Low-Iron Diet-Induced Fatty Liver Development Is Microbiota Dependent and Exacerbated by Loss of the Mitochondrial Iron Importer Mitoferrin2
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Histology
2.3. Lipidomics
2.4. Analysis of Mass Spectrometry Data
2.5. LION/Web Lipid Ontology Analysis
2.6. Comments on Lipid Annotations
2.7. RNA Isolation and Real-Time Quantitative PCR (qPCR)
2.8. HOMA-IR, Serum Insulin, Blood Glucose
2.9. Insulin ELISA
2.10. Insulin Resistance Test
2.11. Triglyceride Analysis
2.12. Statistical Analysis
3. Results
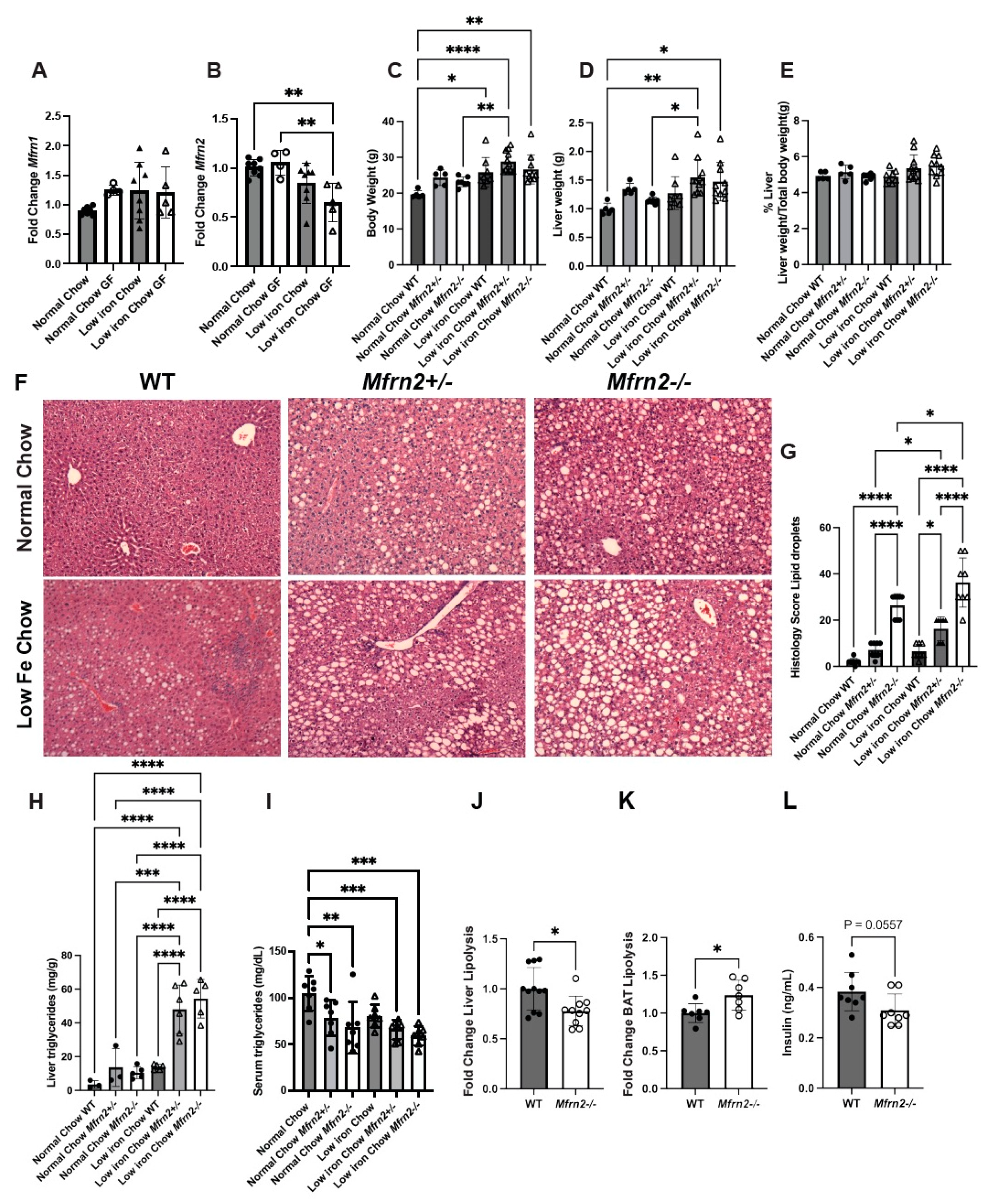
3.1. Decreased Nutritional Iron Availability Results in Increased Fat Deposition in the Liver
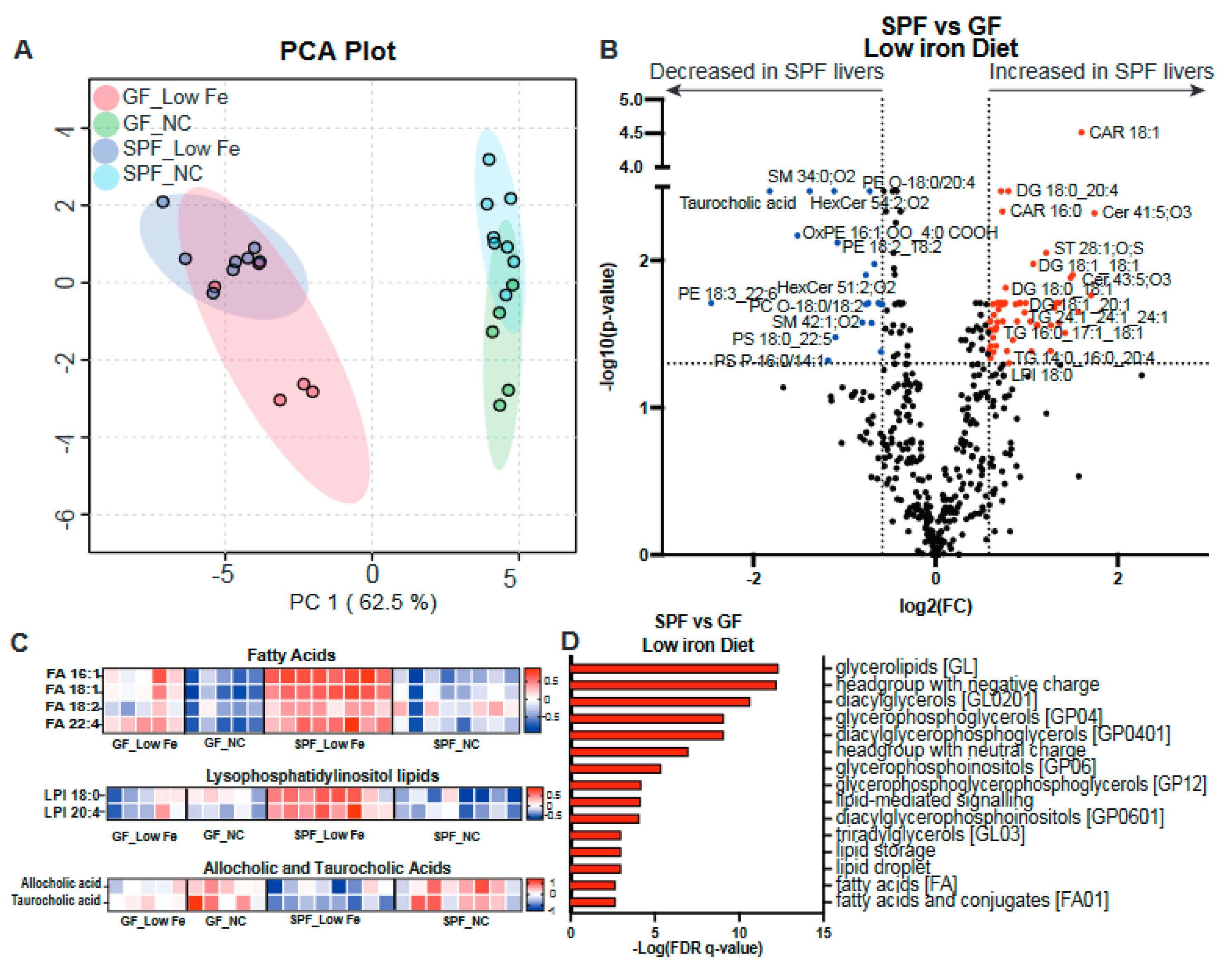
3.2. Decreased Nutritional Iron and the Gut Microbiota Contribute to Altered Lipid Metabolism
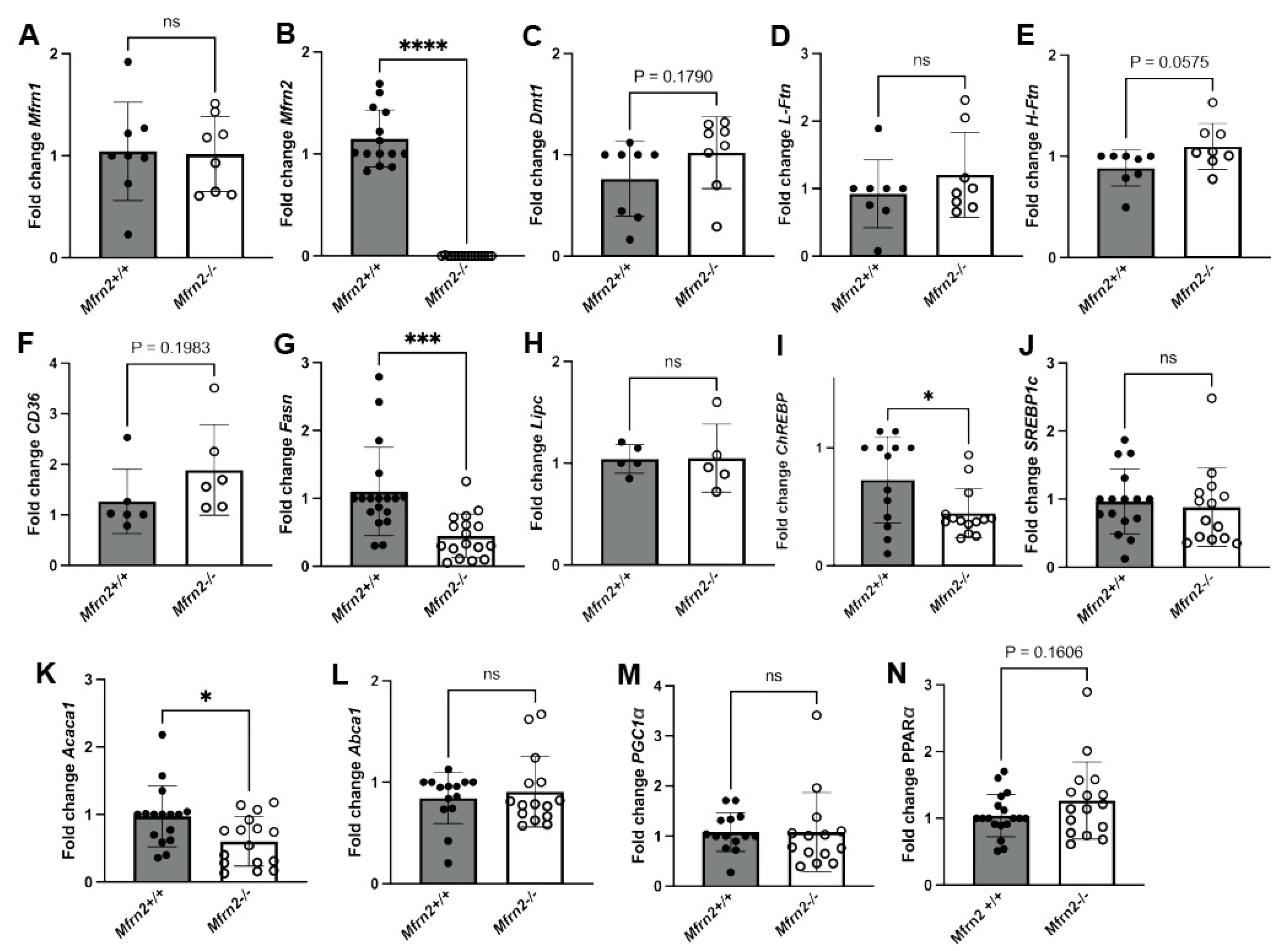
3.3. GF Animals Express Markedly Reduced Levels of Lipogenesis Genes SREBP-1c and Fasn
3.4. Liver Lipid Metabolism Is Altered by Both the Microbiota and Dietary Iron Limitation
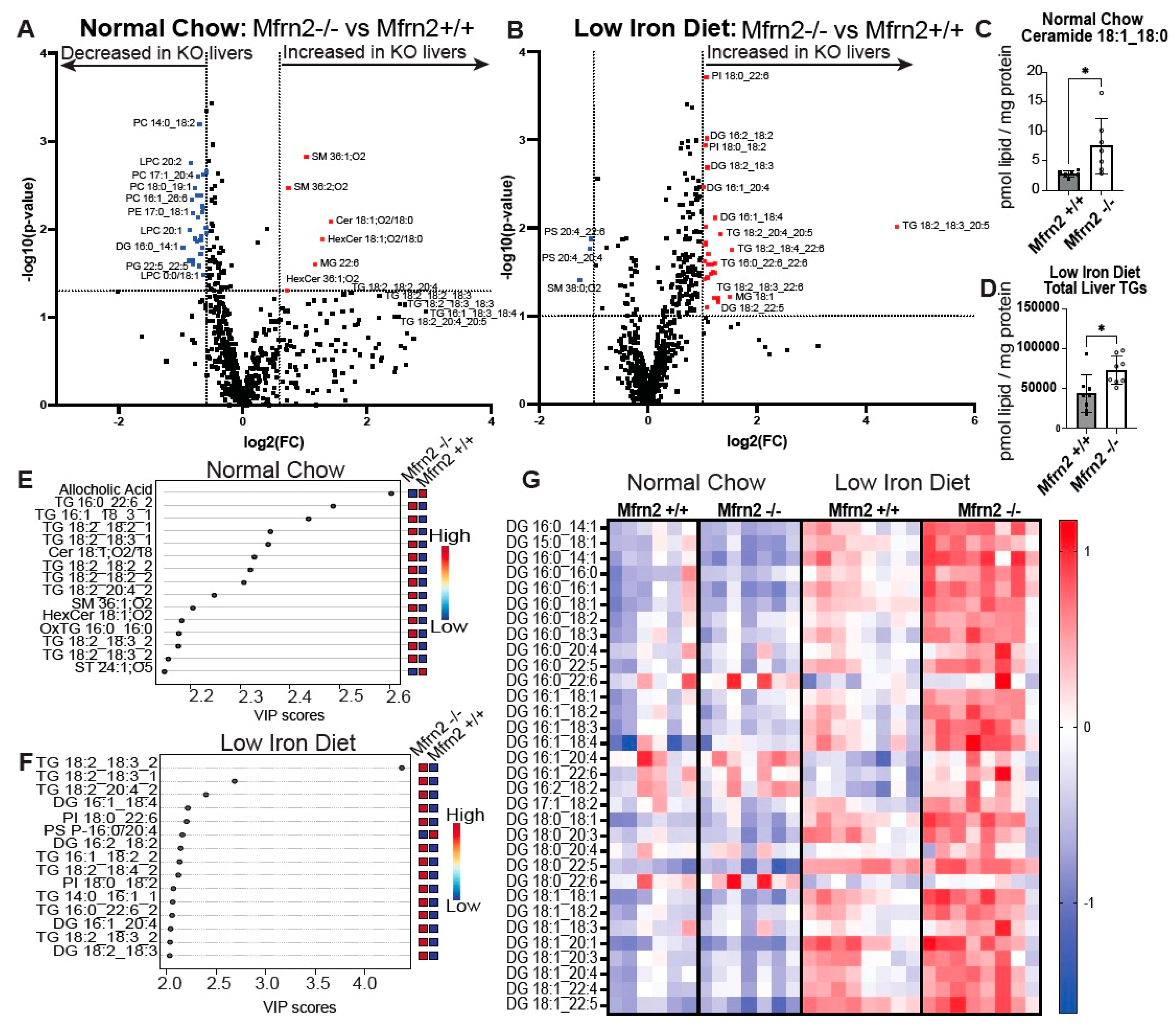
3.5. Loss of Mitochondrial Iron Importer Mitoferrin2 Results in Increased MASLD That Is Exacerbated by a Low-Iron Diet
3.6. Low-Iron Diet and Loss of Mfrn2 Are Additive for Altered Liver Lipid Homeostasis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galy, B.; Conrad, M.; Muckenthaler, M. Mechanisms controlling cellular and systemic iron homeostasis. Nat. Rev. Mol. Cell Biol. 2024, 25, 133–155. [Google Scholar] [CrossRef]
- Ganz, T. Systemic iron homeostasis. Physiol. Rev. 2013, 93, 1721–1741. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. Biometals 2019, 32, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Mayneris-Perxachs, J.; Moreno-Navarrete, J.M.; Fernandez-Real, J.M. The role of iron in host-microbiota crosstalk and its effects on systemic glucose metabolism. Nat. Rev. Endocrinol. 2022, 18, 683–698. [Google Scholar] [CrossRef]
- Puga, A.M.; Samaniego-Vaesken, M.L.; Montero-Bravo, A.; Ruperto, M.; Partearroyo, T.; Varela-Moreiras, G. Iron Supplementation at the Crossroads of Nutrition and Gut Microbiota: The State of the Art. Nutrients 2022, 14, 1926. [Google Scholar] [CrossRef]
- Aksoyalp, Z.S.; Temel, A.; Erdogan, B.R. Iron in infectious diseases friend or foe?: The role of gut microbiota. J. Trace Elem. Med. Biol. 2023, 75, 127093. [Google Scholar] [CrossRef]
- Malesza, I.J.; Bartkowiak-Wieczorek, J.; Winkler-Galicki, J.; Nowicka, A.; Dzieciolowska, D.; Blaszczyk, M.; Gajniak, P.; Slowinska, K.; Niepolski, L.; Walkowiak, J.; et al. The Dark Side of Iron: The Relationship between Iron, Inflammation and Gut Microbiota in Selected Diseases Associated with Iron Deficiency Anaemia-A Narrative Review. Nutrients 2022, 14, 3478. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, X.; Li, H.; Borger, D.K.; Wei, Q.; Yang, E.; Xu, C.; Pinho, S.; Frenette, P.S. The microbiota regulates hematopoietic stem cell fate decisions by controlling iron availability in bone marrow. Cell Stem Cell 2022, 29, 232–247.e7. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhang, J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 2. [Google Scholar] [CrossRef]
