Placental Epigenome Impacts Fetal Development: Effects of Maternal Nutrients and Gut Microbiota
Highlights
- Maternal nutrition and gut microbiota influence placental epigenetic modifications, which affect fetal development and long-term health.
- The maternal gut has an influence on fetal epigenetic programming, including on fetal growth, birth weight, and brain performance.
- The maternal gut microbiome influences placental function via microbial metabolites, linking maternal diet to fetal outcomes, demonstrating the impact of microbiota–placenta interactions.
- Nutrition regulates gene expression, such that specific foods influence placental gene expression, which has ramifications for fetal growth and programming.
- Specific sets of gut metabolites can alter placental function, which is linked to maternal diet and fetal outcomes.
- Integrative investigation of placental epigenomics and maternal microbiota using a novel methodology reveals new information about maternal–fetal health dynamics.
- Synergy between dietary and microbial strategies has clinical implications for improving pregnancy outcomes, such as preventing developmental defects.
Abstract
1. Introduction
2. Heritability of Diet-Induced Epigenetic Changes: Effects on Fetal Development
3. Placental Interactions with Environments: Impacts on Fetoplacental Development
4. Placental Epigenome and Birth Outcomes
5. Maternal Dietary Fats and Placental Epigenome
6. One-Carbon Metabolism-Related Nutrients and Placental Epigenome
7. Vitamin D Levels and Fetoplacental Epigenome
8. Gut Microbiota and Placental Epigenome
9. Maternal Microbiome and Its Impact on Fetal Growth and Development
10. Microbiota and Fetal Immune Development
11. Gut Microbiota and Fetal Brain Development
12. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Duttaroy, A.K. Docosahexaenoic acid supports feto-placental growth and protects cardiovascular and cognitive function: A mini review. Eur. J. Lipid Sci. Technol. 2016, 118, 1439–1449. [Google Scholar] [CrossRef]
- Dimasuay, K.G.; Boeuf, P.; Powell, T.L.; Jansson, T. Placental Responses to Changes in the Maternal Environment Determine Fetal Growth. Front. Physiol. 2016, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Duttaroy, A.K. Influence of Maternal Diet and Environmental Factors on Fetal Development. Nutrients 2023, 15, 4094. [Google Scholar] [CrossRef] [PubMed]
- Duttaroy, A.K. Transport of fatty acids across the human placenta: A review. Prog. Lipid Res. 2009, 48, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Duttaroy, A.K. Maternal PUFAs, Placental Epigenetics, and Their Relevance to Fetal Growth and Brain Development. Reprod. Sci. 2023, 30, 408–427. [Google Scholar] [CrossRef] [PubMed]
- Kuzawa, C.W. Fetal origins of developmental plasticity: Are fetal cues reliable predictors of future nutritional environments? Am. J. Hum. Biol. 2005, 17, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Chandak, G.R.; Silver, M.J.; Saffari, A.; Lillycrop, K.A.; Shrestha, S.; Sahariah, S.A.; Di Gravio, C.; Goldberg, G.; Tomar, A.S.; Betts, M.; et al. Protocol for the EMPHASIS study; epigenetic mechanisms linking maternal pre-conceptional nutrition and children’s health in India and Sub-Saharan Africa. BMC Nutr. 2017, 3, 81. [Google Scholar] [CrossRef] [PubMed]
- Yajnik, C.S.; Deshpande, S.S.; Jackson, A.A.; Refsum, H.; Rao, S.; Fisher, D.J.; Bhat, D.S.; Naik, S.S.; Coyaji, K.J.; Joglekar, C.V.; et al. Vitamin B12 and folate concentrations during pregnancy and insulin resistance in the offspring: The Pune Maternal Nutrition Study. Diabetologia 2008, 51, 29–38. [Google Scholar] [CrossRef]
- Burdge, G.C.; Hanson, M.A.; Slater-Jefferies, J.L.; Lillycrop, K.A. Epigenetic regulation of transcription: A mechanism for inducing variations in phenotype (fetal programming) by differences in nutrition during early life? Br. J. Nutr. 2007, 97, 1036–1046. [Google Scholar] [CrossRef]
- Burdge, G.C.; Lillycrop, K.A. Fatty acids and epigenetics. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, V.; Molangiri, A.; Mallepogu, A.; Kona, S.R.; Ibrahim, A.; Duttaroy, A.K.; Basak, S. Maternal n-3 PUFA deficiency alters uterine artery remodeling and placental epigenome in the mice. J. Nutr. Biochem. 2021, 96, 108784. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, V.; Varma, S.; Kona, S.R.; Ibrahim, A.; Duttaroy, A.K.; Basak, S. Dietary omega-3 fatty acid deficiency from pre-pregnancy to lactation affects expression of genes involved in hippocampal neurogenesis of the offspring. Prostaglandins Leukot. Essent. Fat. Acids 2023, 191, 102566. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, M.D.; Craciunescu, C.N.; Zeisel, S.H. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006, 20, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Craciunescu, C.N.; Albright, C.D.; Mar, M.H.; Song, J.; Zeisel, S.H. Choline availability during embryonic development alters progenitor cell mitosis in developing mouse hippocampus. J. Nutr. 2003, 133, 3614–3618. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Yan, J.; West, A.A.; Perry, C.A.; Malysheva, O.V.; Devapatla, S.; Pressman, E.; Vermeylen, F.; Caudill, M.A. Maternal choline intake alters the epigenetic state of fetal cortisol-regulating genes in humans. FASEB J. 2012, 26, 3563–3574. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Chan, M.M.; Humm, K.C.; Karnik, R.; Mekhoubad, S.; Regev, A.; Eggan, K.; Meissner, A. DNA methylation dynamics of the human preimplantation embryo. Nature 2014, 511, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Okae, H.; Chiba, H.; Hiura, H.; Hamada, H.; Sato, A.; Utsunomiya, T.; Kikuchi, H.; Yoshida, H.; Tanaka, A.; Suyama, M.; et al. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014, 10, e1004868. [Google Scholar] [CrossRef] [PubMed]
- Slieker, R.C.; Bos, S.D.; Goeman, J.J.; Bovee, J.V.; Talens, R.P.; van der Breggen, R.; Suchiman, H.E.; Lameijer, E.W.; Putter, H.; van den Akker, E.B.; et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 2013, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Slieker, R.C.; Roost, M.S.; van Iperen, L.; Suchiman, H.E.; Tobi, E.W.; Carlotti, F.; de Koning, E.J.; Slagboom, P.E.; Heijmans, B.T.; Chuva de Sousa Lopes, S.M. DNA Methylation Landscapes of Human Fetal Development. PLoS Genet. 2015, 11, e1005583. [Google Scholar] [CrossRef]
- Faulk, C.; Dolinoy, D.C. Timing is everything: The when and how of environmentally induced changes in the epigenome of animals. Epigenetics 2011, 6, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, K.; Ito, K.; Saer, B.; Taylor, G.; Ye, S.; Yamano, M.; Toriba, Y.; Hayes, A.; Okamura, H.; Fustin, J.-M. Excess S-adenosylmethionine inhibits methylation via catabolism to adenine. Commun. Biol. 2022, 5, 313. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Honma, K.; Mochizuki, K.; Goda, T. Induction of histone H3K4 methylation at the promoter, enhancer, and transcribed regions of the Si and Sglt1 genes in rat jejunum in response to a high-starch/low-fat diet. Nutrition 2015, 31, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Chen, D.; Yang, C.S. Dietary polyphenols may affect DNA methylation. J. Nutr. 2007, 137, 223S–228S. [Google Scholar] [CrossRef] [PubMed]
- Dolinoy, D.C.; Weidman, J.R.; Waterland, R.A.; Jirtle, R.L. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006, 114, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Day, J.K.; Bauer, A.M.; DesBordes, C.; Zhuang, Y.; Kim, B.E.; Newton, L.G.; Nehra, V.; Forsee, K.M.; MacDonald, R.S.; Besch-Williford, C.; et al. Genistein alters methylation patterns in mice. J. Nutr. 2002, 132, 2419S–2423S. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.N.; Khulan, B.; Owens, S.; Elks, C.E.; Seidel, V.; Prentice, A.M.; Belteki, G.; Ong, K.K.; Affara, N.A.; Constancia, M.; et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: Results of a pilot randomized controlled trial. FASEB J. 2012, 26, 1782–1790. [Google Scholar] [CrossRef] [PubMed]
