A Prebiotic Diet Containing Galactooligosaccharides and Polydextrose Produces Dynamic and Reproducible Changes in the Gut Microbial Ecosystem in Male Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.1.1. Northwestern (NW) Study
2.1.2. University of Colorado Boulder (CU) Study
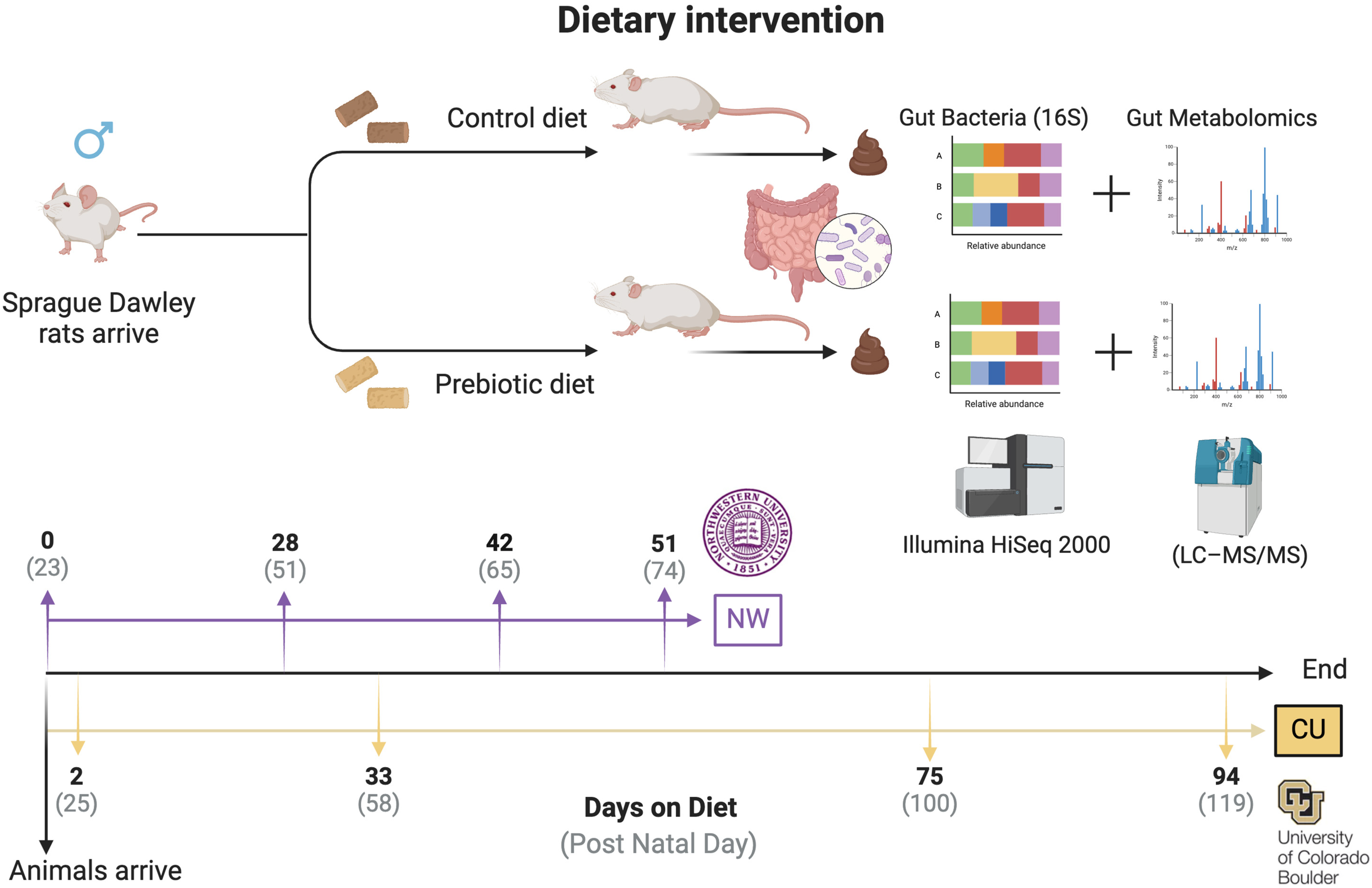
2.2. Experimental Design
2.3. Diets
2.4. Fecal Sample Collection Procedures
2.5. The 16S rRNA Gene Sequencing
2.6. LC–MS/MS Metabolomics
2.7. Statistical Analysis
3. Results
3.1. Microbiome
3.2. Metabolome—Bile Acids
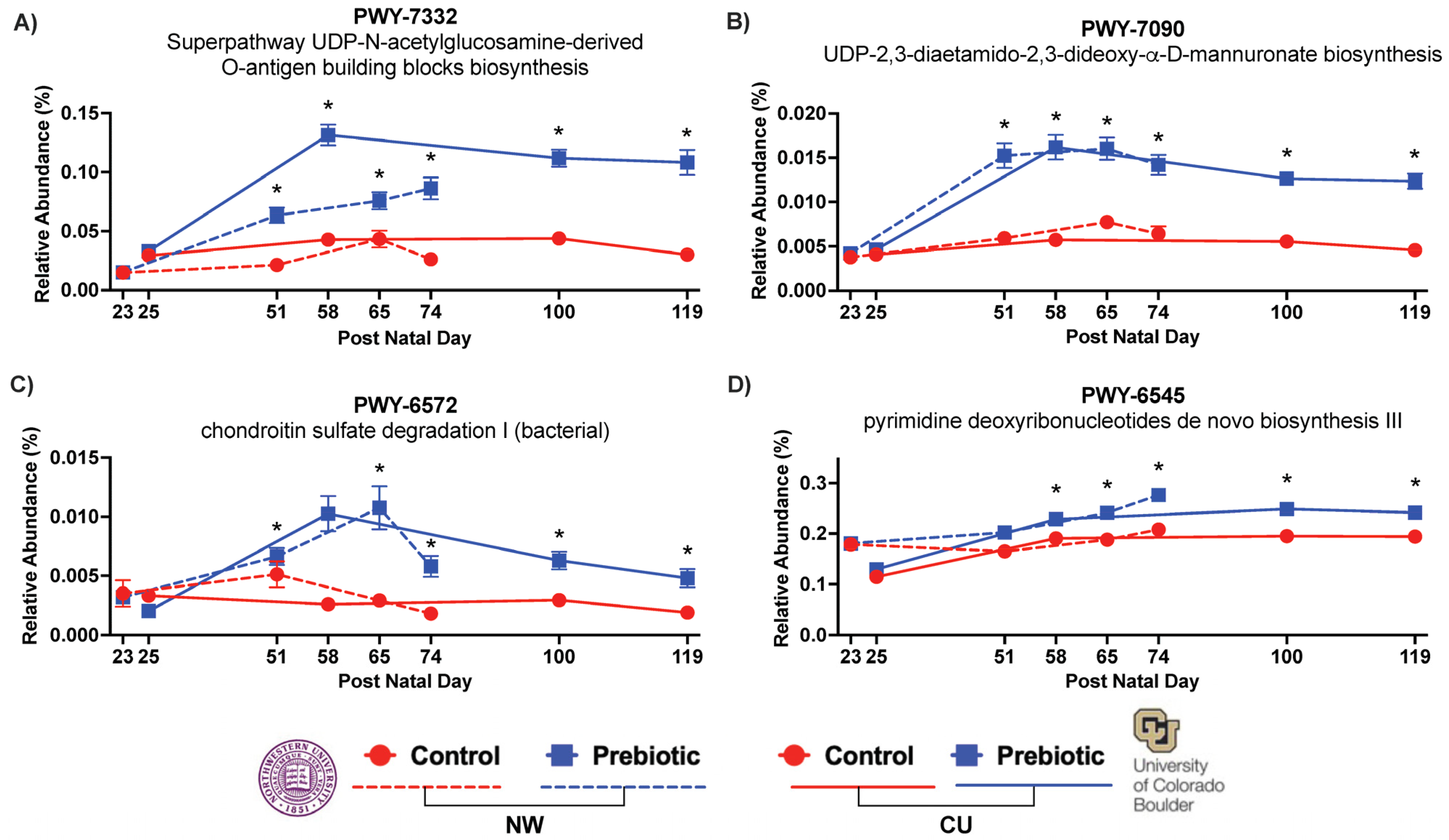
3.3. PICRUSt2—Pathways
3.4. Correlation Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Wang, R. The human gut phageome: Composition, development, and alterations in disease. Front. Microbiol. 2023, 14, 1213625. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, H.; Hu, X.; Yuan, Z. Diversity and functional profile of gut symbiotic bacteria between Lysinibacillus sphaericus C(3)-41 susceptible and resistant Culex quinquefasciatus Say as revealed by 16S rRNA gene high-throughput sequencing. Front. Microbiol. 2022, 13, 991105. [Google Scholar] [CrossRef]
- Naya-Catala, F.; Piazzon, M.C.; Calduch-Giner, J.A.; Sitja-Bobadilla, A.; Perez-Sanchez, J. Diet and Host Genetics Drive the Bacterial and Fungal Intestinal Metatranscriptome of Gilthead Sea Bream. Front. Microbiol. 2022, 13, 883738. [Google Scholar] [CrossRef]
- Vincenot, C.E.; Giannino, F.; Rietkerk, M.; Moriya, K.; Mazzoleni, S. Theoretical considerations on the combined use of System Dynamics and individual-based modeling in ecology. Ecol. Model. 2011, 222, 210–218. [Google Scholar] [CrossRef]
- Molly, K.; Vandewoestyne, M.; Desmet, I.; Verstraete, W. Validation of the Simulator of the Human Intestinal Microbial Ecosystem (Shime) Reactor Using Microorganism-Associated Activities. Microb. Ecol. Health D 1994, 7, 191–200. [Google Scholar] [CrossRef]
- Liu, S.; Qin, P.; Wang, J. High-Fat Diet Alters the Intestinal Microbiota in Streptozotocin-Induced Type 2 Diabetic Mice. Microorganisms 2019, 7, 176. [Google Scholar] [CrossRef]
- Liu, X.; Blouin, J.M.; Santacruz, A.; Lan, A.; Andriamihaja, M.; Wilkanowicz, S.; Benetti, P.H.; Tome, D.; Sanz, Y.; Blachier, F.; et al. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: The increased luminal bulk connection. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G459–G470. [Google Scholar] [CrossRef]
- Barra, N.G.; Anhe, F.F.; Cavallari, J.F.; Singh, A.M.; Chan, D.Y.; Schertzer, J.D. Micronutrients impact the gut microbiota and blood glucose. J. Endocrinol. 2021, 250, R1–R21. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Schloss, P.D. Identifying and Overcoming Threats to Reproducibility, Replicability, Robustness, and Generalizability in Microbiome Research. mBio 2018, 9, e00525-18. [Google Scholar] [CrossRef]
- Vitek, J.; Kalibera, T. R-3—Repeatability, Reproducibility and Rigor. Acm Sigplan Not. 2012, 47, 30–36. [Google Scholar] [CrossRef]
- Eaton, K.; Pirani, A.; Snitkin, E.S.; Reproducibility Project: Cancer Biology. Replication Study: Intestinal inflammation targets cancer-inducing activity of the microbiota. eLife 2018, 7, e34364. [Google Scholar] [CrossRef]
- Repass, J.; Reproducibility Project: Cancer Biology. Replication Study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. eLife 2018, 7, e25801. [Google Scholar] [CrossRef]