- Kirundi, J.; Moghadamrad, S.; Urbaniak, C. Microbiome-liver crosstalk: A multihit therapeutic target for liver disease. World J. Gastroenterol. 2023, 29, 1651–1668. [Google Scholar] [CrossRef]
- Choi, K.J.; Yoon, M.Y.; Kim, J.E.; Yoon, S.S. Gut commensal Kineothrix alysoides mitigates liver dysfunction by restoring lipid metabolism and gut microbial balance. Sci. Rep. 2023, 13, 14668. [Google Scholar] [CrossRef] [PubMed]
- Britton, L.J.; Subramaniam, V.N.; Crawford, D.H. Iron and non-alcoholic fatty liver disease. World J. Gastroenterol. 2016, 22, 8112–8122. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Fracanzani, A.L.; Fargion, S.; Valenti, L. Iron in fatty liver and in the metabolic syndrome: A promising therapeutic target. J. Hepatol. 2011, 55, 920–932. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.H.; Chen, G.C.; Li, D.M.; Lan, L.; Chen, L.H.; Xu, J.Y.; Qin, L.Q. Serum iron and risk of nonalcoholic fatty liver disease and advanced hepatic fibrosis in US adults. Sci. Rep. 2021, 11, 10387. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wu, J.; Li, X.; Xie, D.; Wang, Y.; Yang, T. Association between dietary iron intake and the prevalence of nonalcoholic fatty liver disease: A cross-sectional study. Medicine 2019, 98, e17613. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Zheng, D.; Shibolet, O.; Elinav, E. The role of the microbiome in NAFLD and NASH. EMBO Mol. Med. 2019, 11, e9302. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Schnabl, B. Microbiota and Fatty Liver Disease-the Known, the Unknown, and the Future. Cell Host Microbe 2020, 28, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Mayneris-Perxachs, J.; Cardellini, M.; Hoyles, L.; Latorre, J.; Davato, F.; Moreno-Navarrete, J.M.; Arnoriaga-Rodriguez, M.; Serino, M.; Abbott, J.; Barton, R.H.; et al. Iron status influences non-alcoholic fatty liver disease in obesity through the gut microbiome. Microbiome 2021, 9, 104. [Google Scholar] [CrossRef]
- Bellanti, F.; Lo Buglio, A.; Vendemiale, G. Hepatic Mitochondria-Gut Microbiota Interactions in Metabolism-Associated Fatty Liver Disease. Metabolites 2023, 13, 322. [Google Scholar] [CrossRef]
- Hilton, C.; Sabaratnam, R.; Drakesmith, H.; Karpe, F. Iron, glucose and fat metabolism and obesity: An intertwined relationship. Int. J. Obes. 2023, 47, 554–563. [Google Scholar] [CrossRef]