- Li, C.C.; Cropley, J.E.; Cowley, M.J.; Preiss, T.; Martin, D.I.; Suter, C.M. A sustained dietary change increases epigenetic variation in isogenic mice. PLoS Genet. 2011, 7, e1001380. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Epigenetic Mechanisms Link Maternal Diets and Gut Microbiome to Obesity in the Offspring. Front. Genet. 2018, 9, 342. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Trivino, J.; Alvarez, D.; Cadavid, J.A.; Alvarez, A.M. From gut to placenta: Understanding how the maternal microbiome models life-long conditions. Front. Endocrinol. 2023, 14, 1304727. [Google Scholar] [CrossRef]
- Basak, S.; Das, R.K.; Banerjee, A.; Paul, S.; Pathak, S.; Duttaroy, A.K. Maternal Obesity and Gut Microbiota Are Associated with Fetal Brain Development. Nutrients 2022, 14, 4515. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, J.; Wang, Y. The role of short-chain fatty acids produced by gut microbiota in the regulation of pre-eclampsia onset. Front. Cell. Infect. Microbiol. 2023, 13, 1177768. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Habu, S.; Kasai, M.; Okumura, K.; Ishikawa, D.; Shibuya, T.; Kobayashi, O.; Osada, T.; Ohkusa, T.; Watanabe, S.; et al. Impact of maternal dietary gut microbial metabolites on an offspring’s systemic immune response in mouse models. Biosci. Microbiota Food Health 2020, 39, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Toth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed]
- Miko, E.; Csaszar, A.; Bodis, J.; Kovacs, K. The Maternal-Fetal Gut Microbiota Axis: Physiological Changes, Dietary Influence, and Modulation Possibilities. Life 2022, 12, 424. [Google Scholar] [CrossRef] [PubMed]
- Millership, S.J.; Van de Pette, M.; Withers, D.J. Genomic imprinting and its effects on postnatal growth and adult metabolism. Cell. Mol. Life Sci. 2019, 76, 4009–4021. [Google Scholar] [CrossRef] [PubMed]
- Anway, M.D.; Cupp, A.S.; Uzumcu, M.; Skinner, M.K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005, 308, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Cropley, J.E.; Suter, C.M.; Beckman, K.B.; Martin, D.I. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc. Natl. Acad. Sci. USA 2006, 103, 17308–17312. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Beck, S. Epigenetic variation and inheritance in mammals. Curr. Opin. Genet. Dev. 2006, 16, 573–577. [Google Scholar] [CrossRef]
- Ng, S.F.; Lin, R.C.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef]
- Lesch, B.J.; Tothova, Z.; Morgan, E.A.; Liao, Z.; Bronson, R.T.; Ebert, B.L.; Page, D.C. Intergenerational epigenetic inheritance of cancer susceptibility in mammals. eLife 2019, 8, e39380. [Google Scholar] [CrossRef] [PubMed]
- Burdge, G.C.; Hoile, S.P.; Uller, T.; Thomas, N.A.; Gluckman, P.D.; Hanson, M.A.; Lillycrop, K.A. Progressive, Transgenerational Changes in Offspring Phenotype and Epigenotype following Nutritional Transition. PLoS ONE 2011, 6, e28282. [Google Scholar] [CrossRef] [PubMed]
- Nowacka-Woszuk, J.; Szczerbal, I.; Malinowska, A.M.; Chmurzynska, A. Transgenerational effects of prenatal restricted diet on gene expression and histone modifications in the rat. PLoS ONE 2018, 13, e0193464. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Fu, H.; Dong, H.; Lu, Y.; Lu, Y.; Qi, K. Maternal n-3 polyunsaturated fatty acid deprivation during pregnancy and lactation affects neurogenesis and apoptosis in adult offspring: Associated with DNA methylation of brain-derived neurotrophic factor transcripts. Nutr. Res. 2016, 36, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Ly, L.; Chan, D.; Landry, M.; Angle, C.; Martel, J.; Trasler, J. Impact of mothers’ early life exposure to low or high folate on progeny outcome and DNA methylation patterns. Environ. Epigenetics 2020, 6, dvaa018. [Google Scholar] [CrossRef] [PubMed]
- Sable, P.; Randhir, K.; Kale, A.; Chavan-Gautam, P.; Joshi, S. Maternal micronutrients and brain global methylation patterns in the offspring. Nutr. Neurosci. 2015, 18, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Sapehia, D.; Bagga, R.; Kaur, J. Different dietary combinations of folic acid and vitamin B12 in parental diet results in epigenetic reprogramming of IGF2R and KCNQ1OT1 in placenta and fetal tissues in mice. Mol. Reprod. Dev. 2021, 88, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.; Goyal, D.; Leitzke, A.; Gheorghe, C.P.; Longo, L.D. Brain Renin-Angiotensin System: Fetal Epigenetic Programming by Maternal Protein Restriction during Pregnancy. Reprod. Sci. 2010, 17, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Strakovsky, R.S.; Zhang, X.; Zhou, D.; Pan, Y.-X. Gestational high fat diet programs hepatic phosphoenolpyruvate carboxykinase gene expression and histone modification in neonatal offspring rats. J. Physiol. 2011, 589, 2707–2717. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-Y.; Ganguly, A.; Rubbi, L.; Orozco, L.D.; Morselli, M.; Ashraf, D.; Jaroszewicz, A.; Feng, S.; Jacobsen, S.E.; Nakano, A.; et al. Intrauterine calorie restriction affects placental DNA methylation and gene expression. Physiol. Genom. 2013, 45, 565–576. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, F.; Didelot, X.; Bruce, K.D.; Cagampang, F.R.; Vatish, M.; Hanson, M.; Lehnert, H.; Ceriello, A.; Byrne, C.D. Maternal high fat diet during pregnancy and lactation alters hepatic expression of insulin like growth factor-2 and key microRNAs in the adult offspring. BMC Genom. 2009, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Maloyan, A.; Muralimanoharan, S.; Huffman, S.; Cox, L.A.; Nathanielsz, P.W.; Myatt, L.; Nijland, M.J. Identification and comparative analyses of myocardial miRNAs involved in the fetal response to maternal obesity. Physiol. Genom. 2013, 45, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Wehbe, N.; Nasser, S.A.; Pintus, G.; Badran, A.; Eid, A.H.; Baydoun, E. MicroRNAs in Cardiac Hypertrophy. Int. J. Mol. Sci. 2019, 20, 4714. [Google Scholar] [CrossRef] [PubMed]
- Colpaert, R.M.W.; Calore, M. MicroRNAs in Cardiac Diseases. Cells 2019, 8, 737. [Google Scholar] [CrossRef] [PubMed]
- Duisters, R.F.; Tijsen, A.J.; Schroen, B.; Leenders, J.J.; Lentink, V.; van der Made, I.; Herias, V.; van Leeuwen, R.E.; Schellings, M.W.; Barenbrug, P.; et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009, 104, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Khorram, O.; Chuang, T.D.; Pearce, W.J. Long-term effects of maternal undernutrition on offspring carotid artery remodeling: Role of miR-29c. J. Dev. Orig. Health Dis. 2015, 6, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Plagemann, A. ‘Fetal programming’ and ‘functional teratogenesis’: On epigenetic mechanisms and prevention of perinatally acquired lasting health risks. J. Perinat. Med. 2004, 32, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, K.M.; Lillycrop, K.A.; Burdge, G.C.; Gluckman, P.D.; Hanson, M.A. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr. Res. 2007, 61, 5R–10R. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Gianotti, T.F.; Burgueno, A.L.; Pirola, C.J. Fetal metabolic programming and epigenetic modifications: A systems biology approach. Pediatr. Res. 2013, 73, 531–542. [Google Scholar] [CrossRef]
- Dunlop, A.L.; Mulle, J.G.; Ferranti, E.P.; Edwards, S.; Dunn, A.B.; Corwin, E.J. Maternal Microbiome and Pregnancy Outcomes That Impact Infant Health: A Review. Adv. Neonatal. Care 2015, 15, 377–385. [Google Scholar] [CrossRef]
- Enstad, S.; Cheema, S.; Thomas, R.; Fichorova, R.N.; Martin, C.R.; O’Tierney-Ginn, P.; Wagner, C.L.; Sen, S. The impact of maternal obesity and breast milk inflammation on developmental programming of infant growth. Eur. J. Clin. Nutr. 2021, 75, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Calabuig-Navarro, V.; Puchowicz, M.; Glazebrook, P.; Haghiac, M.; Minium, J.; Catalano, P.; Hauguel deMouzon, S.; O’Tierney-Ginn, P. Effect of omega-3 supplementation on placental lipid metabolism in overweight and obese women. Am. J. Clin. Nutr. 2016, 103, 1064–1072. [Google Scholar] [CrossRef] [PubMed]