- Panek, M.; Cipcic Paljetak, H.; Baresic, A.; Peric, M.; Matijasic, M.; Lojkic, I.; Vranesic Bender, D.; Krznaric, Z.; Verbanac, D. Methodology challenges in studying human gut microbiota—Effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018, 8, 5143. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Kumari, M.; Bhushan, B.; Eslavath, M.R.; Srivastava, A.K.; Meena, R.C.; Varshney, R.; Ganju, L. Impact of high altitude on composition and functional profiling of oral microbiome in Indian male population. Sci. Rep. 2023, 13, 4038. [Google Scholar] [CrossRef]
- Dong, W.; Ma, L.; Huang, Q.; Yang, X.; Mei, Z.; Kong, M.; Sun, Z.; Zhang, Z.; Li, J.; Zou, J.; et al. Gut microbiome alterations in pulmonary hypertension in highlanders and lowlanders. ERJ Open Res. 2023, 9, 00617-2022. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Franklin, C.L. The gut microbiome of laboratory mice: Considerations and best practices for translational research. Mamm. Genome 2021, 32, 239–250. [Google Scholar] [CrossRef]
- Bowers, S.J.; Summa, K.C.; Thompson, R.S.; Gonzalez, A.; Vargas, F.; Olker, C.; Jiang, P.; Lowry, C.A.; Dorrestein, P.C.; Knight, R.; et al. A Prebiotic Diet Alters the Fecal Microbiome and Improves Sleep in Response to Sleep Disruption in Rats. Front. Neurosci. 2022, 16, 889211. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.S.; Gaffney, M.; Hopkins, S.; Kelley, T.; Gonzalez, A.; Bowers, S.J.; Vitaterna, M.H.; Turek, F.W.; Foxx, C.L.; Lowry, C.A.; et al. Ruminiclostridium 5, Parabacteroides distasonis, and bile acid profile are modulated by prebiotic diet and associate with facilitated sleep/clock realignment after chronic disruption of rhythms. Brain Behav. Immun. 2021, 97, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.D.; Guo, Y.S.; Huang, J.S.; Gao, Y.F.; Peng, F.; Xu, R.Y.; Su, H.H.; Zhang, P.J. Isomaltulose Exhibits Prebiotic Activity, and Modulates Gut Microbiota, the Production of Short Chain Fatty Acids, and Secondary Bile Acids in Rats. Molecules 2021, 26, 2464. [Google Scholar] [CrossRef] [PubMed]
- Phungviwatnikul, T.; Lee, A.H.; Belchik, S.E.; Suchodolski, J.S.; Swanson, K.S. Weight loss and high-protein, high-fiber diet consumption impact blood metabolite profiles, body composition, voluntary physical activity, fecal microbiota, and fecal metabolites of adult dogs. J. Anim. Sci. 2022, 100, skab379. [Google Scholar] [CrossRef]
- McMillin, M.; DeMorrow, S. Effects of bile acids on neurological function and disease. FASEB J. 2016, 30, 3658–3668. [Google Scholar] [CrossRef] [PubMed]
- Perino, A.; Demagny, H.; Velazquez-Villegas, L.A.; Schoonjans, K. Molecular Physiology of Bile Acid Signaling in Health, Disease and Aging. Physiol. Rev. 2020, 101, 683–731. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.S.; Roller, R.; Mika, A.; Greenwood, B.N.; Knight, R.; Chichlowski, M.; Berg, B.M.; Fleshner, M. Dietary Prebiotics and Bioactive Milk Fractions Improve NREM Sleep, Enhance REM Sleep Rebound and Attenuate the Stress-Induced Decrease in Diurnal Temperature and Gut Microbial Alpha Diversity. Front. Behav. Neurosci. 2017, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Gaffney, M.; Roller, R.; Hills, A.; Bouchet, C.A.; Hulen, K.A.; Thompson, R.S.; Chichlowski, M.; Berg, B.M.; Fleshner, M. Feeding the developing brain: Juvenile rats fed diet rich in prebiotics and bioactive milk fractions exhibit reduced anxiety-related behavior and modified gene expression in emotion circuits. Neurosci. Lett. 2018, 677, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Day, H.E.; Martinez, A.; Rumian, N.L.; Greenwood, B.N.; Chichlowski, M.; Berg, B.M.; Fleshner, M. Early life diets with prebiotics and bioactive milk fractions attenuate the impact of stress on learned helplessness behaviours and alter gene expression within neural circuits important for stress resistance. Eur. J. Neurosci. 2016, 45, 342–357. [Google Scholar] [CrossRef]