- Zhang, Z.; Funcke, J.B.; Zi, Z.; Zhao, S.; Straub, L.G.; Zhu, Y.; Zhu, Q.; Crewe, C.; An, Y.A.; Chen, S.; et al. Adipocyte iron levels impinge on a fat-gut crosstalk to regulate intestinal lipid absorption and mediate protection from obesity. Cell Metab. 2021, 33, 1624–1639.e9. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Yoon, S.Y.; Ul-Haq, A.; Jo, S.; Kim, S.; Rahim, M.A.; Park, H.A.; Ghorbanian, F.; Kim, M.J.; Lee, M.Y.; et al. The Effects of Iron Deficiency on the Gut Microbiota in Women of Childbearing Age. Nutrients 2023, 15, 691. [Google Scholar] [CrossRef] [PubMed]
- Stangl, G.I.; Kirchgessner, M. Different degrees of moderate iron deficiency modulate lipid metabolism of rats. Lipids 1998, 33, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Rockfield, S.; Chhabra, R.; Robertson, M.; Rehman, N.; Bisht, R.; Nanjundan, M. Links Between Iron and Lipids: Implications in Some Major Human Diseases. Pharmaceuticals 2018, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Oexle, H.; Gnaiger, E.; Weiss, G. Iron-dependent changes in cellular energy metabolism: Influence on citric acid cycle and oxidative phosphorylation. Biochim. Biophys. Acta 1999, 1413, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Volani, C.; Doerrier, C.; Demetz, E.; Haschka, D.; Paglia, G.; Lavdas, A.A.; Gnaiger, E.; Weiss, G. Dietary iron loading negatively affects liver mitochondrial function. Metallomics 2017, 9, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.B.; Knutson, M.D.; Paler-Martinez, A.; Lee, S.; Xu, Y.; Viteri, F.E.; Ames, B.N. Iron deficiency and iron excess damage mitochondria and mitochondrial DNA in rats. Proc. Natl. Acad. Sci. USA 2002, 99, 2264–2269. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, U.; Latham, P.S.; Oates, P.S. Interactions between hepatic iron and lipid metabolism with possible relevance to steatohepatitis. World J. Gastroenterol. 2012, 18, 4651–4658. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef]
- Seguin, A.; Jia, X.; Earl, A.M.; Li, L.; Wallace, J.; Qiu, A.; Bradley, T.; Shrestha, R.; Troadec, M.B.; Hockin, M.; et al. The mitochondrial metal transporters mitoferrin1 and mitoferrin2 are required for liver regeneration and cell proliferation in mice. J. Biol. Chem. 2020, 295, 11002–11020. [Google Scholar] [CrossRef]
- Shaw, G.C.; Cope, J.J.; Li, L.; Corson, K.; Hersey, C.; Ackermann, G.E.; Gwynn, B.; Lambert, A.J.; Wingert, R.A.; Traver, D.; et al. Mitoferrin is essential for erythroid iron assimilation. Nature 2006, 440, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; van de Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/web: A web-based ontology enrichment tool for lipidomic data analysis. Gigascience 2019, 8, giz061. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.R.; Haaker, M.W.; Vaandrager, A.B.; Houweling, M.; Helms, J.B. Lipidomic profiling of rat hepatic stellate cells during activation reveals a two-stage process accompanied by increased levels of lysosomal lipids. J. Biol. Chem. 2023, 299, 103042. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.C.; Barton, J.C.; Acton, R.T. Non-alcoholic fatty liver disease in hemochromatosis probands with iron overload and HFE p.C282Y/p.C282Y. BMC Gastroenterol. 2023, 23, 137. [Google Scholar] [CrossRef] [PubMed]