- Khanal, P.; Duttaroy, A.K. Prospect of potential intrauterine programming impacts associated with COVID-19. Front. Public Health 2022, 10, 986162. [Google Scholar] [CrossRef] [PubMed]
- Lassance, L.; Haghiac, M.; Leahy, P.; Basu, S.; Minium, J.; Zhou, J.; Reider, M.; Catalano, P.M.; Hauguel-de Mouzon, S. Identification of early transcriptome signatures in placenta exposed to insulin and obesity. Am. J. Obstet. Gynecol. 2015, 212, 647.e1–647.e11. [Google Scholar] [CrossRef] [PubMed]
- Apicella, C.; Ruano, C.S.M.; Mehats, C.; Miralles, F.; Vaiman, D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int. J. Mol. Sci. 2019, 20, 2837. [Google Scholar] [CrossRef] [PubMed]
- Troncoso, F.; Acurio, J.; Herlitz, K.; Aguayo, C.; Bertoglia, P.; Guzman-Gutierrez, E.; Loyola, M.; Gonzalez, M.; Rezgaoui, M.; Desoye, G.; et al. Gestational diabetes mellitus is associated with increased pro-migratory activation of vascular endothelial growth factor receptor 2 and reduced expression of vascular endothelial growth factor receptor 1. PLoS ONE 2017, 12, e0182509. [Google Scholar] [CrossRef]
- Devarshi, P.P.; Grant, R.W.; Ikonte, C.J.; Hazels Mitmesser, S. Maternal Omega-3 Nutrition, Placental Transfer and Fetal Brain Development in Gestational Diabetes and Preeclampsia. Nutrients 2019, 11, 1107. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Vilasagaram, S.; Duttaroy, A.K. Maternal dietary deficiency of n-3 fatty acids affects metabolic and epigenetic phenotypes of the developing fetus. Prostaglandins Leukot. Essent. Fat. Acids 2020, 158, 102109–102120. [Google Scholar] [CrossRef]
- Johnsen, G.M.; Basak, S.; Weedon-Fekjaer, M.S.; Staff, A.C.; Duttaroy, A.K. Docosahexaenoic acid stimulates tube formation in first trimester trophoblast cells, HTR8/SVneo. Placenta 2011, 32, 626–632. [Google Scholar] [CrossRef]
- Duttaroy, A.K.; Basak, S. Maternal Fatty Acid Metabolism in Pregnancy and Its Consequences in the Feto-Placental Development. Front. Physiol. 2022, 12, 787848. [Google Scholar] [CrossRef]
- Basak, S.; Das, M.K.; Duttaroy, A.K. Fatty acid-induced angiogenesis in first trimester placental trophoblast cells: Possible roles of cellular fatty acid-binding proteins. Life Sci. 2013, 93, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.J.; O’Neill, K.; Condon, D.; Sasson, I.; Sen, P.; Xia, Y.; Simmons, R.A. Diet-induced obesity alters the maternal metabolome and early placenta transcriptome and decreases placenta vascularity in the mouse. Biol. Reprod. 2018, 98, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Qadri, S.S.Y.; Mir, I.A.; Kondapalli, N.B.; Basak, S.; Rajkumar, H. Fructooligosaccharide ameliorates high-fat induced intrauterine inflammation and improves lipid profile in the hamster offspring. J. Nutr. Biochem. 2022, 101, 108925. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Sun, B.; Boersma, G.J.; Cordner, Z.A.; Yan, J.; Moran, T.H.; Tamashiro, K.L.K. Prenatal high-fat diet alters placental morphology, nutrient transporter expression, and mtorc1 signaling in rat. Obesity 2017, 25, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Saben, J.; Lindsey, F.; Zhong, Y.; Thakali, K.; Badger, T.M.; Andres, A.; Gomez-Acevedo, H.; Shankar, K. Maternal obesity is associated with a lipotoxic placental environment. Placenta 2014, 35, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, D.; Workalemahu, T.; Tekola-Ayele, F. Maternal dyslipidemia during early pregnancy and epigenetic ageing of the placenta. Epigenetics 2019, 14, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Dube, E.; Gravel, A.; Martin, C.; Desparois, G.; Moussa, I.; Ethier-Chiasson, M.; Forest, J.C.; Giguere, Y.; Masse, A.; Lafond, J. Modulation of fatty acid transport and metabolism by maternal obesity in the human full-term placenta. Biol. Reprod. 2012, 87, 11. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.T.; Demmelmair, H.; Krauss-Etschmann, S.; Nathan, P.; Dehmel, S.; Padilla, M.C.; Rueda, R.; Koletzko, B.; Campoy, C. Maternal BMI and gestational diabetes alter placental lipid transporters and fatty acid composition. Placenta 2017, 57, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Dutta-Roy, A.K. Transport mechanisms for long-chain polyunsaturated fatty acids in the human placenta. Am. J. Clin. Nutr. 2000, 71, 315S–322S. [Google Scholar] [CrossRef] [PubMed]
- Gazquez, A.; Prieto-Sanchez, M.T.; Blanco-Carnero, J.E.; Ruiz-Palacios, M.; Nieto, A.; van Harskamp, D.; Oosterink, J.E.; Schierbeek, H.; van Goudoever, J.B.; Demmelmair, H.; et al. Altered materno-fetal transfer of 13C-polyunsaturated fatty acids in obese pregnant women. Clin. Nutr. 2020, 39, 1101–1107. [Google Scholar] [CrossRef]
- Hirschmugl, B.; Desoye, G.; Catalano, P.; Klymiuk, I.; Scharnagl, H.; Payr, S.; Kitzinger, E.; Schliefsteiner, C.; Lang, U.; Wadsack, C.; et al. Maternal obesity modulates intracellular lipid turnover in the human term placenta. Int. J. Obes. 2017, 41, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Laskewitz, A.; van Benthem, K.L.; Kieffer, T.E.C.; Faas, M.M.; Verkaik-Schakel, R.N.; Plosch, T.; Scherjon, S.A.; Prins, J.R. The influence of maternal obesity on macrophage subsets in the human decidua. Cell. Immunol. 2019, 336, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Nogues, P.; Dos Santos, E.; Jammes, H.; Berveiller, P.; Arnould, L.; Vialard, F.; Dieudonne, M.N. Maternal obesity influences expression and DNA methylation of the adiponectin and leptin systems in human third-trimester placenta. Clin. Epigenetics 2019, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Gauster, M.; Hiden, U.; van Poppel, M.; Frank, S.; Wadsack, C.; Hauguel-de Mouzon, S.; Desoye, G. Dysregulation of placental endothelial lipase in obese women with gestational diabetes mellitus. Diabetes 2011, 60, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Herrera, E.; Ortega-Senovilla, H. Lipid metabolism during pregnancy and its implications for fetal growth. Curr. Pharm. Biotechnol. 2014, 15, 24–31. [Google Scholar] [CrossRef]
- Davis, C.D.; Ross, S.A. Dietary components impact histone modifications and cancer risk. Nutr. Rev. 2007, 65, 88–94. [Google Scholar] [CrossRef]
- Jansson, N.; Rosario, F.J.; Gaccioli, F.; Lager, S.; Jones, H.N.; Roos, S.; Jansson, T.; Powell, T.L. Activation of placental mTOR signaling and amino acid transporters in obese women giving birth to large babies. J. Clin. Endocrinol. Metab. 2013, 98, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zhao, W.; Zhang, L.; Wang, H.; Yang, X. Low folate levels are associated with methylation-mediated transcriptional repression of miR-203 and miR-375 during cervical carcinogenesis. Oncol. Lett. 2016, 11, 3863–3869. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhang, Q.; Mul, J.D.; Yu, M.; Xu, J.; Qi, C.; Wang, T.; Xiao, X. Maternal high-calorie diet is associated with altered hepatic microRNA expression and impaired metabolic health in offspring at weaning age. Endocrine 2016, 54, 70–80. [Google Scholar] [CrossRef]
- Cao, C.; Zhang, H.; Zhao, L.; Zhou, L.; Zhang, M.; Xu, H.; Han, X.; Li, G.; Yang, X.; Jiang, Y. miR-125b targets DNMT3b and mediates p53 DNA methylation involving in the vascular smooth muscle cells proliferation induced by homocysteine. Exp. Cell Res. 2016, 347, 95–104. [Google Scholar] [CrossRef]
- Koturbash, I.; Melnyk, S.; James, S.J.; Beland, F.A.; Pogribny, I.P. Role of epigenetic and miR-22 and miR-29b alterations in the downregulation of Mat1a and Mthfr genes in early preneoplastic livers in rats induced by 2-acetylaminofluorene. Mol. Carcinog. 2013, 52, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Y.; Liu, A.; Tang, X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem. Biophys. Res. Commun. 2016, 477, 768–773. [Google Scholar] [CrossRef]