- Bowers, S.J.; Vargas, F.; Gonzalez, A.; He, S.; Jiang, P.; Dorrestein, P.C.; Knight, R.; Wright, K.P., Jr.; Lowry, C.A.; Fleshner, M.; et al. Repeated sleep disruption in mice leads to persistent shifts in the fecal microbiome and metabolome. PLoS ONE 2020, 15, e0229001. [Google Scholar] [CrossRef]
- Krauth, S.J.; Coulibaly, J.T.; Knopp, S.; Traore, M.; N’Goran, E.K.; Utzinger, J. An in-depth analysis of a piece of shit: Distribution of Schistosoma mansoni and hookworm eggs in human stool. PLoS Negl. Trop. Dis. 2012, 6, e1969. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Corrected in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glockner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Faith, D.P. Phylogenetic pattern and the quantification of organismal biodiversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994, 345, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbo, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Melnik, A.V.; da Silva, R.R.; Hyde, E.R.; Aksenov, A.A.; Vargas, F.; Bouslimani, A.; Protsyuk, I.; Jarmusch, A.K.; Tripathi, A.; Alexandrov, T.; et al. Coupling Targeted and Untargeted Mass Spectrometry for Metabolome-Microbiome-Wide Association Studies of Human Fecal Samples. Anal. Chem. 2017, 89, 7549–7559. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.S.; Vargas, F.; Dorrestein, P.C.; Chichlowski, M.; Berg, B.M.; Fleshner, M. Dietary prebiotics alter novel microbial dependent fecal metabolites that improve sleep. Sci. Rep. 2020, 10, 3848. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.J.; Gross, R.; Bittinger, K.; Sherrill-Mix, S.; Lewis, J.D.; Collman, R.G.; Bushman, F.D.; Li, H. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 2015, 31, 2461–2468. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.Z.; Chen, G.; Alekseyenko, A.V. PERMANOVA-S: Association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics 2016, 32, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Almela, I.; Romani-Perez, M.; Bullich-Vilarrubias, C.; Benitez-Paez, A.; Gomez Del Pulgar, E.M.; Frances, R.; Liebisch, G.; Sanz, Y. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Kano, C.; Ishii, C.; Kagata, N.; Ishikawa, T.; Hirayama, A.; Uchiyama, Y.; Hara, S.; Nakamura, T.; Fukuda, S. Bacteroides uniformis and its preferred substrate, alpha-cyclodextrin, enhance endurance exercise performance in mice and human males. Sci. Adv. 2023, 9, eadd2120. [Google Scholar] [CrossRef]
- Carbajo-Pescador, S.; Porras, D.; Garcia-Mediavilla, M.V.; Martinez-Florez, S.; Juarez-Fernandez, M.; Cuevas, M.J.; Mauriz, J.L.; Gonzalez-Gallego, J.; Nistal, E.; Sanchez-Campos, S. Beneficial effects of exercise on gut microbiota functionality and barrier integrity, and gut-liver crosstalk in an in vivo model of early obesity and non-alcoholic fatty liver disease. Dis. Model. Mech. 2019, 12, dmm039206. [Google Scholar] [CrossRef]