- Britton, L.; Jaskowski, L.; Bridle, K.; Santrampurwala, N.; Reiling, J.; Musgrave, N.; Subramaniam, V.N.; Crawford, D. Heterozygous Hfe gene deletion leads to impaired glucose homeostasis, but not liver injury in mice fed a high-calorie diet. Physiol. Rep. 2016, 4, e12837. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.H.G.; Ross, D.G.F.; Jaskowski, L.A.; Burke, L.J.; Britton, L.J.; Musgrave, N.; Briskey, D.; Rishi, G.; Bridle, K.R.; Subramaniam, V.N. Iron depletion attenuates steatosis in a mouse model of non-alcoholic fatty liver disease: Role of iron-dependent pathways. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166142. [Google Scholar] [CrossRef] [PubMed]
- Radicheva, M.P.; Andonova, A.N.; Milcheva, H.T.; Ivanova, N.G.; Kyuchukova, S.G.; Nikolova, M.S.; Platikanova, A.S. Serum Markers of Iron Metabolism in Chronic Liver Diseases. Open Access Maced. J. Med. Sci. 2018, 6, 1010–1016. [Google Scholar] [CrossRef]
- Saremi, L.; Lotfipanah, S.; Mohammadi, M.; Hosseinzadeh, H.; Sayad, A.; Saltanatpour, Z. Association of HFE gene mutations with nonalcoholic fatty liver disease in the Iranian population. Cell. Mol. Biol. 2016, 62, 123–128. [Google Scholar] [CrossRef]
- Sikorska, K.; Stalke, P.; Romanowski, T.; Rzepko, R.; Bielawski, K.P. Liver steatosis correlates with iron overload but not with HFE gene mutations in chronic hepatitis C. Hepatobiliary Pancreat. Dis. Int. 2013, 12, 377–384. [Google Scholar] [CrossRef]
- Britton, L.; Bridle, K.; Reiling, J.; Santrampurwala, N.; Wockner, L.; Ching, H.; Stuart, K.; Subramaniam, V.N.; Jeffrey, G.; St Pierre, T.; et al. Hepatic iron concentration correlates with insulin sensitivity in nonalcoholic fatty liver disease. Hepatol. Commun. 2018, 2, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Spruss, A.; Henkel, J.; Kanuri, G.; Blank, D.; Puschel, G.P.; Bischoff, S.C.; Bergheim, I. Female mice are more susceptible to nonalcoholic fatty liver disease: Sex-specific regulation of the hepatic AMP-activated protein kinase-plasminogen activator inhibitor 1 cascade, but not the hepatic endotoxin response. Mol. Med. 2012, 18, 1346–1355. [Google Scholar] [CrossRef] [PubMed]
- Kaden-Volynets, V.; Basic, M.; Neumann, U.; Pretz, D.; Rings, A.; Bleich, A.; Bischoff, S.C. Lack of liver steatosis in germ-free mice following hypercaloric diets. Eur. J. Nutr. 2019, 58, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Cellular iron: Ferroportin is the only way out. Cell Metab. 2005, 1, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.; Ganz, T. Regulation of iron metabolism by hepcidin. Annu. Rev. Nutr. 2006, 26, 323–342. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, G.; Bennoun, M.; Devaux, I.; Beaumont, C.; Grandchamp, B.; Kahn, A.; Vaulont, S. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. USA 2001, 98, 8780–8785. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, G.; Viatte, L.; Bennoun, M.; Beaumont, C.; Kahn, A.; Vaulont, S. Hepcidin, a new iron regulatory peptide. Blood Cells Mol. Dis. 2002, 29, 327–335. [Google Scholar] [CrossRef]
