi, M.; Iannone, A.; Ierardi, E.; Di Leo, A. The Interaction between Celiac Disease and Intestinal Microbiota. J. Clin. Gastroenterol. 2016, 50 (Suppl. S2), S145–S147. [Google Scholar] [CrossRef] [PubMed]
- Leonard, M.M.; Camhi, S.; Huedo-Medina, T.B.; Fasano, A. Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease. Nutrients 2015, 7, 9325–9336. [Google Scholar] [CrossRef] [PubMed]
- Dieli-Crimi, R.; Cenit, M.C.; Nunez, C. The genetics of celiac disease: A comprehensive review of clinical implications. J. Autoimmun. 2015, 64, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Krishnareddy, S. The Microbiome in Celiac Disease. Gastroenterol. Clin. N. Am. 2019, 48, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, M.F.; Hinks, T.S.C. MAIT cells and the microbiome. Front. Immunol. 2023, 14, 1127588. [Google Scholar] [CrossRef]
- Wu, X.; Qian, L.; Liu, K.; Wu, J.; Shan, Z. Gastrointestinal microbiome and gluten in celiac disease. Ann. Med. 2021, 53, 1797–1805. [Google Scholar] [CrossRef]
- Simon, E.; Molero-Luis, M.; Fueyo-Diaz, R.; Costas-Batlle, C.; Crespo-Escobar, P.; Montoro-Huguet, M.A. The Gluten-Free Diet for Celiac Disease: Critical Insights to Better Understand Clinical Outcomes. Nutrients 2023, 15, 4013. [Google Scholar] [CrossRef]
- Tian, N.; Faller, L.; Leffler, D.A.; Kelly, C.P.; Hansen, J.; Bosch, J.A.; Wei, G.; Paster, B.J.; Schuppan, D.; Helmerhorst, E.J. Salivary Gluten Degradation and Oral Microbial Profiles in Healthy Individuals and Celiac Disease Patients. Appl. Environ. Microbiol. 2017, 83, e03330-16. [Google Scholar] [CrossRef]
- Fernandez-Feo, M.; Wei, G.; Blumenkranz, G.; Dewhirst, F.E.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin. Microbiol. Infect. 2013, 19, E386–E394. [Google Scholar] [CrossRef] [PubMed]
- Akobeng, A.K.; Singh, P.; Kumar, M.; Al Khodor, S. Role of the gut microbiota in the pathogenesis of coeliac disease and potential therapeutic implications. Eur. J. Nutr. 2020, 59, 3369–3390. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; McCarville, J.L.; Galipeau, H.J.; Deraison, C.; Bernier, S.P.; Constante, M.; Rolland, C.; Meisel, M.; Murray, J.A.; Yu, X.B.; et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat. Commun. 2019, 10, 1198. [Google Scholar] [CrossRef]
- Abadie, V.; Kim, S.M.; Lejeune, T.; Palanski, B.A.; Ernest, J.D.; Tastet, O.; Voisine, J.; Discepolo, V.; Marietta, E.V.; Hawash, M.B.F.; et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature 2020, 578, 600–604. [Google Scholar] [CrossRef]
- Sacchetti, L.; Nardelli, C. Gut microbiome investigation in celiac disease: From methods to its pathogenetic role. Clin. Chem. Lab. Med. 2020, 58, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.; Donat, E.; Ribes-Koninckx, C.; Fernandez-Murga, M.L.; Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013, 79, 5472–5479. [Google Scholar] [CrossRef] [PubMed]
- Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult humans. Gut Microbes 2010, 1, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Cristofori, F.; Indrio, F.; Miniello, V.L.; De Angelis, M.; Francavilla, R. Probiotics in Celiac Disease. Nutrients 2018, 10, 1824. [Google Scholar] [CrossRef]
- Yemula, N. Gut microbiota in celiac disease. Ann. Gastroenterol. 2024, 37, 125–132. [Google Scholar] [CrossRef]
- Sellitto, M.; Bai, G.; Serena, G.; Fricke, W.F.; Sturgeon, C.; Gajer, P.; White, J.R.; Koenig, S.S.; Sakamoto, J.; Boothe, D.; et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 2012, 7, e33387. [Google Scholar] [CrossRef]