- Ezeji, J.C.; Sarikonda, D.K.; Hopperton, A.; Erkkila, H.L.; Cohen, D.E.; Martinez, S.P.; Cominelli, F.; Kuwahara, T.; Dichosa, A.E.K.; Good, C.E.; et al. Parabacteroides distasonis: Intriguing aerotolerant gut anaerobe with emerging antimicrobial resistance and pathogenic and probiotic roles in human health. Gut Microbes 2021, 13, 1922241. [Google Scholar] [CrossRef] [PubMed]
- Berding, K.; Long-Smith, C.M.; Carbia, C.; Bastiaanssen, T.F.S.; van de Wouw, M.; Wiley, N.; Strain, C.R.; Fouhy, F.; Stanton, C.; Cryan, J.F.; et al. A specific dietary fibre supplementation improves cognitive performance-an exploratory randomised, placebo-controlled, crossover study. Psychopharmacology 2021, 238, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Jiang, Y.; Tan, A.; Ye, J.; Xian, X.; Xie, Y.; Wang, Q.; Yao, Z.; Mo, Z. 16S rRNA gene sequencing reveals altered composition of gut microbiota in individuals with kidney stones. Urolithiasis 2018, 46, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, C.; Wang, J.; Zhou, H.; Lu, Y.; Lou, L.; Zheng, J.; Tian, L.; Wang, X.; Cao, Z.; et al. Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS ONE 2017, 12, e0176583. [Google Scholar] [CrossRef]
- Verheggen, R.; Konstanti, P.; Smidt, H.; Hermus, A.; Thijssen, D.H.J.; Hopman, M.T.E. Eight-week exercise training in humans with obesity: Marked improvements in insulin sensitivity and modest changes in gut microbiome. Obesity 2021, 29, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Toya, T.; Corban, M.T.; Marrietta, E.; Horwath, I.E.; Lerman, L.O.; Murray, J.A.; Lerman, A. Coronary artery disease is associated with an altered gut microbiome composition. PLoS ONE 2020, 15, e0227147. [Google Scholar] [CrossRef] [PubMed]
- Babacan Yildiz, G.; Kayacan, Z.C.; Karacan, I.; Sumbul, B.; Elibol, B.; Gelisin, O.; Akgul, O. Altered gut microbiota in patients with idiopathic Parkinson’s disease: An age-sex matched case-control study. Acta Neurol. Belg. 2023, 123, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, E.; Ilina, L.; Laptev, G.; Filippova, V.; Brazhnik, E.; Dunyashev, T.; Dubrovin, A.; Novikova, N.; Tiurina, D.; Tarlavin, N.; et al. The structure and functional profile of ruminal microbiota in young and adult reindeers (Rangifer tarandus) consuming natural winter-spring and summer-autumn seasonal diets. PeerJ 2021, 9, e12389. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ni, Y.; Wang, Z.; Tu, W.; Ni, L.; Zhuge, F.; Zheng, A.; Hu, L.; Zhao, Y.; Zheng, L.; et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 2020, 12, 1–19. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, T.; Guo, R.; Cui, W.; Yu, W.; Wang, Z.; Jiang, Y.; Jiang, M.; Wang, X.; Liu, C.; et al. Variation of Serum Uric Acid Is Associated With Gut Microbiota in Patients With Diabetes Mellitus. Front. Cell Infect. Microbiol. 2021, 11, 761757. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Tang, G.; Deng, P.; Qin, Y.; Han, J.; Wang, S.; Sun, X.; Li, D.; Chen, Z. The causal relationship between gut microbiota and bone mineral density: A Mendelian randomization study. Front. Microbiol. 2023, 14, 1268935. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shen, H.; Liu, T.; Pan, B.; De Alwis, S.; Zhang, W.; Luo, X.; Li, Z.; Wang, N.; Ma, W.; et al. Effects of three different mannans on obesity and gut microbiota in high-fat diet-fed C57BL/6J mice. Food Funct. 2021, 12, 4606–4620. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Qi, L.; Zhao, L.; Liu, J.; Guo, Y.; Zhang, C. Degradation of chondroitin sulfate: Mechanism of degradation, influence factors, structure-bioactivity relationship and application. Carbohydr. Polym. 2023, 301, 120361. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, N.; Li, Z.; Wang, X.; Shi, H.; Xue, C.; Li, R.W.; Tang, Q. Chondroitin sulfate disaccharides modified the structure and function of the murine gut microbiome under healthy and stressed conditions. Sci. Rep. 2017, 7, 6783. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gong, Z.; Zhang, X.; Zhu, F.; Liu, Y.; Jin, C.; Du, X.; Xu, C.; Chen, Y.; Cai, W.; et al. Gut microbial bile acid metabolite skews macrophage polarization and contributes to high-fat diet-induced colonic inflammation. Gut Microbes 2020, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Tache, Y.; Hofmann, A.F. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig. Liver Dis. 2014, 46, 302–312. [Google Scholar] [CrossRef]
- Pols, T.W.; Noriega, L.G.; Nomura, M.; Auwerx, J.; Schoonjans, K. The bile acid membrane receptor TGR5: A valuable metabolic target. Dig. Dis. 2011, 29, 37–44. [Google Scholar] [CrossRef]
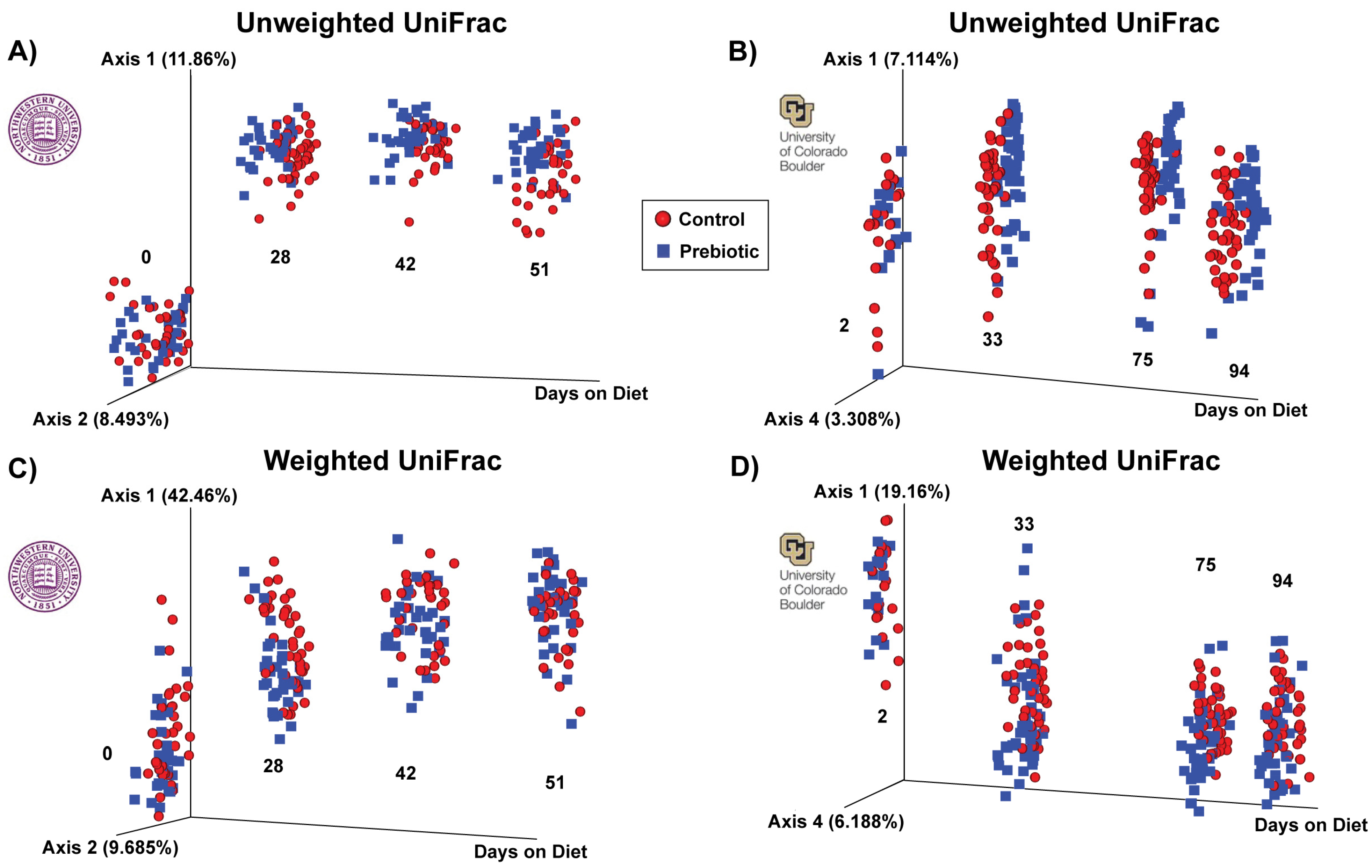
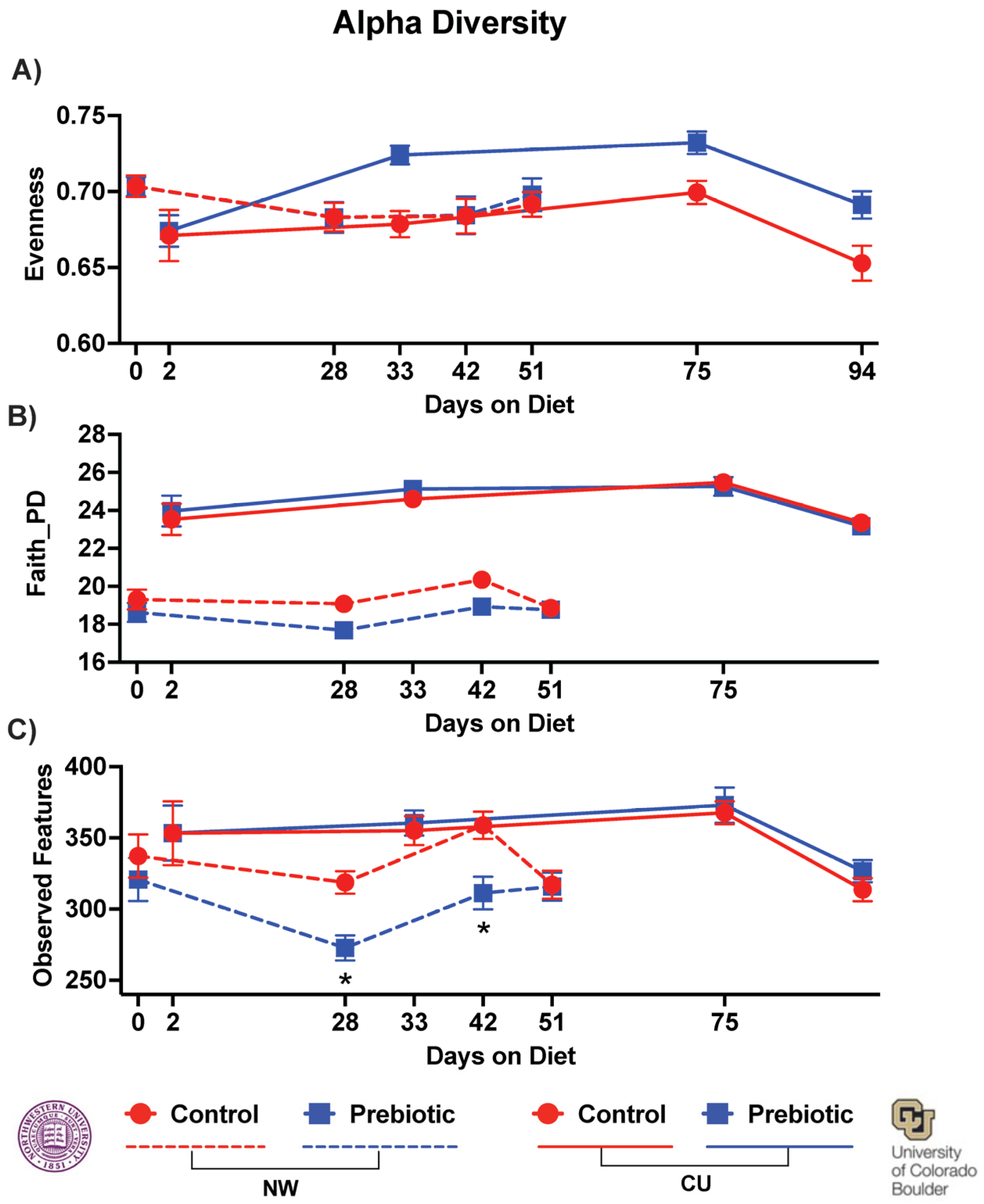


| PERMANOVAs (Pseudo-F) | ||||
|---|---|---|---|---|
| Northwestern | ||||
| 0 | 28 | 42 | 51 | |
| Unweighted | F(2,68) = 1.24; p = 0.154 | F(2,80) = 7.68; p = 0.001 | F(2,66) = 5.60; p = 0.001 | F(2,69) = 5.87; p = 0.001 |
| Weighted | F(2,68) = 2.19; p = 0.053 | F(2,80) = 9.31; p = 0.001 | F(2,66) = 4.26; p = 0.001 | F(2,66) = 4.34; p = 0.001 |
| University of Colorado Boulder | ||||
| 2 | 33 | 75 | 94 | |
| Unweighted | F(2,48) = 1.31; p = 0.053 | F(2,78) = 4.89; p = 0.001 | F(2,83) = 3.84; p = 0.001 | F(2,84) = 4.16; p = 0.001 |
| Weighted | F(2,48) = 3.97; p = 0.006 | F(2,78) = 10.99; p = 0.001 | F(2,83) = 7.39; p = 0.001 | F(2,84) = 3.93; p = 0.001 |
| Nonparametric Longitudinal Data (nparLD) Table: ANOVA-Type Statistics (ATSs) | |||||||
|---|---|---|---|---|---|---|---|
| Genera (Relative Abundance) | |||||||
| Diet—F-Value; p-Value | Time—F-Value; p-Value | Diet × Time | |||||
| Higher in Prebiotic Diet, color indicates consistent effect across study site | |||||||
| High Relative Abundance (2–20%) | |||||||
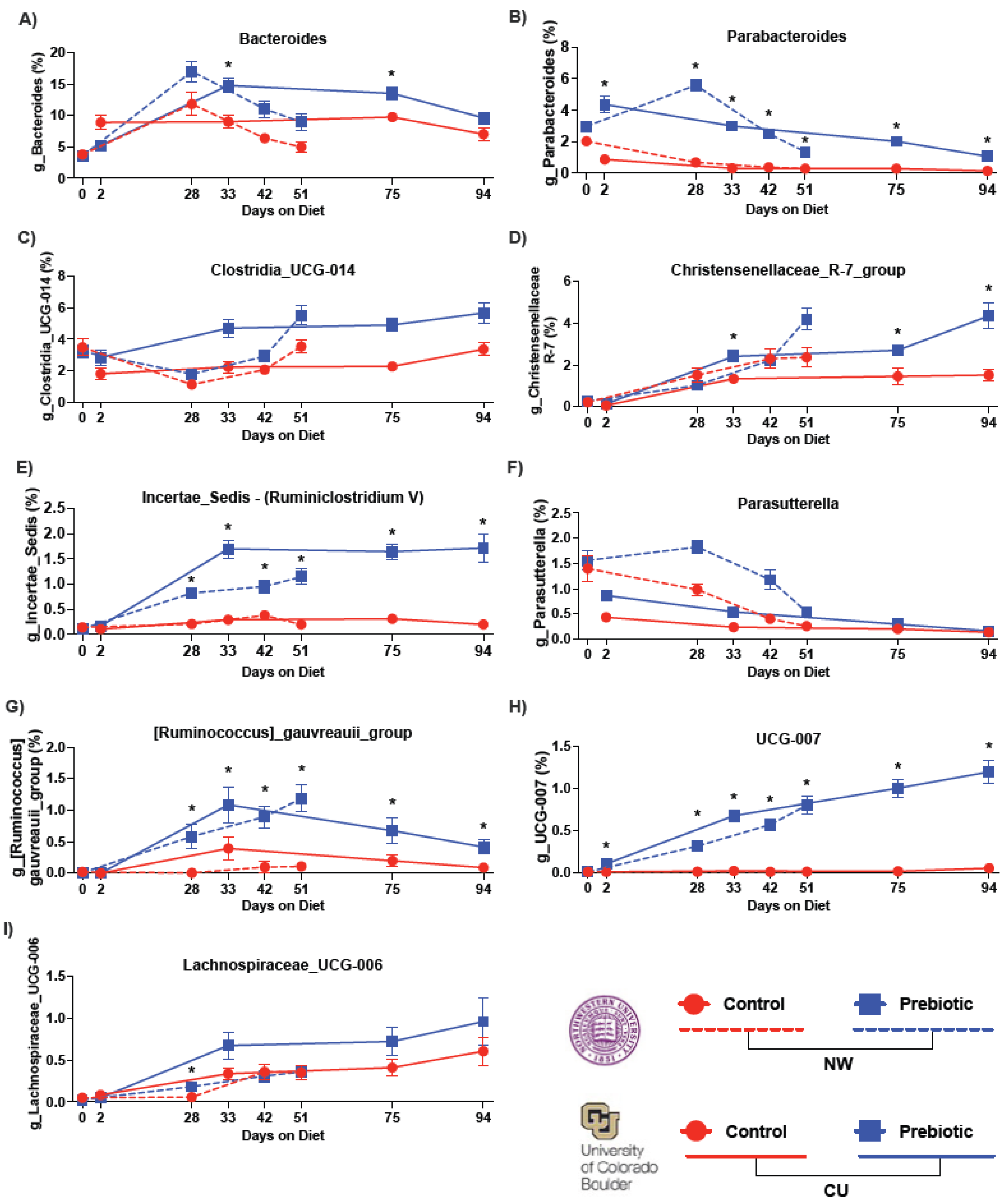
| Bacteroides (Figure 3A) | NW | F(1,2.74) = 12.62 | p = 0.00038 | F(2.74,59.91) = 41.26 | p = 1.47 × 1024 | F(2.74,59.91) = 2.78 | p = 0.044 |
| CU | F(1,2.76) = 7.42 | p = 0.0064 | F(2.76,70.42) = 17.71 | p = 9.39 × 1011 | F(2.76,70.42) = 9.90 | p = 3.63 × 106 | |
| Clostridia_UCG-014 (Figure 3C) | NW | F(1,2.65) = 4.72 | p = 0.029 | F(2.65,55.97) = 38.09 | p = 4.85 × 1022 | F(2.65,55.97) = 1.01 | p = 0.379 |
| CU | F(1,2.71) = 35.93 | p = 2.05 × 109 | F(2.71,67.05) = 12.88 | p = 8.44 × 108 | F(2.71,67.05) = 2.18 | p = 0.095 | |
| Christensenellaceae_R7_group (Figure 3D) | NW | F(1,2.82) = 3.70 | p = 0.054 | F(2.82,53.48) = 79.63 | p = 1.30 × 1048 | F(2.82,53.48) = 2.59 | p = 0.055 |
| CU | F(1,2.81) = 48.65 | p = 3.06 × 1012 | F(2.81,69.23) = 197.98 | p = 2.63 × 10120 | F(2.81,69.23) = 9.16 | p = 8.53 × 106 | |
| Incertae_Sedis (Ruminiclostridium V) (Figure 3E) | NW | F(1,2.81) = 76.70 | p = 1.99 × 1018 | F(2.81,56.25) = 46.03 | p = 4.62 × 1028 | F(2.81,56.25) = 16.85 | p = 2.09 × 1010 |
| CU | F(1,2.51) = 210.79 | p = 9.26 × 1048 | F(2.51,71.88) = 85.26 | p = 1.03 × 1046 | F(2.51,71.88) = 19.26 | p = 7.86 × 1011 | |