- Wilting, S.M.; Miok, V.; Jaspers, A.; Boon, D.; Sorgard, H.; Lando, M.; Snoek, B.C.; van Wieringen, W.N.; Meijer, C.J.; Lyng, H.; et al. Aberrant methylation-mediated silencing of microRNAs contributes to HPV-induced anchorage independence. Oncotarget 2016, 7, 43805–43819. [Google Scholar] [CrossRef]
- Du, J.; Cheng, X.; Shen, L.; Tan, Z.; Luo, J.; Wu, X.; Liu, C.; Yang, Q.; Jiang, Y.; Tang, G.; et al. Methylation of miR-145a-5p promoter mediates adipocytes differentiation. Biochem. Biophys. Res. Commun. 2016, 475, 140–148. [Google Scholar] [CrossRef]
- Tekola-Ayele, F.; Zeng, X.; Ouidir, M.; Workalemahu, T.; Zhang, C.; Delahaye, F.; Wapner, R. DNA methylation loci in placenta associated with birthweight and expression of genes relevant for early development and adult diseases. Clin. Epigenetics 2020, 12, 78. [Google Scholar] [CrossRef]
- Sun, L.; Sun, S. Within-sample co-methylation patterns in normal tissues. BioData Min. 2019, 12, 9. [Google Scholar] [CrossRef]
- Clark, J.; Avula, V.; Ring, C.; Eaves, L.A.; Howard, T.; Santos, H.P.; Smeester, L.; Bangma, J.T.; O’Shea, T.M.; Fry, R.C.; et al. Comparing the Predictivity of Human Placental Gene, microRNA, and CpG Methylation Signatures in Relation to Perinatal Outcomes. Toxicol. Sci. 2021, 183, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Morriseau, T.S.; Kereliuk, S.M.; Doucette, C.A.; Wicklow, B.A.; Dolinsky, V.W. Maternal obesity, diabetes during pregnancy and epigenetic mechanisms that influence the developmental origins of cardiometabolic disease in the offspring. Crit. Rev. Clin. Lab. Sci. 2018, 55, 71–101. [Google Scholar] [CrossRef]
- Sinha, T.; Brushett, S.; Prins, J.; Zhernakova, A. The maternal gut microbiome during pregnancy and its role in maternal and infant health. Curr. Opin. Microbiol. 2023, 74, 102309. [Google Scholar] [CrossRef]
- McGovern, N.; Shin, A.; Low, G.; Low, D.; Duan, K.; Yao, L.J.; Msallam, R.; Low, I.; Shadan, N.B.; Sumatoh, H.R.; et al. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature 2017, 546, 662–666. [Google Scholar] [CrossRef]
- Innis, S.M. Essential fatty acids in growth and development. Prog. Lipid Res. 1991, 30, 39–103. [Google Scholar] [CrossRef]
- Basak, S.; Mallick, R.; Banerjee, A.; Pathak, S.; Duttaroy, A.K. Maternal Supply of Both Arachidonic and Docosahexaenoic Acids Is Required for Optimal Neurodevelopment. Nutrients 2021, 13, 2061. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Mallick, R.; Duttaroy, A.K. Maternal Docosahexaenoic Acid Status during Pregnancy and Its Impact on Infant Neurodevelopment. Nutrients 2020, 12, 3615. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, J.T.; Gordon, M.J.; Campbell, F.M.; Dutta-Roy, A.K. Differential distribution and metabolism of arachidonic acid and docosahexaenoic acid by human placental choriocarcinoma (BeWo) cells. Mol. Cell. Biochem. 1998, 185, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, V.; Molangiri, A.; Varma, S.; Mallepogu, A.; Kona, S.R.; Ibrahim, A.; Duttaroy, A.K.; Basak, S. Maternal omega-3 fatty acid deficiency affects fetal thermogenic development and postnatal musculoskeletal growth in mice. J. Nutr. Biochem. 2023, 112, 109218. [Google Scholar] [CrossRef] [PubMed]
- DeCapo, M.; Thompson, J.R.; Dunn, G.; Sullivan, E.L. Perinatal Nutrition and Programmed Risk for Neuropsychiatric Disorders: A Focus on Animal Models. Biol. Psychiatry 2019, 85, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mendez, E.; Quintero-Fabian, S.; Fernandez-Mejia, C.; Lazo-de-la-Vega-Monroy, M.L. Early-life programming of adipose tissue. Nutr. Res. Rev. 2020, 33, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Barraza-Villarreal, A.; Biessy, C.; Duarte-Salles, T.; Sly, P.D.; Ramakrishnan, U.; Rivera, J.; Herceg, Z.; Romieu, I. Dietary supplementation with polyunsaturated fatty acid during pregnancy modulates DNA methylation at IGF2/H19 imprinted genes and growth of infants. Physiol. Genom. 2014, 46, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Barraza-Villarreal, A.; Hernandez-Vargas, H.; Sly, P.D.; Biessy, C.; Ramakrishnan, U.; Romieu, I.; Herceg, Z. Modulation of DNA methylation states and infant immune system by dietary supplementation with omega-3 PUFA during pregnancy in an intervention study. Am. J. Clin. Nutr. 2013, 98, 480–487. [Google Scholar] [CrossRef]
- Refsum, H.; Yajnik, C.S.; Gadkari, M.; Schneede, J.; Vollset, S.E.; Orning, L.; Guttormsen, A.B.; Joglekar, A.; Sayyad, M.G.; Ulvik, A.; et al. Hyperhomocysteinemia and elevated methylmalonic acid indicate a high prevalence of cobalamin deficiency in Asian Indians. Am. J. Clin. Nutr. 2001, 74, 233–241. [Google Scholar] [CrossRef]
- Fryer, A.A.; Nafee, T.M.; Ismail, K.M.; Carroll, W.D.; Emes, R.D.; Farrell, W.E. LINE-1 DNA methylation is inversely correlated with cord plasma homocysteine in man: A preliminary study. Epigenetics 2009, 4, 394–398. [Google Scholar] [CrossRef]
- Haggarty, P.; Hoad, G.; Campbell, D.M.; Horgan, G.W.; Piyathilake, C.; McNeill, G. Folate in pregnancy and imprinted gene and repeat element methylation in the offspring. Am. J. Clin. Nutr. 2013, 97, 94–99. [Google Scholar] [CrossRef]
- Chen, G.; Broseus, J.; Hergalant, S.; Donnart, A.; Chevalier, C.; Bolanos-Jimenez, F.; Gueant, J.L.; Houlgatte, R. Identification of master genes involved in liver key functions through transcriptomics and epigenomics of methyl donor deficiency in rat: Relevance to nonalcoholic liver disease. Mol. Nutr. Food Res. 2015, 59, 293–302. [Google Scholar] [CrossRef]
- Gueant, J.L.; Namour, F.; Gueant-Rodriguez, R.M.; Daval, J.L. Folate and fetal programming: A play in epigenomics? Trends Endocrinol. Metab. 2013, 24, 279–289. [Google Scholar] [CrossRef]
- Mehedint, M.G.; Craciunescu, C.N.; Zeisel, S.H. Maternal dietary choline deficiency alters angiogenesis in fetal mouse hippocampus. Proc. Natl. Acad. Sci. USA 2010, 107, 12834–12839. [Google Scholar] [CrossRef]
- Kovacheva, V.P.; Mellott, T.J.; Davison, J.M.; Wagner, N.; Lopez-Coviella, I.; Schnitzler, A.C.; Blusztajn, J.K. Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J. Biol. Chem. 2007, 282, 31777–31788. [Google Scholar] [CrossRef]
- Medici, V.; Shibata, N.M.; Kharbanda, K.K.; Islam, M.S.; Keen, C.L.; Kim, K.; Tillman, B.; French, S.W.; Halsted, C.H.; LaSalle, J.M. Maternal choline modifies fetal liver copper, gene expression, DNA methylation, and neonatal growth in the tx-j mouse model of Wilson disease. Epigenetics 2014, 9, 286–296. [Google Scholar] [CrossRef]
- Aon, M.A.; Cortassa, S.; Juhaszova, M.; Sollott, S.J. Mitochondrial health, the epigenome and healthspan. Clin. Sci. 2016, 130, 1285–1305. [Google Scholar] [CrossRef]
- Pauwels, S.; Duca, R.C.; Devlieger, R.; Freson, K.; Straetmans, D.; Van Herck, E.; Huybrechts, I.; Koppen, G.; Godderis, L. Maternal Methyl-Group Donor Intake and Global DNA (Hydroxy)Methylation before and during Pregnancy. Nutrients 2016, 8, 474. [Google Scholar] [CrossRef]
- Hollis, B.W.; Wagner, C.L. Vitamin D supplementation during pregnancy: Improvements in birth outcomes and complications through direct genomic alteration. Mol. Cell. Endocrinol. 2017, 453, 113–130. [Google Scholar] [CrossRef]
- Sablok, A.; Batra, A.; Thariani, K.; Batra, A.; Bharti, R.; Aggarwal, A.R.; Kabi, B.C.; Chellani, H. Supplementation of vitamin D in pregnancy and its correlation with feto-maternal outcome. Clin. Endocrinol. 2015, 83, 536–541. [Google Scholar] [CrossRef]
- Smolders, J.; Menheere, P.; Kessels, A.; Damoiseaux, J.; Hupperts, R. Association of vitamin D metabolite levels with relapse rate and disability in multiple sclerosis. Mult. Scler. 2008, 14, 1220–1224. [Google Scholar] [CrossRef]
- Fetahu, I.S.; Hobaus, J.; Kallay, E. Vitamin D and the epigenome. Front. Physiol. 2014, 5, 164. [Google Scholar] [CrossRef]
- Dominguez-Salas, P.; Moore, S.E.; Baker, M.S.; Bergen, A.W.; Cox, S.E.; Dyer, R.A.; Fulford, A.J.; Guan, Y.; Laritsky, E.; Silver, M.J.; et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 2014, 5, 3746. [Google Scholar] [CrossRef]