- Leonard, M.M.; Karathia, H.; Pujolassos, M.; Troisi, J.; Valitutti, F.; Subramanian, P.; Camhi, S.; Kenyon, V.; Colucci, A.; Serena, G.; et al. Multi-omics analysis reveals the influence of genetic and environmental risk factors on developing gut microbiota in infants at risk of celiac disease. Microbiome 2020, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Leonard, M.M.; Serena, G.; Sturgeon, C.; Fasano, A. Genetics and celiac disease: The importance of screening. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Kaukinen, K.; Tuovinen, E.; Collin, P.; Lindfors, K.; Partanen, J.; Maki, M.; Matto, J. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 2013, 19, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Tohumcu, E.; Raoul, P.; Fiorani, M.; Cintoni, M.; Mele, M.C.; Cammarota, G.; Gasbarrini, A.; Ianiro, G. The role of diet in shaping human gut microbiota. Best Pract. Res. Clin. Gastroenterol. 2023, 62–63, 101828. [Google Scholar] [CrossRef] [PubMed]
- Namatovu, F.; Olsson, C.; Lindkvist, M.; Myleus, A.; Hogberg, U.; Ivarsson, A.; Sandstrom, O. Maternal and perinatal conditions and the risk of developing celiac disease during childhood. BMC Pediatr. 2016, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Pes, G.M.; Bibbo, S.; Dore, M.P. Coeliac disease: Beyond genetic susceptibility and gluten. A narrative review. Ann. Med. 2019, 51, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tanpowpong, P.; Li, S.; Espinola, J.A.; Santos, L.C.; James, K.E.; Powe, C.E.; Camargo, C.A., Jr. Pregnancy- and birth-related risk factors for the development of childhood celiac disease. Acta Paediatr. 2023, 112, 1029–1034. [Google Scholar] [CrossRef]
- Cenit, M.C.; Olivares, M.; Codoner-Franch, P.; Sanz, Y. Intestinal Microbiota and Celiac Disease: Cause, Consequence or Co-Evolution? Nutrients 2015, 7, 6900–6923. [Google Scholar] [CrossRef]
- Lionetti, E.; Castellaneta, S.; Francavilla, R.; Pulvirenti, A.; Tonutti, E.; Amarri, S.; Barbato, M.; Barbera, C.; Barera, G.; Bellantoni, A.; et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 2014, 371, 1295–1303. [Google Scholar] [CrossRef]
- Szajewska, H.; Shamir, R.; Chmielewska, A.; Piescik-Lech, M.; Auricchio, R.; Ivarsson, A.; Kolacek, S.; Koletzko, S.; Korponay-Szabo, I.; Mearin, M.L.; et al. Systematic review with meta-analysis: Early infant feeding and coeliac disease--update 2015. Aliment. Pharmacol. Ther. 2015, 41, 1038–1054. [Google Scholar] [CrossRef]
- Aversa, Z.; Atkinson, E.J.; Schafer, M.J.; Theiler, R.N.; Rocca, W.A.; Blaser, M.J.; LeBrasseur, N.K. Association of Infant Antibiotic Exposure With Childhood Health Outcomes. Mayo Clin. Proc. 2021, 96, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Dydensborg Sander, S.; Nybo Andersen, A.M.; Murray, J.A.; Karlstad, O.; Husby, S.; Stordal, K. Association Between Antibiotics in the First Year of Life and Celiac Disease. Gastroenterology 2019, 156, 2217–2229. [Google Scholar] [CrossRef]
- Fenneman, A.C.; Weidner, M.; Chen, L.A.; Nieuwdorp, M.; Blaser, M.J. Antibiotics in the pathogenesis of diabetes and inflammatory diseases of the gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Walker, A.W.; Capilla, A.; Benitez-Paez, A.; Palau, F.; Parkhill, J.; Castillejo, G.; Sanz, Y. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome 2018, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Bouziat, R.; Hinterleitner, R.; Brown, J.J.; Stencel-Baerenwald, J.E.; Ikizler, M.; Mayassi, T.; Meisel, M.; Kim, S.M.; Discepolo, V.; Pruijssers, A.J.; et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 2017, 356, 44–50. [Google Scholar] [CrossRef]