| Parabacteroides (Figure 3B) | NW | F(1,2.74) = 158.1 | p = 2.96 × 1036 | F(2.74,59.18) = 44.69 | p = 1.32 × 1026 | F(2.74,59.18) = 21.04 | p = 1.19 × 1012 |
| CU | F(1,2.74) = 467.75 | p = 9.91 × 10104 | F(2.74,71.88) = 71.88 | p = 6.98 × 1032 | F(2.74,71.88) = 4.99 | p = 0.00258 | |
| Low Relative Abundance (1–2%) | |||||||
| Parasutterella (Figure 3F) | NW | F(1,78) = 29.19 | p =6.57 × 108 | F(2.78,59.45) = 40.76 | p = 1.46 × 1024 | F(2.78,59.45) = 1.09 | p = 0.127 |
| CU | F(1,2.78) = 9.15 | p = 0.0025 | F(2.78,71.92) = 63.79 | p = 1.78 × 1038 | F(2.78,71.92) = 2.25 | p = 0.052 | |
| Ruminococcus_gauvreauii_group (Figure 3G) | NW | F(1,2.71) = 104.03 | p = 1.99 × 1024 | F(2.71,59.31) = 27.24 | p = 3.73 × 1016 | F(2.71,59.31) = 17.95 | p = 9.61 × 1011 |
| CU | F(1,2.33) = 16.93 | p = 0.000039 | F(2.33,69.45) = 19.51 | p = 2.48 × 1010 | F(2.33,69.45) = 4.79 | p = 0.00018 | |
| UCG-007 (Figure 3H) | NW | F(1,2.84) = 289.83 | p = 5.42 × 1065 | F(2.84,55.73) = 40.50 | p = 7.18 × 1025 | F(2.84,55.73) = 31.13 | p = 3.74 × 1019 |
| CU | F(1,2.66) = 140.28 | p = 2.31 × 1032 | F(2.66,57.78) = 32.24 | p = 9.19 × 1019 | F(2.66,57.78) = 10.11 | p = 3.89 × 106 | |
| Lachnospiraceae_UCG-006 (Figure 3I) | NW | F(1,2.77) = 1.76 | p = 0.184 | F(2.77,59.93) = 42.81 | p = 9.83 × 1026 | F(2.77,59.93) = 4.89 | p = 0.0028 |
| CU | F(1,2.77) = 6.33 | p = 0.0118 | F(2.77,65.94) = 51.97 | p = 3.48 × 1031 | F(2.77,65.94) = 4.61 | p = 0.00410 | |
| Higher in Control Diet, color indicates consistent effect across study site | |||||||
| High Relative Abundance (2–20%) | |||||||
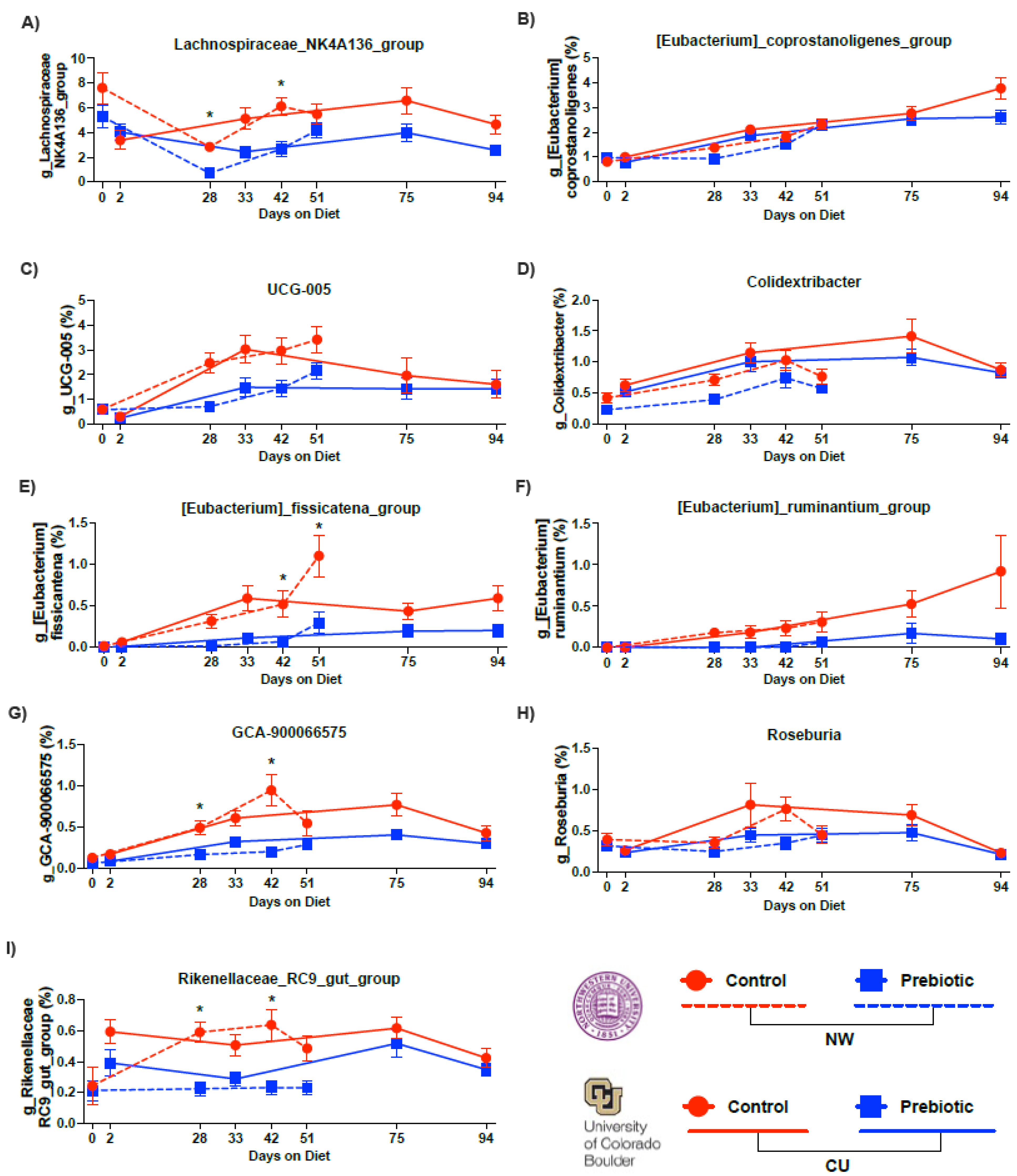
| Lachnospiraceae_NK4A136_group (Figure 4A) | NW | F(1,2.81) = 36.70 | p = 1.38 × 109 | F(2.81,59.99) = 20.53 | p = 1.34 × 1012 | F(2.81,59.99) = 2.81 | p = 0.020 |
| CU | F(1,2.83) = 13.13 | p = 0.0003 | F(2.83,71.45) = 2.30 | p = 0.079 | F(2.83,71.45) = 3.99 | p = 0.0087 | |
| Eubacterium_coprostanoligenes_group (Figure 4B) | NW | F(1,2.48) = 1.34 | p = 0.247 | F(2.48,57.50) = 30.14 | p = 1.56 × 1016 | F(2.48,57.50) = 1.66 | p = 0.183 |
| CU | F(1,2.87) = 3.64 | p = 0.056 | F(2.87,71.52) = 56.35 | p = 5.72 × 1035 | F(2.87,71.52) = 0.55 | p = 0.638 | |
| UCG-005 (Figure 4C) | NW | F(1,2.82) = 8.07 | p = 0.0451 | F(2.82,59.10) = 21.62 | p = 2.59 × 1013 | F(2.82,59.10) = 1.62 | p = 0.184 |
| CU | F(1,2.37) = 0.841 | p = 0.359 | F(2.37,71.81) = 18.66 | p = 4.57 × 1010 | F(2.37,71.81) = 1.73 | p = 0.171 | |
| Low Relative Abundance (1–2%) | |||||||
| Colidextribacter (Figure 4D) | NW | F(1,2.73) = 13.95 | p = 0.00019 | F(2.73,59.62) = 18.55 | p = 3.78 × 1011 | F(2.73,59.62) = 0.816 | p = 0.475 |
| CU | F(1,2.78) = 0.013 | p = 0.911 | F(2.78,71.96) = 32.26 | p = 1.64 × 108 | F(2.78,71.96) = 0.328 | p = 0.790 | |
| Eubacterium_fissicatena_group (Figure 4E) | NW | F(1,2.41) = 9.51 | p = 0.002 | F(2.41,54.79) = 25.73 | p = 7.31 × 1014 | F(2.41,54.79) = 7.08 | p = 0.00034 |
| CU | F(1,2.26) = 4.64 | p = 0.031 | F(2.26,68.41) = 18.53 | p = 1.32 × 109 | F(2.26,68.41) = 3.09 | p = 0.039 | |
| Eubacterium_ruminantium_group (Figure 4F) | NW | F(1,2.62) = 17.80 | p = 0.00002 | F(2.62,38.97) = 7.97 | p = 0.00006 | F(2.62,38.97) = 11.57 | p = 6.82 × 107 |
| CU | F(1,2.44) = 6.22 | p = 0.013 | F(2.44,63.83) = 8.63 | p = 0.00005 | F(2.44,63.83) = 6.31 | p = 0.0008 | |
| GCA-900066575 (Figure 4G) | NW | F(1,2.92) = 20.93 | p = 0.000005 | F(2.92,58.16) = 24.09 | p = 3.24 × 1015 | F(2.92,58.16) = 5.18 | p = 0.0016 |