- Xue, J.; Schoenrock, S.A.; Valdar, W.; Tarantino, L.M.; Ideraabdullah, F.Y. Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clin. Epigenetics 2016, 8, 107. [Google Scholar] [CrossRef]
- Junge, K.M.; Bauer, T.; Geissler, S.; Hirche, F.; Thurmann, L.; Bauer, M.; Trump, S.; Bieg, M.; Weichenhan, D.; Gu, L.; et al. Increased vitamin D levels at birth and in early infancy increase offspring allergy risk-evidence for involvement of epigenetic mechanisms. J. Allergy Clin. Immunol. 2016, 137, 610–613. [Google Scholar] [CrossRef]
- Suderman, M.; Stene, L.C.; Bohlin, J.; Page, C.M.; Holvik, K.; Parr, C.L.; Magnus, M.C.; Haberg, S.E.; Joubert, B.R.; Wu, M.C.; et al. 25-Hydroxyvitamin D in pregnancy and genome wide cord blood DNA methylation in two pregnancy cohorts (MoBa and ALSPAC). J. Steroid Biochem. Mol. Biol. 2016, 159, 102–109. [Google Scholar] [CrossRef]
- Benjamin Neelon, S.E.; White, A.J.; Vidal, A.C.; Schildkraut, J.M.; Murtha, A.P.; Murphy, S.K.; Kullman, S.W.; Hoyo, C. Maternal vitamin D, DNA methylation at imprint regulatory regions and offspring weight at birth, 1 year and 3 years. Int. J. Obes. 2018, 42, 587–593. [Google Scholar] [CrossRef]
- Reichetzeder, C.; Dwi Putra, S.E.; Li, J.; Hocher, B. Developmental Origins of Disease-Crisis Precipitates Change. Cell. Physiol. Biochem. 2016, 39, 919–938. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, L.J.; Xu, X.; Ye, A.; Travers-Gustafson, D.; Zhou, B.; Wang, H.W.; Zhang, W.; Lee Hamm, L.; Deng, H.W.; et al. DNA methylation levels of CYP2R1 and CYP24A1 predict vitamin D response variation. J. Steroid Biochem. Mol. Biol. 2014, 144 Pt A, 207–214. [Google Scholar] [CrossRef]
- Adams, J.S.; Hewison, M. Extrarenal expression of the 25-hydroxyvitamin D-1-hydroxylase. Arch. Biochem. Biophys. 2012, 523, 95–102. [Google Scholar] [CrossRef]
- Fransen, F.; van Beek, A.A.; Borghuis, T.; Meijer, B.; Hugenholtz, F.; van der Gaast-de Jongh, C.; Savelkoul, H.F.; de Jonge, M.I.; Faas, M.M.; Boekschoten, M.V.; et al. The Impact of Gut Microbiota on Gender-Specific Differences in Immunity. Front. Immunol. 2017, 8, 754. [Google Scholar] [CrossRef]
- Korpela, K.; Kallio, S.; Salonen, A.; Hero, M.; Kukkonen, A.K.; Miettinen, P.J.; Savilahti, E.; Kohva, E.; Kariola, L.; Suutela, M.; et al. Gut microbiota develop towards an adult profile in a sex-specific manner during puberty. Sci. Rep. 2021, 11, 23297. [Google Scholar] [CrossRef]
- Wilmanski, T.; Diener, C.; Rappaport, N.; Patwardhan, S.; Wiedrick, J.; Lapidus, J.; Earls, J.C.; Zimmer, A.; Glusman, G.; Robinson, M.; et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 2021, 3, 274–286. [Google Scholar] [CrossRef]
- Yuan, X.; Chen, R.; Zhang, Y.; Lin, X.; Yang, X. Gut microbiota: Effect of pubertal status. BMC Microbiol. 2020, 20, 334. [Google Scholar] [CrossRef]
- Pelzer, E.; Gomez-Arango, L.F.; Barrett, H.L.; Nitert, M.D. Review: Maternal health and the placental microbiome. Placenta 2017, 54, 30–37. [Google Scholar] [CrossRef]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Yim, Y.S.; Ha, S.; Atarashi, K.; Tan, T.G.; Longman, R.S.; Honda, K.; Littman, D.R.; Choi, G.B.; et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 2017, 549, 528–532. [Google Scholar] [CrossRef]
- Vuong, H.E.; Pronovost, G.N.; Williams, D.W.; Coley, E.J.L.; Siegler, E.L.; Qiu, A.; Kazantsev, M.; Wilson, C.J.; Rendon, T.; Hsiao, E.Y. The maternal microbiome modulates fetal neurodevelopment in mice. Nature 2020, 586, 281–286. [Google Scholar] [CrossRef]
- Dawson, S.L.; O’Hely, M.; Jacka, F.N.; Ponsonby, A.L.; Symeonides, C.; Loughman, A.; Collier, F.; Moreno-Betancur, M.; Sly, P.; Burgner, D.; et al. Maternal prenatal gut microbiota composition predicts child behaviour. eBioMedicine 2021, 68, 103400. [Google Scholar] [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra265. [Google Scholar] [CrossRef]
- Solt, I. The human microbiome and the great obstetrical syndromes: A new frontier in maternal-fetal medicine. Best Pract. Res. Clin. Obstet. Gynaecol. 2015, 29, 165–175. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Romero, R.; Amogan, H.P.; Kusanovic, J.P.; Bik, E.M.; Gotsch, F.; Kim, C.J.; Erez, O.; Edwin, S.; Relman, D.A. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: A molecular and culture-based investigation. PLoS ONE 2008, 3, e3056. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Romero, R.; Kusanovic, J.P.; Gomez, R.; Kim, C.J.; Seok, K.S.; Gotsch, F.; Mazaki-Tovi, S.; Vaisbuch, E.; Sanders, K.; et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 2010, 64, 38–57. [Google Scholar] [CrossRef]
- Onderdonk, A.B.; Hecht, J.L.; McElrath, T.F.; Delaney, M.L.; Allred, E.N.; Leviton, A.; Investigators, E.S. Colonization of second-trimester placenta parenchyma. Am. J. Obstet. Gynecol. 2008, 199, 52.e1–52.e10. [Google Scholar] [CrossRef]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef]
- Chen, J.J.; Zeng, B.H.; Li, W.W.; Zhou, C.J.; Fan, S.H.; Cheng, K.; Zeng, L.; Zheng, P.; Fang, L.; Wei, H.; et al. Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behav. Brain Res. 2017, 322, 34–41. [Google Scholar] [CrossRef]
- Bale, T.L. Epigenetic and transgenerational reprogramming of brain development. Nat. Rev. Neurosci. 2015, 16, 332–344. [Google Scholar] [CrossRef]
- Mallick, R.; Duttaroy, A.K. Epigenetic modification impacting brain functions: Effects of physical activity, micronutrients, caffeine, toxins, and addictive substances. Neurochem. Int. 2023, 171, 105627. [Google Scholar] [CrossRef]
- Di Simone, N.; Santamaria Ortiz, A.; Specchia, M.; Tersigni, C.; Villa, P.; Gasbarrini, A.; Scambia, G.; D’Ippolito, S. Recent Insights on the Maternal Microbiota: Impact on Pregnancy Outcomes. Front. Immunol. 2020, 11, 528202. [Google Scholar] [CrossRef]
- Bhatia, P.; Chhabra, S. Physiological and anatomical changes of pregnancy: Implications for anaesthesia. Indian J. Anaesth. 2018, 62, 651–657. [Google Scholar] [CrossRef]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Backhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Prochazkova, N.; Falony, G.; Dragsted, L.O.; Licht, T.R.; Raes, J.; Roager, H.M. Advancing human gut microbiota research by considering gut transit time. Gut 2023, 72, 180–191. [Google Scholar] [CrossRef]
- Zakaria, Z.Z.; Al-Rumaihi, S.; Al-Absi, R.S.; Farah, H.; Elamin, M.; Nader, R.; Bouabidi, S.; Suleiman, S.E.; Nasr, S.; Al-Asmakh, M. Physiological Changes and Interactions Between Microbiome and the Host During Pregnancy. Front. Cell. Infect. Microbiol. 2022, 12, 824925. [Google Scholar] [CrossRef]
- Rubini, E.; Schenkelaars, N.; Rousian, M.; Sinclair, K.D.; Wekema, L.; Faas, M.M.; Steegers-Theunissen, R.P.M.; Schoenmakers, S. Maternal obesity during pregnancy leads to derangements in one-carbon metabolism and the gut microbiota: Implications for fetal development and offspring wellbeing. Am. J. Obstet. Gynecol. 2022, 227, 392–400. [Google Scholar] [CrossRef]
- Manrique-Corredor, E.J.; Orozco-Beltran, D.; Lopez-Pineda, A.; Quesada, J.A.; Gil-Guillen, V.F.; Carratala-Munuera, C. Maternal periodontitis and preterm birth: Systematic review and meta-analysis. Community Dent. Oral Epidemiol. 2019, 47, 243–251. [Google Scholar] [CrossRef]
- Zakis, D.R.; Paulissen, E.; Kornete, L.; Kaan, A.M.M.; Nicu, E.A.; Zaura, E. The evidence for placental microbiome and its composition in healthy pregnancies: A systematic review. J. Reprod. Immunol. 2022, 149, 103455. [Google Scholar] [CrossRef]
- Mousa, W.K.; Chehadeh, F.; Husband, S. Microbial dysbiosis in the gut drives systemic autoimmune diseases. Front. Immunol. 2022, 13, 906258. [Google Scholar] [CrossRef]
- Wang, J.; Gu, X.; Yang, J.; Wei, Y.; Zhao, Y. Gut Microbiota Dysbiosis and Increased Plasma LPS and TMAO Levels in Patients With Preeclampsia. Front. Cell. Infect. Microbiol. 2019, 9, 409. [Google Scholar] [CrossRef] [PubMed]