- Olshan, K.L.; Leonard, M.M.; Serena, G.; Zomorrodi, A.R.; Fasano, A. Gut microbiota in Celiac Disease: Microbes, metabolites, pathways and therapeutics. Expert Rev. Clin. Immunol. 2020, 16, 1075–1092. [Google Scholar] [CrossRef]
- Lindfors, K.; Lin, J.; Lee, H.S.; Hyoty, H.; Nykter, M.; Kurppa, K.; Liu, E.; Koletzko, S.; Rewers, M.; Hagopian, W.; et al. Metagenomics of the faecal virome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: The TEDDY study. Gut 2020, 69, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Belei, O.; Juganaru, I.; Basaca, D.G.; Munteanu, A.I.; Marginean, O. The Role of Intestinal Microbiota in Celiac Disease and Further Therapeutic Perspectives. Life 2023, 13, 2039. [Google Scholar] [CrossRef]
- Chibbar, R.; Dieleman, L.A. The Gut Microbiota in Celiac Disease and Probiotics. Nutrients 2019, 11, 2375. [Google Scholar] [CrossRef]
- Marasco, G.; Cirota, G.G.; Rossini, B.; Lungaro, L.; Di Biase, A.R.; Colecchia, A.; Volta, U.; De Giorgio, R.; Festi, D.; Caio, G. Probiotics, Prebiotics and Other Dietary Supplements for Gut Microbiota Modulation in Celiac Disease Patients. Nutrients 2020, 12, 2674. [Google Scholar] [CrossRef]
- Valitutti, F.; Cucchiara, S.; Fasano, A. Celiac Disease and the Microbiome. Nutrients 2019, 11, 2403. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Benitez-Paez, A.; de Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Marcos, A.; Garrote, J.A.; Polanco, I.; et al. Increased prevalence of pathogenic bacteria in the gut microbiota of infants at risk of developing celiac disease: The PROFICEL study. Gut Microbes 2018, 9, 551–558. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Del Vecchio Blanco, G.; et al. Metagenomics Reveals Dysbiosis and a Potentially Pathogenic N. flavescens Strain in Duodenum of Adult Celiac Patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol. 2008, 8, 232. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Hernandez-Galan, L.; Galipeau, H.J.; Constante, M.; Clarizio, A.; Jury, J.; Breyner, N.M.; Caminero, A.; Rueda, G.; Hayes, C.L.; et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci. Transl. Med. 2020, 12, eaba0624. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Fu, J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Vannini, L.; Di Cagno, R.; Cavallo, N.; Minervini, F.; Francavilla, R.; Ercolini, D.; Gobbetti, M. Salivary and fecal microbiota and metabolome of celiac children under gluten-free diet. Int. J. Food Microbiol. 2016, 239, 125–132. [Google Scholar] [CrossRef]
- Poddighe, D.; Kushugulova, A. Salivary Microbiome in Pediatric and Adult Celiac Disease. Front. Cell Infect. Microbiol. 2021, 11, 625162. [Google Scholar] [CrossRef]
- Tuganbaev, T.; Yoshida, K.; Honda, K. The effects of oral microbiota on health. Science 2022, 376, 934–936. [Google Scholar] [CrossRef]
- Peng, X.; Cheng, L.; You, Y.; Tang, C.; Ren, B.; Li, Y.; Xu, X.; Zhou, X. Oral microbiota in human systematic diseases. Int. J. Oral Sci. 2022, 14, 14. [Google Scholar] [CrossRef]
- Lindfors, K.; Ciacci, C.; Kurppa, K.; Lundin, K.E.A.; Makharia, G.K.; Mearin, M.L.; Murray, J.A.; Verdu, E.F.; Kaukinen, K. Coeliac disease. Nat. Rev. Dis. Primers 2019, 5, 3. [Google Scholar] [CrossRef]
- Bonder, M.J.; Tigchelaar, E.F.; Cai, X.; Trynka, G.; Cenit, M.C.; Hrdlickova, B.; Zhong, H.; Vatanen, T.; Gevers, D.; Wijmenga, C.; et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Saviano, A.; Petruzziello, C.; Brigida, M.; Morabito Loprete, M.R.; Savioli, G.; Migneco, A.; Ojetti, V. Gut Microbiota Alteration and Its Modulation with Probiotics in Celiac Disease. Biomedicines 2023, 11, 2638. [Google Scholar] [CrossRef] [PubMed]