| CU | F(1,2.91) = 9.67 | p = 0.0019 | F(2.91,71.91) = 29.78 | p = 49.98 × 1019 | F(2.91,71.91) = 0.937 | p = 0.420 | |
| Roseburia (Figure 4H) | NW | F(1,2.80) = 6.48 | p = 0.0109 | F(2.80,59.66) = 2.90 | p = 0.037 | F(2.80,59.66) = 1.73 | p = 0.161 |
| CU | F(1,2.74) = 4.79 | p = 0.029 | F(2.74,72.00) = 8.71 | p = 0.000019 | F(2.74,72.00) = 0.776 | p = 0.50 | |
| Rikenellaceae_RC9_gut_group (Figure 4I) | NW | F(1,2.74) = 25.70 | p = 3.99 × 107 | F(2.74,59.55) = 9.20 | p = 0.00006 | F(2.74,59.55) = 5.55 | p = 0.0012 |
| CU | F(1,2.72) = 10.90 | p = 0.00096 | F(2.72,70.925) = 2.88 | p = 0.040 | F(2.72,70.925) = 1.42 | p = 0.236 | |
| Nonparametric Longitudinal Data (naprLD) Table: ANOVA-Type Statistics (ATSs) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bile Acids | ||||||||||
| Diet—F-Value; p-Value | p-adj. (Holm) | Time—F-Value; p-Value | p-adj. (Holm) | Diet × Time | p-adj. (Holm) | |||||
| Color indicates consistent effect across study site | ||||||||||
| Primary Bile Acids | ||||||||||
| Cholic Acid | NW | F(1,2.846) = 0.190 | p = 0.663 | n/a | F(2.846,58.12) = 9.534 | p = 4.493 × 106 | p = 8.98 × 106 | F(2.846,58.12) = 0.921 | p = 0.426 | ns |
| CU | F(1,2.605) = 4.0759 | p = 0.0435 | p = 0.136 | F(2.605,36.68) = 4.934 | p = 0.0033 | p = 0.0066 | F(2.605,36.68) = 0.2756 | p = 0.815 | n/a | |
| Muricholic_alpha | NW | F(1,2.72) = 0.188 | p = 0.665 | n/a | F(2.72,58.508) = 35.817 | p = 3.20 × 1021 | p = 1.92 × 1020 | F(2.72,58.508) = 0.755 | p = 0.507 | ns |
| CU | F(1,2.81) = 2.24 | p = 0.135 | ns | F(2.81,77.408) = 57.30 | p = 7.157 × 1035 | p = 1.0024 × 1033 | F(2.81,77.408) = 0.397 | p = 0.742 | n/a | |
| Muricholic_beta (Figure 6A) | NW | F(1,2.911) = 2.623 | p = 0.105 | n/a | F(2.911,54.878,) = 36.129 | p = 1.011 × 1022 | p = 8.08 × 1022 | F(2.911,54.878,) = 2.706 | p = 0.0453 | p = 0.0453 |
| CU | F(1,2.68) = 9.452 | p = 0.0021 | p = 0.019 | F(2.68,78.32) = 81.99 | p = 1.00 × 1047 | p = 1.9 × 1046 | F(2.68,78.32) = 0.272 | p = 0.823 | n/a | |
| Conjugated Bile Acids | ||||||||||
| Glycochenodeoxycholic Acid | NW | F(1,2.819) = 0.578 | p = 0.447 | n/a | F(2.819,60.355) = 59.90 | p = 1.503 × 1036 | p = 1.95 × 1035 | F(2.819,60.355) = 1.784 | p = 0.151 | ns |
| CU | F(1,2.917) = 2.459 | p = 0.116 | ns | F(2.917,78.25) = 17.47 | p = 4.35 × 1011 | p = 3.48 × 1010 | F(2.917,78.25) = 0.508 | p = 0.671 | n/a | |
| Glycocholic Acid | NW | F(1,2.627) = 0.146 | p = 0.701 | n/a | F(2.627,60.03) = 108.142 | p = 1.084 × 1061 | p = 1.728 × 1060 | F(2.627,60.03) = 0.257 | p = 0.831 | ns |
| CU | F(1,2.744) = 4.479 | p = 0.0343 | p = 0.136 | F(2.744,75.63) = 41.22 | p = 1.38 × 1024 | p = 1.794 × 1023 | F(2.744,75.63) = 0.109 | p = 0.274 | n/a | |
| Glycohyocholic Acid | NW | F(1,2.913) = 0.092 | p = 0.762 | n/a | F(2.913,60.943) = 28.238 | p = 8.523 × 1018 | p = 4.26 × 1017 | F(2.913,60.943) = 0.514 | p = 0.667 | ns |
| CU | F(1,2.706) = 0.543 | p = 0.4611 | ns | F(2.706,79.81) = 29.44 | p = 2.083 × 1017 | p = 1.872 × 1016 | F(2.706,79.81) = 0.146 | p = 0.917 | n/a | |
| Taurochenodeoxycholic Acid | NW | F(1,2.57) = 0.453 | p = 0.501 | n/a | F(2.57,58.52) = 43.784 | p = 1.290 × 1024 | p = 1.161 × 1023 | F(2.57,58.52) = 1.378 | p = 0.250 | ns |
| CU | F(1,2.82) = 3.133 | p = 0.0688 | ns | F(2.82,76.881) = 34.930 | p = 2.46 × 1021 | p = 2.706 × 1020 | F(2.82,76.881) = 0.567 | p = 0.625 | n/a | |
| Taurocholic Acid | NW | F(1,2.83) = 0.417 | p = 0.518 | n/a | F(2.83,60.603) = 15.834 | p = 7.724 × 1010 | p = 2.316 × 109 | F(2.83,60.603) = 0.743 | p = 0.519 | ns |