- Liu, J.; Yang, P.; Zuo, G.; He, S.; Tan, W.; Zhang, X.; Su, C.; Zhao, L.; Wei, L.; Chen, Y.; et al. Long-chain fatty acid activates hepatocytes through CD36 mediated oxidative stress. Lipids Health Dis. 2018, 17, 153. [Google Scholar] [CrossRef]
- Matsukawa, T.; Yagi, T.; Uchida, T.; Sakai, M.; Mitsushima, M.; Naganuma, T.; Yano, H.; Inaba, Y.; Inoue, H.; Yanagida, K.; et al. Hepatic FASN deficiency differentially affects nonalcoholic fatty liver disease and diabetes in mouse obesity models. JCI Insight 2023, 8, e161282. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Qin, H.; Liao, M.; Zheng, E.; Luo, X.; Xiao, A.; Li, Y.; Chen, L.; Wei, L.; Zhao, L.; et al. CD36 promotes de novo lipogenesis in hepatocytes through INSIG2-dependent SREBP1 processing. Mol. Metab. 2022, 57, 101428. [Google Scholar] [CrossRef] [PubMed]
- Cedo, L.; Santos, D.; Roglans, N.; Julve, J.; Pallares, V.; Rivas-Urbina, A.; Llorente-Cortes, V.; Laguna, J.C.; Blanco-Vaca, F.; Escola-Gil, J.C. Human hepatic lipase overexpression in mice induces hepatic steatosis and obesity through promoting hepatic lipogenesis and white adipose tissue lipolysis and fatty acid uptake. PLoS ONE 2017, 12, e0189834. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Todisco, S.; Santarsiero, A.; Convertini, P.; De Stefano, G.; Gilio, M.; Iacobazzi, V.; Infantino, V. PPAR Alpha as a Metabolic Modulator of the Liver: Role in the Pathogenesis of Nonalcoholic Steatohepatitis (NASH). Biology 2022, 11, 792. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, J.; Feng, J.; Ji, J.; Yu, Q.; Li, Y.; Zheng, Y.; Dai, W.; Wu, J.; Guo, C. Crosstalk between PPARs and gut microbiota in NAFLD. Biomed. Pharmacother. 2021, 136, 111255. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Transcriptional mediators of lipid homeostasis. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.A.; Liang, G.; Xie, X.; Frank-Kamenetsky, M.; Fitzgerald, K.; Koteliansky, V.; Brown, M.S.; Goldstein, J.L.; Horton, J.D. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab. 2012, 15, 240–246. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef]
- Weber, M.; Mera, P.; Casas, J.; Salvador, J.; Rodriguez, A.; Alonso, S.; Sebastian, D.; Soler-Vazquez, M.C.; Montironi, C.; Recalde, S.; et al. Liver CPT1A gene therapy reduces diet-induced hepatic steatosis in mice and highlights potential lipid biomarkers for human NAFLD. FASEB J. 2020, 34, 11816–11837. [Google Scholar] [CrossRef] [PubMed]