- Pecora, F.; Persico, F.; Gismondi, P.; Fornaroli, F.; Iuliano, S.; de’Angelis, G.L.; Esposito, S. Gut Microbiota in Celiac Disease: Is There Any Role for Probiotics? Front. Immunol. 2020, 11, 957. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, R.; De Angelis, M.; Rizzello, C.G.; Cavallo, N.; Dal Bello, F.; Gobbetti, M. Selected Probiotic Lactobacilli Have the Capacity To Hydrolyze Gluten Peptides during Simulated Gastrointestinal Digestion. Appl. Environ. Microbiol. 2017, 83, e00376-17. [Google Scholar] [CrossRef] [PubMed]
- Jenickova, E.; Andren Aronsson, C.; Mascellani Bergo, A.; Cinek, O.; Havlik, J.; Agardh, D. Effects of Lactiplantibacillus plantarum and Lacticaseibacillus paracasei supplementation on the faecal metabolome in children with coeliac disease autoimmunity: A randomised, double-blinded placebo-controlled clinical trial. Front. Nutr. 2023, 10, 1183963. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Castillejo, G.; Varea, V.; Sanz, Y. Double-blind, randomised, placebo-controlled intervention trial to evaluate the effects of Bifidobacterium longum CECT 7347 in children with newly diagnosed coeliac disease. Br. J. Nutr. 2014, 112, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Laparra, J.M.; Olivares, M.; Gallina, O.; Sanz, Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS ONE 2012, 7, e30744. [Google Scholar] [CrossRef] [PubMed]
- Quagliariello, A.; Aloisio, I.; Bozzi Cionci, N.; Luiselli, D.; D’Auria, G.; Martinez-Priego, L.; Perez-Villarroya, D.; Langerholc, T.; Primec, M.; Micetic-Turk, D.; et al. Effect of Bifidobacterium breve on the Intestinal Microbiota of Coeliac Children on a Gluten Free Diet: A Pilot Study. Nutrients 2016, 8, 660. [Google Scholar] [CrossRef]
- Primec, M.; Klemenak, M.; Di Gioia, D.; Aloisio, I.; Bozzi Cionci, N.; Quagliariello, A.; Gorenjak, M.; Micetic-Turk, D.; Langerholc, T. Clinical intervention using Bifidobacterium strains in celiac disease children reveals novel microbial modulators of TNF-alpha and short-chain fatty acids. Clin. Nutr. 2019, 38, 1373–1381. [Google Scholar] [CrossRef]
- McCarville, J.L.; Dong, J.; Caminero, A.; Bermudez-Brito, M.; Jury, J.; Murray, J.A.; Duboux, S.; Steinmann, M.; Delley, M.; Tangyu, M.; et al. A Commensal Bifidobacterium longum Strain Prevents Gluten-Related Immunopathology in Mice through Expression of a Serine Protease Inhibitor. Appl. Environ. Microbiol. 2017, 83, e01323-17. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, R.; Piccolo, M.; Francavilla, A.; Polimeno, L.; Semeraro, F.; Cristofori, F.; Castellaneta, S.; Barone, M.; Indrio, F.; Gobbetti, M.; et al. Clinical and Microbiological Effect of a Multispecies Probiotic Supplementation in Celiac Patients With Persistent IBS-type Symptoms: A Randomized, Double-Blind, Placebo-controlled, Multicenter Trial. J. Clin. Gastroenterol. 2019, 53, e117–e125. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, A.; Xu, X.; Colee, J.; Tompkins, T.A. Efficacy of a Multi-Strain Probiotic Formulation in Pediatric Populations: A Comprehensive Review of Clinical Studies. Nutrients 2021, 13, 1908. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, E.; Dominijanni, V.; Iasevoli, M.; Cimadamore, E.; Acquaviva, I.; Gatti, S.; Monachesi, C.; Catassi, G.; Pino, A.; Faragalli, A.; et al. Effects of the supplementation with a multispecies probiotic on clinical and laboratory recovery of children with newly diagnosed celiac disease: A randomized, placebo-controlled trial. Dig. Liver Dis. 2023, 55, 1328–1337. [Google Scholar] [CrossRef]