- Nyangahu, D.D.; Jaspan, H.B. Influence of maternal microbiota during pregnancy on infant immunity. Clin. Exp. Immunol. 2019, 198, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jin, P.; Peng, J.; Zhang, X.; Wong, F.S.; Wen, L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J. Autoimmun. 2016, 72, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Holladay, S.D.; Smialowicz, R.J. Development of the murine and human immune system: Differential effects of immunotoxicants depend on time of exposure. Environ. Health Perspect. 2000, 108 (Suppl. S3), 463–473. [Google Scholar] [CrossRef] [PubMed]
- Eberl, G.; Lochner, M. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2009, 2, 478–485. [Google Scholar] [CrossRef]
- Westrom, B.; Arevalo Sureda, E.; Pierzynowska, K.; Pierzynowski, S.G.; Perez-Cano, F.J. The Immature Gut Barrier and Its Importance in Establishing Immunity in Newborn Mammals. Front. Immunol. 2020, 11, 1153. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Li, J.V.; Marchesi, J.R.; Nicholson, J.K. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012, 16, 559–564. [Google Scholar] [CrossRef] [PubMed]
- McElroy, S.J.; Weitkamp, J.H. Innate Immunity in the Small Intestine of the Preterm Infant. Neoreviews 2011, 12, e517–e526. [Google Scholar] [CrossRef]
- Timm, S.; Schlunssen, V.; Olsen, J.; Ramlau-Hansen, C.H. Prenatal antibiotics and atopic dermatitis among 18-month-old children in the Danish National Birth Cohort. Clin. Exp. Allergy 2017, 47, 929–936. [Google Scholar] [CrossRef]
- Ortqvist, A.K.; Lundholm, C.; Halfvarson, J.; Ludvigsson, J.F.; Almqvist, C. Fetal and early life antibiotics exposure and very early onset inflammatory bowel disease: A population-based study. Gut 2019, 68, 218–225. [Google Scholar] [CrossRef]
- Joly, A.; Leulier, F.; De Vadder, F. Microbial Modulation of the Development and Physiology of the Enteric Nervous System. Trends Microbiol. 2021, 29, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Millischer, V.; Rodin, S.; MacFabe, D.F.; Villaescusa, J.C.; Lavebratt, C. Enteric short-chain fatty acids promote proliferation of human neural progenitor cells. J. Neurochem. 2020, 154, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Pessa-Morikawa, T.; Husso, A.; Karkkainen, O.; Koistinen, V.; Hanhineva, K.; Iivanainen, A.; Niku, M. Maternal microbiota-derived metabolic profile in fetal murine intestine, brain and placenta. BMC Microbiol. 2022, 22, 46. [Google Scholar] [CrossRef] [PubMed]
- Garzoni, L.; Faure, C.; Frasch, M.G. Fetal cholinergic anti-inflammatory pathway and necrotizing enterocolitis: The brain-gut connection begins in utero. Front. Integr. Neurosci. 2013, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Kwok, L.Y.; Xi, X.; Zhong, Z.; Ma, T.; Xu, H.; Meng, H.; Zhao, F.; Zhang, H. The meconium microbiota shares more features with the amniotic fluid microbiota than the maternal fecal and vaginal microbiota. Gut Microbes 2020, 12, 1794266. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.M.; Hill, D.R.; Aurora, M.; Spence, J.R. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin. Cell Dev. Biol. 2017, 66, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Toure, A.M.; Landry, M.; Souchkova, O.; Kembel, S.W.; Pilon, N. Gut microbiota-mediated Gene-Environment interaction in the TashT mouse model of Hirschsprung disease. Sci. Rep. 2019, 9, 492. [Google Scholar] [CrossRef] [PubMed]
- Schneider, U.; Bode, F.; Schmidt, A.; Nowack, S.; Rudolph, A.; Dolker, E.M.; Schlattmann, P.; Gotz, T.; Hoyer, D. Developmental milestones of the autonomic nervous system revealed via longitudinal monitoring of fetal heart rate variability. PLoS ONE 2018, 13, e0200799. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F.; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Martinez, K.A., 2nd; Romano-Keeler, J.; Zackular, J.P.; Moore, D.J.; Brucker, R.M.; Hooper, C.; Meng, S.; Brown, N.; Mallal, S.; Reese, J.; et al. Bacterial DNA is present in the fetal intestine and overlaps with that in the placenta in mice. PLoS ONE 2018, 13, e0197439. [Google Scholar] [CrossRef] [PubMed]
- Ostrea, E.M., Jr.; Bielawski, D.M.; Posecion, N.C., Jr. Meconium analysis to detect fetal exposure to neurotoxicants. Arch. Dis. Child. 2006, 91, 628–629. [Google Scholar] [CrossRef] [PubMed]
- Jasarevic, E.; Howard, C.D.; Misic, A.M.; Beiting, D.P.; Bale, T.L. Stress during pregnancy alters temporal and spatial dynamics of the maternal and offspring microbiome in a sex-specific manner. Sci. Rep. 2017, 7, 44182. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Miyamoto, J.; Ohue-Kitano, R.; Watanabe, K.; Yamada, T.; Onuki, M.; Aoki, R.; Isobe, Y.; Kashihara, D.; Inoue, D.; et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 2020, 367, eaaw8429. [Google Scholar] [CrossRef] [PubMed]
- Segarra, M.; Aburto, M.R.; Acker-Palmer, A. Blood-Brain Barrier Dynamics to Maintain Brain Homeostasis. Trends Neurosci. 2021, 44, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Goasdoue, K.; Miller, S.M.; Colditz, P.B.; Bjorkman, S.T. Review: The blood-brain barrier; protecting the developing fetal brain. Placenta 2017, 54, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Saunders, N.R.; Liddelow, S.A.; Dziegielewska, K.M. Barrier mechanisms in the developing brain. Front. Pharmacol. 2012, 3, 46. [Google Scholar] [CrossRef]
- Yu, K.; Rodriguez, M.; Paul, Z.; Gordon, E.; Gu, T.; Rice, K.; Triplett, E.W.; Keller-Wood, M.; Wood, C.E. Transfer of oral bacteria to the fetus during late gestation. Sci. Rep. 2021, 11, 708. [Google Scholar] [CrossRef]
- Rodriguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Guzzardi, M.A.; Ait Ali, L.; D’Aurizio, R.; Rizzo, F.; Saggese, P.; Sanguinetti, E.; Weisz, A.; Pellegrini, M.; Iozzo, P. Fetal cardiac growth is associated with in utero gut colonization. Nutr. Metab. Cardiovasc. Dis. 2019, 29, 170–176. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Mikhailidis, D.P.; Jeremy, J.Y.; Barradas, M.A.; Dandona, P.; Hutton, R.A. Increases in platelet and red cell counts, blood viscosity, and arterial pressure during mild surface cooling. Br. Med. J. 1985, 290, 74–75. [Google Scholar] [CrossRef][Green Version]
- Ratsika, A.; Codagnone, M.C.; O’Mahony, S.; Stanton, C.; Cryan, J.F. Priming for Life: Early Life Nutrition and the Microbiota-Gut-Brain Axis. Nutrients 2021, 13, 423. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef]
- Jena, A.; Montoya, C.A.; Mullaney, J.A.; Dilger, R.N.; Young, W.; McNabb, W.C.; Roy, N.C. Gut-Brain Axis in the Early Postnatal Years of Life: A Developmental Perspective. Front. Integr. Neurosci. 2020, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Sajdel-Sulkowska, E.M. The Impact of Maternal Gut Microbiota during Pregnancy on Fetal Gut-Brain Axis Development and Life-Long Health Outcomes. Microorganisms 2023, 11, 2199. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, A.; Goeden, N.; Chen, K.; Wilson, M.L.; King, J.; Shih, J.C.; Blakely, R.D.; Deneris, E.S.; Levitt, P. A transient placental source of serotonin for the fetal forebrain. Nature 2011, 472, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, A.; Levitt, P. Fetal, maternal, and placental sources of serotonin and new implications for developmental programming of the brain. Neuroscience 2011, 197, 1–7. [Google Scholar] [CrossRef]
- Dicks, L.M.T. Gut Bacteria and Neurotransmitters. Microorganisms 2022, 10, 1838. [Google Scholar] [CrossRef]
- Momose-Sato, Y.; Sato, K. Development of synaptic networks in the mouse vagal pathway revealed by optical mapping with a voltage-sensitive dye. Eur. J. Neurosci. 2016, 44, 1906–1918. [Google Scholar] [CrossRef] [PubMed]
- Sachis, P.N.; Armstrong, D.L.; Becker, L.E.; Bryan, A.C. Myelination of the human vagus nerve from 24 weeks postconceptional age to adolescence. J. Neuropathol. Exp. Neurol. 1982, 41, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar] [PubMed]