- Kadenbach, B. Complex IV–The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion 2021, 58, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Arifin, S.A.; Falasca, M. Lysophosphatidylinositol Signalling and Metabolic Diseases. Metabolites 2016, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Du, J.; Wei, T.T.; Chen, L.Y.; Yang, X.X.; Bo, T.; Liu, H.Y.; Xie, M.Z.; Zhao, T.S.; Yang, J.L.; et al. Alterations in bile acids as metabolic signatures in the patients with human adenovirus type 7 infection. Front. Med. 2022, 9, 896409. [Google Scholar] [CrossRef] [PubMed]
- Foury, F.; Roganti, T. Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem. 2002, 277, 24475–24483. [Google Scholar] [CrossRef] [PubMed]
- Muhlenhoff, U.; Stadler, J.A.; Richhardt, N.; Seubert, A.; Eickhorst, T.; Schweyen, R.J.; Lill, R.; Wiesenberger, G. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 2003, 278, 40612–40620. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.M.; Cloonan, S.M. Mitochondrial Iron in Human Health and Disease. Annu. Rev. Physiol. 2019, 81, 453–482. [Google Scholar] [CrossRef] [PubMed]
- Troadec, M.B.; Warner, D.; Wallace, J.; Thomas, K.; Spangrude, G.J.; Phillips, J.; Khalimonchuk, O.; Paw, B.H.; Ward, D.M.; Kaplan, J. Targeted deletion of the mouse Mitoferrin1 gene: From anemia to protoporphyria. Blood 2011, 117, 5494–5502. [Google Scholar] [CrossRef] [PubMed]
- Caligiuri, A.; Gentilini, A.; Marra, F. Molecular Pathogenesis of NASH. Int. J. Mol. Sci. 2016, 17, 1575. [Google Scholar] [CrossRef]
- Kakisaka, K.; Cazanave, S.C.; Fingas, C.D.; Guicciardi, M.E.; Bronk, S.F.; Werneburg, N.W.; Mott, J.L.; Gores, G.J. Mechanisms of lysophosphatidylcholine-induced hepatocyte lipoapoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G77–G84. [Google Scholar] [CrossRef]
- Davis, M.R.; Hester, K.K.; Shawron, K.M.; Lucas, E.A.; Smith, B.J.; Clarke, S.L. Comparisons of the iron deficient metabolic response in rats fed either an AIN-76 or AIN-93 based diet. Nutr. Metab. 2012, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xin, W.; Anderson, G.J.; Li, R.; Gao, L.; Chen, S.; Zhao, J.; Liu, S. Double-edge sword roles of iron in driving energy production versus instigating ferroptosis. Cell Death Dis. 2022, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Sherman, A.R. Lipogenesis in iron-deficient adult rats. Lipids 1978, 13, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Gorden, D.L.; Ivanova, P.T.; Myers, D.S.; McIntyre, J.O.; VanSaun, M.N.; Wright, J.K.; Matrisian, L.M.; Brown, H.A. Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS ONE 2011, 6, e22775. [Google Scholar] [CrossRef]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Hliwa, A.; Ramos-Molina, B.; Laski, D.; Mika, A.; Sledzinski, T. The Role of Fatty Acids in Non-Alcoholic Fatty Liver Disease Progression: An Update. Int. J. Mol. Sci. 2021, 22, 6900. [Google Scholar] [CrossRef] [PubMed]