- Hou, Q.; Ye, L.; Liu, H.; Huang, L.; Yang, Q.; Turner, J.R.; Yu, Q. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2018, 25, 1657–1670. [Google Scholar] [CrossRef]
- Tran, T.; Senger, S.; Baldassarre, M.; Brosnan, R.A.; Cristofori, F.; Crocco, M.; De Santis, S.; Elli, L.; Faherty, C.S.; Francavilla, R.; et al. Novel Bacteroides vulgatus strain protects against gluten-induced break of human celiac gut epithelial homeostasis: A pre-clinical proof-of-concept study. Pediatr. Res. 2024, 95, 1254–1264. [Google Scholar] [CrossRef]
- Freire, R.; Ingano, L.; Serena, G.; Cetinbas, M.; Anselmo, A.; Sapone, A.; Sadreyev, R.I.; Fasano, A.; Senger, S. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 2019, 9, 7029. [Google Scholar] [CrossRef]
- Conte, M.; Nigro, F.; Porpora, M.; Bellomo, C.; Furone, F.; Budelli, A.L.; Nigro, R.; Barone, M.V.; Nanayakkara, M. Gliadin Peptide P31-43 Induces mTOR/NFkbeta Activation and Reduces Autophagy: The Role of Lactobacillus paracasei CBA L74 Postbiotc. Int. J. Mol. Sci. 2022, 23, 3655. [Google Scholar] [CrossRef]
- Olivares, M.; Neef, A.; Castillejo, G.; Palma, G.D.; Varea, V.; Capilla, A.; Palau, F.; Nova, E.; Marcos, A.; Polanco, I.; et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut 2015, 64, 406–417. [Google Scholar] [CrossRef]
- Leonard, M.M.; Valitutti, F.; Karathia, H.; Pujolassos, M.; Kenyon, V.; Fanelli, B.; Troisi, J.; Subramanian, P.; Camhi, S.; Colucci, A.; et al. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc. Natl. Acad. Sci. USA 2021, 118, e2020322118. [Google Scholar] [CrossRef]
| (A) Clinical Trials | |
|---|---|
| Probiotics Tested to Treat Celiac Disease | Clinical Trials |
| Lactobacillus casei BGP93, Lactobacillus delbrueckii subsp. bulgaricus SP5, Lactobacillus paracasei LPC01 and BGP2, Lactobacillus plantarum BGP12, LP27, LP35, LP40, LP47 and SP1 | [55] |
| Lactiplantibacillus plantarum HEAL9 and Lacticaseibacillus paracasei 8700:2 | [56] |
| Bifidobacterium longum CECT 7347 (ES1) | [57] |
| [58] | |
| Bifidobacterium breve (B632 and BR03) | [59] |
| [60] | |
| B. longum NCC2705 | [61] |
| Lactobacillus casei LMG 101/37 P-17504, Lactobacillus plantarum CECT 4528, Bifidobacterium animalis subsp. lactis Bi1 LMG P-17502, Bifidobacterium breve Bbr8 LMG P-17501 and Bl. breve Bl10 LMG P-17500 | [62] |
| Lactobacillus helveticus Rosell-52, Bifidobacterium infantis Rosell-33 and Bifidobacterium bifidum Rosell-71 with fructooligosaccharides | [63] |
| (Lactobacillus paracasei 101/37 LMG P-17504, Lactobacillus plantarum 14D CECT 4528, Bifidobacterium animalis subsp. lactis Bi1 LMG P-17502, Bifidobacterium breve Bbr8 LMG P-17501, Bifidobacterium breve BL10 LMG P-17500) | [64] |
| (B) In vitro studies | |
| Probiotics tested to treat Celiac Disease | In vitro studies |
| L. reuteri D8 | [65] |
| B. vulgatus (20220303-A2) | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matera, M.; Guandalini, S. How the Microbiota May Affect Celiac Disease and What We Can Do. Nutrients 2024, 16, 1882. https://doi.org/10.3390/nu16121882
Matera M, Guandalini S. How the Microbiota May Affect Celiac Disease and What We Can Do. Nutrients. 2024; 16(12):1882. https://doi.org/10.3390/nu16121882
Chicago/Turabian StyleMatera, Mariarosaria, and Stefano Guandalini. 2024. "How the Microbiota May Affect Celiac Disease and What We Can Do" Nutrients 16, no. 12: 1882. https://doi.org/10.3390/nu16121882
APA StyleMatera, M., & Guandalini, S. (2024). How the Microbiota May Affect Celiac Disease and What We Can Do. Nutrients, 16(12), 1882. https://doi.org/10.3390/nu16121882