- Chaudhry, T.S.; Senapati, S.G.; Gadam, S.; Mannam, H.; Voruganti, H.V.; Abbasi, Z.; Abhinav, T.; Challa, A.B.; Pallipamu, N.; Bheemisetty, N.; et al. The Impact of Microbiota on the Gut-Brain Axis: Examining the Complex Interplay and Implications. J. Clin. Med. 2023, 12, 5231. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Syed, Y.A.; Khan, M.R. Understanding the Role of the Gut Microbiome in Brain Development and Its Association With Neurodevelopmental Psychiatric Disorders. Front. Cell Dev. Biol. 2022, 10, 880544. [Google Scholar] [CrossRef]
- Rusch, J.A.; Layden, B.T.; Dugas, L.R. Signalling cognition: The gut microbiota and hypothalamic-pituitary-adrenal axis. Front. Endocrinol. 2023, 14, 1130689. [Google Scholar] [CrossRef]
- Farzi, A.; Frohlich, E.E.; Holzer, P. Gut Microbiota and the Neuroendocrine System. Neurotherapeutics 2018, 15, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Dunn, E.; Kostaki, A.; Andrews, M.H.; Matthews, S.G. Fetal programming of hypothalamo-pituitary-adrenal function: Prenatal stress and glucocorticoids. J. Physiol. 2006, 572, 31–44. [Google Scholar] [CrossRef]
- Condon, J.; Gosden, C.; Gardener, D.; Nickson, P.; Hewison, M.; Howie, A.J.; Stewart, P.M. Expression of type 2 11beta-hydroxysteroid dehydrogenase and corticosteroid hormone receptors in early human fetal life. J. Clin. Endocrinol. Metab. 1998, 83, 4490–4497. [Google Scholar] [CrossRef][Green Version]
- Howland, M.A.; Sandman, C.A.; Glynn, L.M. Developmental origins of the human hypothalamic-pituitary-adrenal axis. Expert Rev. Endocrinol. Metab. 2017, 12, 321–339. [Google Scholar] [CrossRef]
- Sheng, J.A.; Bales, N.J.; Myers, S.A.; Bautista, A.I.; Roueinfar, M.; Hale, T.M.; Handa, R.J. The Hypothalamic-Pituitary-Adrenal Axis: Development, Programming Actions of Hormones, and Maternal-Fetal Interactions. Front. Behav. Neurosci. 2020, 14, 601939. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C. The fetal and neonatal hypothalamic-pituitary-adrenal axis. Arch. Dis. Child Fetal. Neonatal Ed. 2000, 82, F250–F254. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, S.J.; Zhou, J.; Peters, T.J.; Buckley, M.; Sutcliffe, B.; Oytam, Y.; Gibson, R.A.; McPhee, A.; Yelland, L.N.; Makrides, M.; et al. Effect of prenatal DHA supplementation on the infant epigenome: Results from a randomized controlled trial. Clin. Epigenetics 2016, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.L.; Mumford, S.L.; Guan, W.; Zeng, X.; Kim, K.; Radoc, J.G.; Trinh, M.-H.; Flannagan, K.; Schisterman, E.F.; Yeung, E. Maternal fatty acid concentrations and newborn DNA methylation. Am. J. Clin. Nutr. 2020, 111, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Saffari, A.; Shrestha, S.; Issarapu, P.; Sajjadi, S.; Betts, M.; Sahariah, S.A.; Tomar, A.S.; James, P.; Dedaniya, A.; Yadav, D.K.; et al. Effect of maternal preconceptional and pregnancy micronutrient interventions on children’s DNA methylation: Findings from the EMPHASIS study. Am. J. Clin. Nutr. 2020, 112, 1099–1113. [Google Scholar] [CrossRef]
- Bianchi, M.; Alisi, A.; Fabrizi, M.; Vallone, C.; Ravà, L.; Giannico, R.; Vernocchi, P.; Signore, F.; Manco, M. Maternal Intake of n-3 Polyunsaturated Fatty Acids During Pregnancy Is Associated with Differential Methylation Profiles in Cord Blood White Cells. Front. Genet. 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Koemel, N.A.; Senior, A.M.; Dissanayake, H.U.; Ross, J.; McMullan, R.L.; Kong, Y.; Phang, M.; Hyett, J.; Raubenheimer, D.; Gordon, A.; et al. Maternal dietary fatty acid composition and newborn epigenetic aging—a geometric framework approach. Am. J. Clin. Nutr. 2022, 115, 118–127. [Google Scholar] [CrossRef]
- Chen, L.; Wagner, C.L.; Dong, Y.; Wang, X.; Shary, J.R.; Huang, Y.; Hollis, B.W.; Zhu, H. Effects of Maternal Vitamin D3 Supplementation on Offspring Epigenetic Clock of Gestational Age at Birth: A Post-hoc Analysis of a Randomized Controlled Trial. Epigenetics 2020, 15, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, A.; Irwin, R.E.; McNulty, H.; Strain, J.J.; Lees-Murdock, D.J.; McNulty, B.A.; Ward, M.; Walsh, C.P.; Pentieva, K. Gene-specific DNA methylation in newborns in response to folic acid supplementation during the second and third trimesters of pregnancy: Epigenetic analysis from a randomized controlled trial. Am. J. Clin. Nutr. 2018, 107, 566–575. [Google Scholar] [CrossRef]
- Küpers, L.K.; Fernández-Barrés, S.; Mancano, G.; Johnson, L.; Ott, R.; Vioque, J.; Colombo, M.; Landgraf, K.; Tobi, E.W.; Körner, A.; et al. Maternal Dietary Glycemic Index and Glycemic Load in Pregnancy and Offspring Cord Blood DNA Methylation. Diabetes Care 2022, 45, 1822–1832. [Google Scholar] [CrossRef]
- Lecorguillé, M.; Navarro, P.; Chen, L.-W.; Murrin, C.; Viljoen, K.; Mehegan, J.; Shivappa, N.; Hébert, J.R.; Kelleher, C.C.; Suderman, M.; et al. Maternal and Paternal Dietary Quality and Dietary Inflammation Associations with Offspring DNA Methylation and Epigenetic Biomarkers of Aging in the Lifeways Cross-Generation Study. J. Nutr. 2023, 153, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Nahm, S.; Mendez, M.; Robinson, W.; Murphy, S.K.; Hoyo, C.; Hogan, V.; Rowley, D. Low maternal adherence to a Mediterranean diet is associated with increase in methylation at the MEG3-IG differentially methylated region in female infants. Environ. Epigenetics 2017, 3, dvx007. [Google Scholar] [CrossRef] [PubMed]
- House, J.S.; Mendez, M.; Maguire, R.L.; Gonzalez-Nahm, S.; Huang, Z.; Daniels, J.; Murphy, S.K.; Fuemmeler, B.F.; Wright, F.A.; Hoyo, C. Periconceptional Maternal Mediterranean Diet Is Associated with Favorable Offspring Behaviors and Altered CpG Methylation of Imprinted Genes. Front. Cell Dev. Biol. 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-H.; Fadadu, R.P.; Gaskins, A.J.; Rifas-Shiman, S.L.; Laue, H.E.; Moley, K.H.; Hivert, M.-F.; Baccarelli, A.; Oken, E.; Chavarro, J.E.; et al. Dietary fat intake during early pregnancy is associated with cord blood DNA methylation at IGF2 and H19 genes in newborns. Environ. Mol. Mutagen. 2021, 62, 388–398. [Google Scholar] [CrossRef]
- Geraghty, A.A.; Sexton-Oates, A.; O’Brien, E.C.; Saffery, R.; McAuliffe, F.M. Epigenetic Patterns in Five-Year-Old Children Exposed to a Low Glycemic Index Dietary Intervention during Pregnancy: Results from the ROLO Kids Study. Nutrients 2020, 12, 3602. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, A.A.; Sexton-Oates, A.; O’Brien, E.C.; Alberdi, G.; Fransquet, P.; Saffery, R.; McAuliffe, F.M. A Low Glycaemic Index Diet in Pregnancy Induces DNA Methylation Variation in Blood of Newborns: Results from the ROLO Randomised Controlled Trial. Nutrients 2018, 10, 455. [Google Scholar] [CrossRef]
- Sasaki, T.; Kawamura, M.; Okuno, C.; Lau, K.; Riel, J.; Lee, M.-J.; Miller, C. Impact of Maternal Mediterranean-Type Diet Adherence on Microbiota Composition and Epigenetic Programming of Offspring. Nutrients 2024, 16, 47. [Google Scholar] [CrossRef]
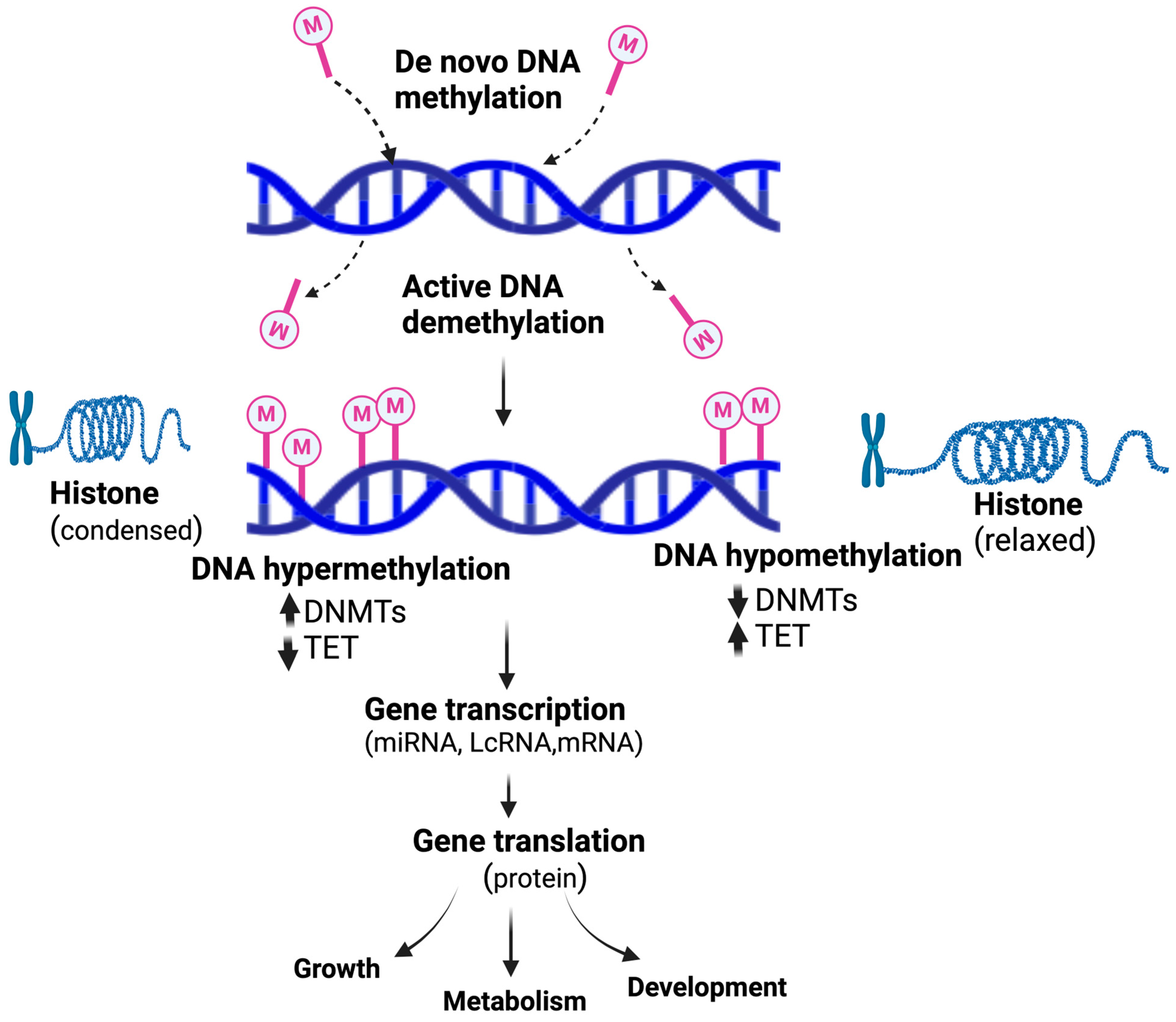
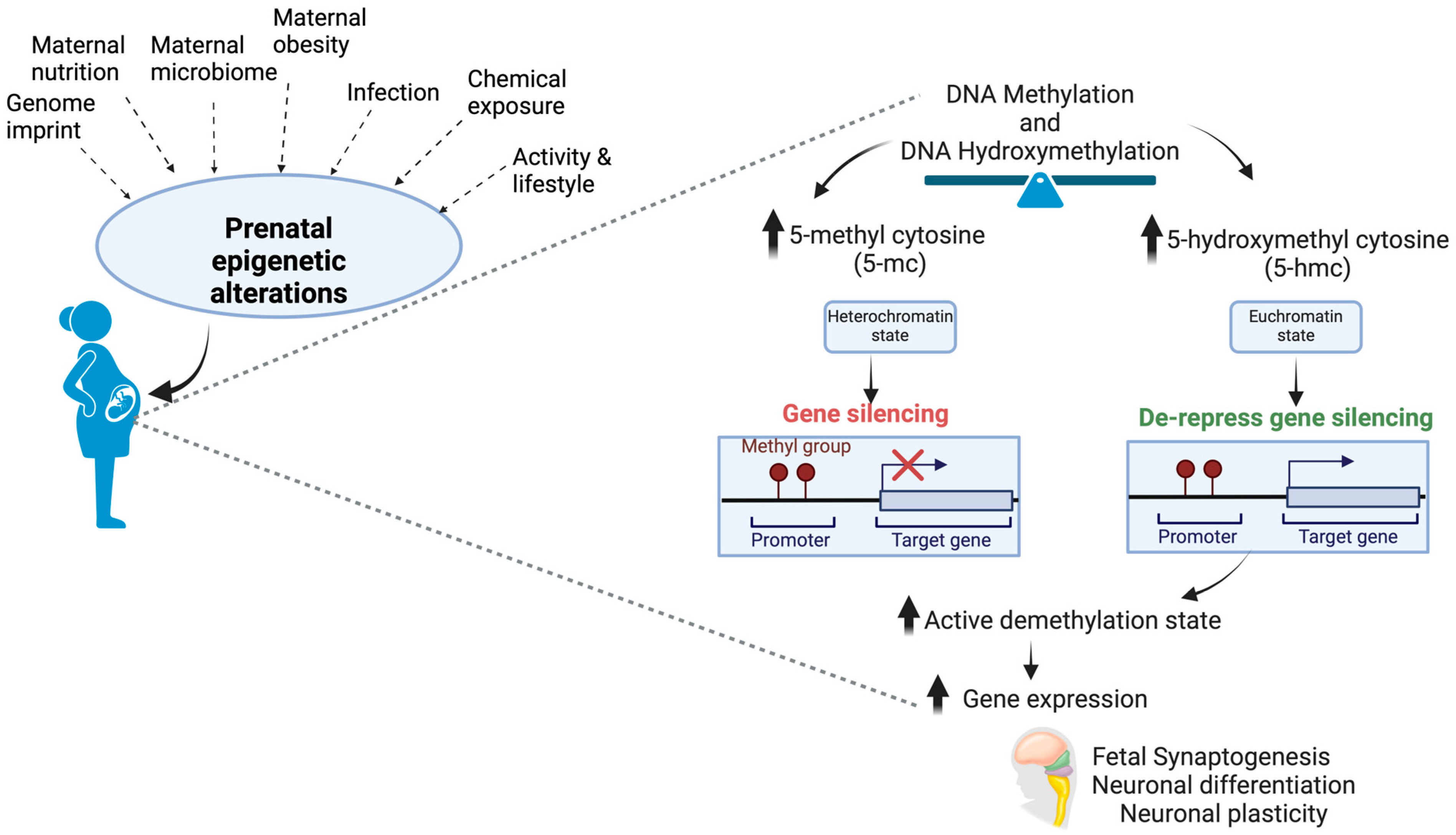
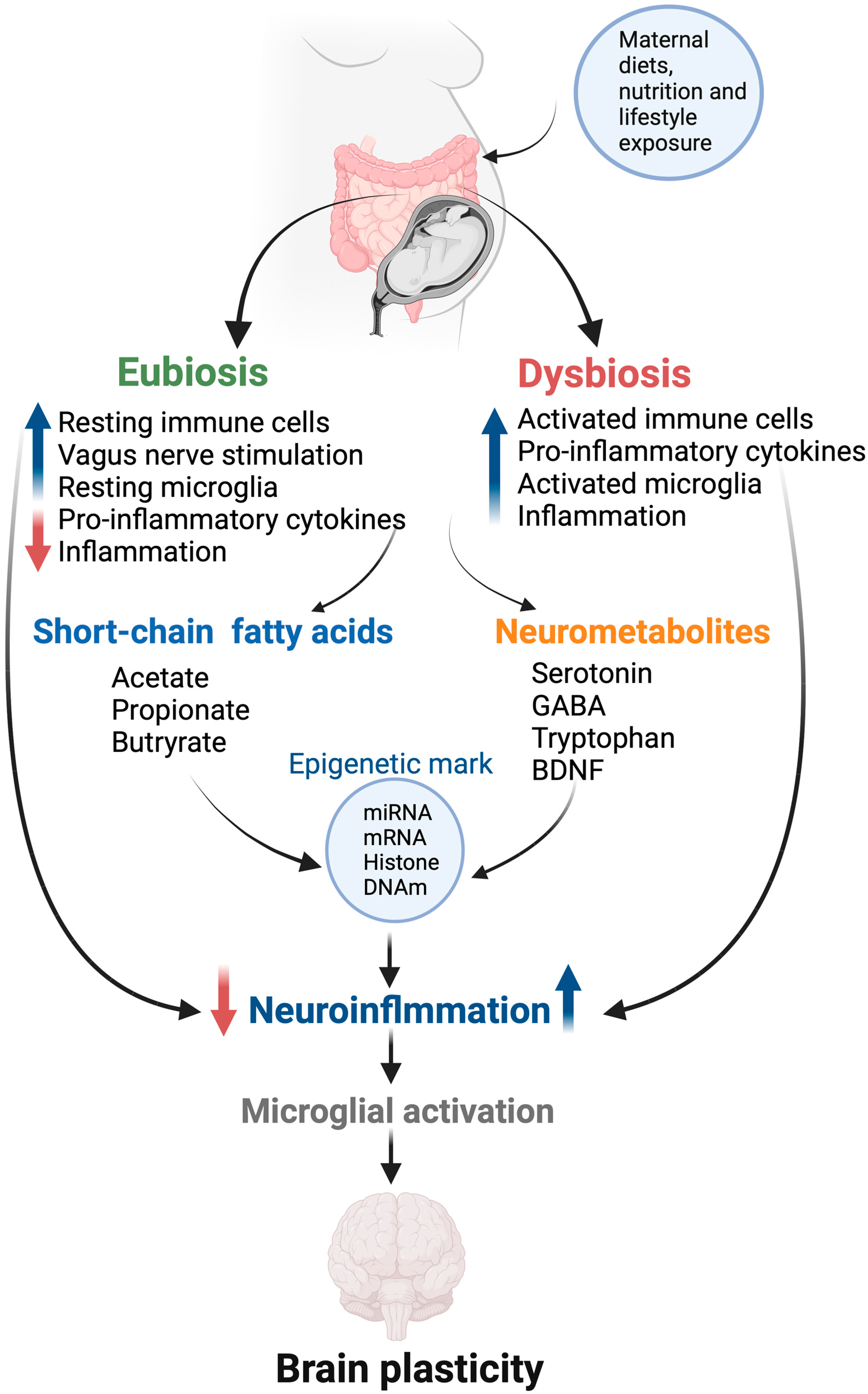
| Aim | Study Design | Primary Outcome | Refs. |
|---|---|---|---|
| To distinguish between direct transmission of epigenetic states and de novo epigenetically induced marks in each generation in response to energy diets | Female Wistar rats, dietary energy increased by 25% at conception in F0 and maintained up to generation F3; epigenetic marks over four generations studied |
| [42] |
| Effect of dietary choline on fetal brain and memory function in deficient mice | Timed-pregnant C57 BL/6 mice, choline diet—1.1 g/kg choline chloride from embryonic day 12 to day 17, brain analysis on day 17 |
| [14] |
| Restricted prenatal diet and transgenerational epigenetic alterations in four successive generations in rats | Wistar rats, control—chow diet, restricted group—50% of their daily intake from the appearance of vaginal plug until parturition |
| [43] |