- Das, N.K.; Schwartz, A.J.; Barthel, G.; Inohara, N.; Liu, Q.; Sankar, A.; Hill, D.R.; Ma, X.; Lamberg, O.; Schnizlein, M.K.; et al. Microbial Metabolite Signaling Is Required for Systemic Iron Homeostasis. Cell Metab. 2020, 31, 115–130.e6. [Google Scholar] [CrossRef] [PubMed]
- Deschemin, J.C.; Noordine, M.L.; Remot, A.; Willemetz, A.; Afif, C.; Canonne-Hergaux, F.; Langella, P.; Karim, Z.; Vaulont, S.; Thomas, M.; et al. The microbiota shifts the iron sensing of intestinal cells. FASEB J. 2016, 30, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Seyoum, Y.; Baye, K.; Humblot, C. Iron homeostasis in host and gut bacteria—A complex interrelationship. Gut Microbes 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Moslehi, A.; Hamidi-Zad, Z. Role of SREBPs in Liver Diseases: A Mini-review. J. Clin. Transl. Hepatol. 2018, 6, 332–338. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 2016, 65, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Soyal, S.M.; Nofziger, C.; Dossena, S.; Paulmichl, M.; Patsch, W. Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol. Sci. 2015, 36, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, S.; Matsushita, Y.; Kurosaki, S.; Tange, M.; Fujiwara, N.; Hayata, Y.; Hayakawa, Y.; Suzuki, N.; Hata, M.; Tsuboi, M.; et al. Inhibiting SCAP/SREBP exacerbates liver injury and carcinogenesis in murine nonalcoholic steatohepatitis. J. Clin. Investig. 2022, 132, e151895. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.H.; Maeng, H.Y.; Sun, Y.K.; Kim, Y.A.; Park, D.W.; Park, T.S.; Lee, S.T.; Choi, J.R. Oxidative status in iron-deficiency anemia. J. Clin. Lab. Anal. 2009, 23, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, F.; Yuan, J.; Liu, H.; Wang, Y. Gut microbiota metabolites, redox status, and the related regulatory effects of probiotics. Heliyon 2023, 9, e21431. [Google Scholar] [CrossRef] [PubMed]
- Sumida, Y.; Yoshikawa, T.; Okanoue, T. Role of hepatic iron in non-alcoholic steatohepatitis. Hepatol. Res. 2009, 39, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Aigner, E.; Theurl, I.; Haufe, H.; Seifert, M.; Hohla, F.; Scharinger, L.; Stickel, F.; Mourlane, F.; Weiss, G.; Datz, C. Copper availability contributes to iron perturbations in human nonalcoholic fatty liver disease. Gastroenterology 2008, 135, 680–688. [Google Scholar] [CrossRef]
- Campo, L.; Eiseler, S.; Apfel, T.; Pyrsopoulos, N. Fatty Liver Disease and Gut Microbiota: A Comprehensive Update. J. Clin. Transl. Hepatol. 2019, 7, 56–60. [Google Scholar] [CrossRef]


| Gene | Primer Sequence | Reference | |
|---|---|---|---|
| Abca1 | Forward | AAAACCGCAGACATCCTTCAG | Origene |
| Reverse | CATACCGAAACTCGTTCACCC | β | |
| Acaca1 | Forward | TGTACAAGCAGTGTGGGCTGGCT | β |
| Reverse | CCACATGGCCTGGCTTGGAGGG | β | |
| ATP5a1 | Forward | TGGTGAAGAGACTGACGGATGC | β |
| Reverse | TCAAAGCGTGCTTGCCGTTGTC | β | |
| β Actin | Forward | GACGGCCAAGTCATCACTATTG | β |
| Reverse | CCACAGGATTCCATACCCAAGA | β | |
| CD36 | Forward | GGACATTGAGATTCTTTTCCTCTG | β |
| Reverse | GCAAAGGCATTGGCTGGAAGAAC | β | |
| ChREBP | Forward | AGATGGAGAACCGACGTATCA | β |
| Reverse | ACTGAGCGTGCTGACAAGTC | β | |
| Cox4i1 | Forward | TCATTGGCTTCACTGCGCTCGT | β |
| Reverse | TCCAGCATTCGCTTGGTCTGCA | β | |
| CPT1a | Forward | GGCATAAACGCAGAGCATTCCTG | β |
| Reverse | CAGTGTCCATCCTCTGAGTAGC | β | |
| Dmt1 | Forward | AGCTAGGGCATGTGGCACTCT | β |
| Reverse | ATGTTGCCACCGCTGGTATC | β | |
| Fasn | Forward | GCTGCGGAAACTTCAGGAAAT | β |
| Reverse | AGAGACGTGTCACTCCTGGACTT | β | |
| Fpn | Forward | CCATAGTCTCTGTCAGCCTGCT | β |
| Reverse | CTTGCAGCAACTGTGTCACCGT | β | |
| L-Ftn | Forward | ATGACCTCTCAGATTCGTCAG | β |
| Reverse | ATTCGCGGAAGAAGTGGCCTA | β | |
| H-Ftn | Forward | CTCCTACGTCTATCTGTCTATG | β |
| Reverse | ATTCGGCCACCTCGCTGGTTCT | β | |
| Hamp | Forward | CAGCACCACCTATCTCCATCAAC | β |
| Reverse | CAGATGGGGAAGTTGGTGTCTC | β | |
| Lipc | Forward | CTTCCAGCCTGGCTGCCACTT | β |
| Reverse | GCAAGGAGTCAATGAAGAGGTGC | β | |
| Mfrn1 | Forward | TTGAATCCAGATCCCAAAGC | β |
| Reverse | GTTTCCTTGGTGGCTGAAAA | β | |
| Mfrn2 | Forward | TCGTCAAGCAGAGGATGCAGAT | β |
| Reverse | GTTAAAGTGCTCTTGCAGGAAC | β | |
| PGC1α | Forward | GAATCAAGCCACTACAGACACCG | β |
| Reverse | CATCCCTCTTGAGCCTTTCGTG | β | |
| Pparα | Forward | ACCACTACGGAGTTCACGCATG | β |
| Reverse | GAATCTTGCAGCTCCGATCACAC | β | |
| SDHB | Forward | TGCGGACCTATGGTGTTGGATG | β |
| Reverse | CCAGAGTATTGCCTCCGTTGATG | β | |
| Srebp-1c | Forward | CGACTACATCCGCTTCTTGCAG | β |
| Reverse | CCTCCATAGACACATCTGTGCC | β |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klag, K.A.; Bell, R.; Jia, X.; Seguin, A.; Maschek, J.A.; Bronner, M.; Cox, J.E.; Round, J.L.; Ward, D.M. Low-Iron Diet-Induced Fatty Liver Development Is Microbiota Dependent and Exacerbated by Loss of the Mitochondrial Iron Importer Mitoferrin2. Nutrients 2024, 16, 1804. https://doi.org/10.3390/nu16121804
Klag KA, Bell R, Jia X, Seguin A, Maschek JA, Bronner M, Cox JE, Round JL, Ward DM. Low-Iron Diet-Induced Fatty Liver Development Is Microbiota Dependent and Exacerbated by Loss of the Mitochondrial Iron Importer Mitoferrin2. Nutrients. 2024; 16(12):1804. https://doi.org/10.3390/nu16121804
Chicago/Turabian StyleKlag, Kendra A., Rickesha Bell, Xuan Jia, Alexandra Seguin, J. Alan Maschek, Mary Bronner, James E. Cox, June L. Round, and Diane M. Ward. 2024. "Low-Iron Diet-Induced Fatty Liver Development Is Microbiota Dependent and Exacerbated by Loss of the Mitochondrial Iron Importer Mitoferrin2" Nutrients 16, no. 12: 1804. https://doi.org/10.3390/nu16121804
APA StyleKlag, K. A., Bell, R., Jia, X., Seguin, A., Maschek, J. A., Bronner, M., Cox, J. E., Round, J. L., & Ward, D. M. (2024). Low-Iron Diet-Induced Fatty Liver Development Is Microbiota Dependent and Exacerbated by Loss of the Mitochondrial Iron Importer Mitoferrin2. Nutrients, 16(12), 1804. https://doi.org/10.3390/nu16121804