| n-3 PUFA deficiency at the time of pregnancy and lactation and epigenetic changes in the brain of adult offspring in mice | C57BL/6J mice, n-3 PUFA deficient diet—sunflower oil with high LA and no n-3 PUFA; n-3 PUFA adequate diet—fish oil, flax seed and sunflower oil containing 0.47% DHA and 5.25% LA |
| [44] |
| Effect of folic acid availability or deficiency on oocyte development and epigenetic effects in next-generation progeny | Female BALB/c mice, deficient in (7-fold) or supplemented with (10-fold) folic acid starting from 4 wks prior to mating throughout gestation and lactation |
| [45] |
| Maternal n-3 PUFA deficiency and changes in the epigenetic modulators of the placenta | Female Swiss albino mice, n-3 PUFA deficient diet—n-6/n-3 PUFA = 50/1, 0.13% energy from ALA; n-3 sufficient diet—n-6/n-3 PUFA = 2/1; 2.26% energy from ALA |
| [12] |
| Effect of maternal micronutrients and omega-3 fatty acids on global methylation patterns in the brain | Pregnant rats fed with folic acid (normal and excess) in presence or absence of Vit B12; omega-3 supplementation in Vit B12-deficient rats |
| [46] |
| Effects of dietary combinations of B12 and folic acid on genome-imprinting modulators | 6-week-old C57BL/6 male mice fed with standard chow diet and females with different levels of folic acid and B12 for two generations | Non-coding RNA expression of IGF2R and KCNQ1OT1 sensitive to different dietary combinations folic acid and B12 in mouse offspring, indicating epigenetic programming | [47] |
| Maternal low protein diet (MLPD) and epigenetic alterations in brain renin–angiotensin system (RAS) in mice | FVB/NJ mice, MLPD—50% protein depletion, from gD10.5 to 17.5 |
| [48] |
| Effect of maternal HFD on fetal gluconeogenic gene expression and regulation of histone modifications | Timed-pregnant obese resistant rats, control—64%, 20%, 16%; HFD—35%, 20%, 45% of carbohydrates, proteins, and fats, respectively, from embryonic day 2 to 20 | In utero exposure to HFD programmed the gluconeogenic capacity of offspring through epigenetic modifications and predisposed offspring with altered insulin sensitivity in adulthood | [49] |
| Maternal calorie restriction (CR) on DNA methylation and placental gene expression | C57/BL6 mice, control—chow diet, CR—50% (w/w) daily intake from gD 10 to 19 | Altered DMRs in CR mice associated with IUGR phenotypes enriched with micro-RNA target genes linked with risk factors of cardiovascular and neurological diseases | [50] |
| Maternal HFD during pregnancy and its effects on regulation of gene functions | Female C57BL6J, HFD—22.6% fat, 48.6% carbohydrate, 23% protein; standard chow diet—10% fat, 68.8% carbohydrate, 18% protein; prior to conception and during pregnancy and lactation | Altered hepatic expression of insulin-like growth factor-2 and key microRNAs in adult offspring | [51] |
| Aim | Subjects and Design | Analysis and Method | Primary Outcome | Refs. |
|---|---|---|---|---|
| In utero DHA supplement and fetal epigenome |
| Blood spots at birth (n = 991) |
| [213] |
| Association between maternal fatty acids (FAs) and newborn DNA methylation (DNAm) |
| Cord blood DNA from singletons |
| [214] |
| Dietary PUFAs during pregnancy and methylation of imprinted genes | DHA (400 mg/day, n = 131) or placebo (n = 130) from gD 18–22 until delivery | DNA methylation of IGF2 promoter 3 (P3), IGF2 DMRs, and H19 DMR in cord blood mononuclear cells |
| [108] |
| Maternal micronutrient supplementation and DNAm in their children | Indian (n = 698) and Gambian (n = 293) pregnant women before and during pregnancy |
|
| [215] |
| Prenatal n-3 PUFA intake and DMRs in cord blood | Pregnant women, n = 577, Italy | Pregnant women: 45–64% of daily calories from carbohydrates, 20–35% of daily calories from fats, and 60 g/day of proteins; 3 portions of fish per week |
| [216] |
| Relationship of maternal dietary FA quality with epigenetic aging and newborn cardiometabolic risk | Healthy mothers and children, n = 224, Australia | Body fat, aortic intima media thickness, heart rate variability, and epigenetic age acceleration in newborn infants |
| [217] |
| Vitamin D supplementation and epigenetic gestational age acceleration (GAA) in newborns | Pregnant women, n = 92, multiethnic population, USA |
|
| [218] |
| Folic acid beyond first trimester and DNAm of genes related to brain development and function | Pregnant women, n = 86, Ireland | 400 µg folic acid/day through the second and third trimesters compared with placebo |
| [219] |
| Effect of maternal choline intake on methylations of cortisol regulating genes in placenta and cord venous blood | Pregnant women, n = 26, USA | Choline supplementation: 480 mg/day and 930 mg/day for 12 weeks from 26–29 gestational weeks until delivery |
| [16] |
| Effect of maternal dietary glycemic index on cord blood DNAm | Mother–offspring pairs, n = 2003, U.K., Netherlands and Spain | Maternal dietary glycemic index calculated by FFQ and correlated with cord blood DNAm | Maternal glycemic index associated with changes in DNAm of genes associated with neurodevelopment and lipid metabolism in overweight/obesity subjects | [220] |
| Parental dietary quality on offspring DNAm | Families, n = 1124, Ireland | Maternal diet in first trimester and paternal diet were assessed by FFQ |
| [221] |
| Mediterranean diet in pregnancy and neonatal DNA methylation at birth | Mother–infant pairs, n= 390, USA | Overall dietary assessment by FFQ in periconception and once in each trimester |
| [222] |
| Mediterranean diet adherence during pregnancy and imprinted gene methylation of brain task | Mother–infant pairs, n = 325, USA | Maternal periconceptional FFQ and correlated with child behavioral outcomes at 2 years of age |
| [223] |
| Dietary fat intake during pregnancy and imprinting gene methylations | Pregnant women, n = 154, Eastern Massachusetts | Maternal fat intake during first and second trimester by FFQ | Maternal total fat and PUFA intake inversely correlated with IGF2-DMR and positively correlated with H19-DMR methylation | [224] |
| Effect of substituted low-glycemic-index (GI) diet or GI diet on epigenetic profile of offspring at 5 years of age | 5-year-old children from ROLO kids’ study, n = 60–63, Australia, Ireland | High-GI diet substituted with low-GI from second trimester until delivery | No association between maternal factors due to substituted GI or low-GI diet on DNAm status of offspring at 5 years of age | [225,226] |
| Mediterranean diet adherence during pregnancy, fetal gut microbiota and offspring epigenetic regulation | Pregnant women, n = 41, USA | Dietary patterns assessed by FFQ, Mediterranean diet adherence scores correlated with neonatal microbiome and fetal epigenetic programming | Adherence to Mediterranean diet results in a greater abundance of Pasteurellaceae, Bacteroidaceae, and other short-chain fatty acid-producing species and is connected with fetal DMRs of in utero development | [227] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basak, S.; Mallick, R.; Navya Sree, B.; Duttaroy, A.K. Placental Epigenome Impacts Fetal Development: Effects of Maternal Nutrients and Gut Microbiota. Nutrients 2024, 16, 1860. https://doi.org/10.3390/nu16121860
Basak S, Mallick R, Navya Sree B, Duttaroy AK. Placental Epigenome Impacts Fetal Development: Effects of Maternal Nutrients and Gut Microbiota. Nutrients. 2024; 16(12):1860. https://doi.org/10.3390/nu16121860
Chicago/Turabian StyleBasak, Sanjay, Rahul Mallick, Boga Navya Sree, and Asim K. Duttaroy. 2024. "Placental Epigenome Impacts Fetal Development: Effects of Maternal Nutrients and Gut Microbiota" Nutrients 16, no. 12: 1860. https://doi.org/10.3390/nu16121860
APA StyleBasak, S., Mallick, R., Navya Sree, B., & Duttaroy, A. K. (2024). Placental Epigenome Impacts Fetal Development: Effects of Maternal Nutrients and Gut Microbiota. Nutrients, 16(12), 1860. https://doi.org/10.3390/nu16121860