| CU | F(1,2.773) = 6.388 | p = 0.0115 | p = 0.069 | F(2.773,36.7228) = 1.232 | p = 0.296 | ns | F(2.773,36.7228) = 0.644 | p = 0.575 | n/a | |
| Taurohyocholic Acid | NW | F(1,2.581) = 2.896 | p = 0.0878 | n/a | F(2.581,58.664) = 57.948 | p = 1.301 × 1032 | p = 1.56 × 1031 | F(2.581,58.664) = 1.256 | p = 0.288 | ns |
| CU | F(1,2.381) = 4.492 | p = 0.0341 | ns | F(2.381,79.613) = 42.977 | p = 1.39 × 1022 | p = 1.668 × 1021 | F(2.381,79.613) = 1.348 | p = 0.259 | n/a | |
| Secondary Bile Acids | ||||||||||
| Deoxycholic Acid (Figure 6B) | NW | F(1,2.2670) = 5.557 | p = 0.0184 | n/a | F(2.2.267,28.994) = 84.80 | p = 3.18 × 1042 | p = 4.77 × 1041 | F(2.267,28.994) = 2.19 | p = 0.104 | ns |
| CU | F(1,2.79) = 12.219 | p = 0.00047 | p = 0.005 | F(2.79,79.83) = 62.44 | p = 8.56 × 1038 | p = 1.3696 × 1036 | F(2.79,79.83) = 2.188 | p = 0.0918 | n/a | |
| Lithocholic Acid (Figure 6C) | NW | F(1,2.832) = 0.240 | p = 0.624 | n/a | F(2.832,60.296) = 123.84 | p = 6.77 × 1076 | p = 1.2186 × 1074 | F(2.832,60.296) = 3.374 | p = 0.0196 | p = 0.0392 |
| CU | F(1,2.90) = 10.84 | p = 0.0009 | p = 0.010 | F(2.90,79.89) = 12.19 | p = 8.72 × 108 | p = 3.49 × 107 | F(2.90,79.89) = 1.476 | p = 0.220 | n/a | |
| Ursodeoxycholic Acid (Figure 6D) | NW | F(1,2.539) = 2.465 | p = 0.164 | n/a | F(2.539,60.446) = 115.64 | p = 7.468 × 1064 | p = 1.2699 × 1062 | F(2.539,60.446) = 2.228 | p = 0.0935 | ns |
| CU | F(1,2.532) = 9.188 | p = 0.00243 | p = 0.019 | F(2.532,78.672) = 4.966 | p = 0.00349 | p = 0.0066 | F(2.532,78.672) = 1.098 | p = 0.343 | n/a | |
| Secondary Conjugated Bile Acids | ||||||||||
| Glycodeoxycholic Acid (Figure 6E) | NW | F(1,2.818) = 0.485 | p = 0.486 | n/a | F(2.818,60.903) = 48.193 | p = 2.045 × 1029 | p = 2.255 × 1028 | F(2.818,60.903) = 5.310 | p = 0.0015 | p = 0.0045 |
| CU | F(1,2.916) = 5.013 | p = 0.0252 | p = 0.126 | F(2.916,79.05) = 31.25 | p = 1.064 × 1019 | p = 1.06 × 1018 | F(2.916,79.05) = 0.972 | p = 0.403 | n/a | |
| Glycolithocholic Acid | NW | F(1,2.513) = 1.268 | p = 0.260 | n/a | F(2.513,59.330) = 72.0 | p = 1.784 × 1039 | p = 2.492 × 1038 | F(2.513,59.330) = 0.534 | p = 0.627 | ns |
| CU | F(1,2.513) = 0.009 | p = 0.923 | ns | F(2.513,78.64) = 76.44 | p = 6.81 × 1042 | p = 1.1577 × 1040 | F(2.513,78.64) = 0.150 | p = 0.903 | n/a | |
| Glycoursodeoxycholic Acid | NW | F(1,2.488) = 0.898 | p = 0.343 | n/a | F(2.488,56.613) = 48.827 | p = 1.26 × 1026 | p = 1.26 × 1025 | F(2.488,56.613) = 0.194 | p = 0.920 | ns |
| CU | F(1,2.911) = 1.24 | p = 0.265 | ns | F(2.911,79.71) = 74.60 | p = 6.64 × 1047 | p = 1.1952 × 1045 | F(2.911,79.71) = 0.214 | p = 0.881 | n/a | |
| Taurodeoxycholic Acid | NW | F(1,2.501) = 0.262 | p = 0.609 | n/a | F(2.501,59.216) = 40.489 | p = 3.021 × 1022 | p = 2.114 × 1021 | F(2.501,59.216) = 0.835 | p = 0.456 | ns |
| CU | F(1,2.768) = 4.484 | p = 0.0342 | ns | F(2.768,78.280) = 16.797 | p = 3.07 × 1010 | p = 1.84 × 109 | F(2.768,78.280) = 0.511 | p = 0.659 | n/a | |
| Taurohyodeoxycholic Acid | NW | F(1,2.817) = 1.459 | p = 0.227 | n/a | F(2.817,60.123) = 150.64 | p = 7.138 × 1092 | p = 1.4994 × 1090 | F(2.817,60.123) = 1.111 | p = 0.341 | ns |
| CU | F(1,2.785) = 7.212 | p = 0.00724 | p = 0.050 | F(2.785,79.746) = 13.787 | p = 1.68 × 108 | p = 8.40 × 108 | F(2.785,79.746) = 0.473 | p = 0.687 | n/a | |
| Taurolithocholic Acid | NW | F(1,2.895) = 0.001 | p = 0.974 | n/a | F(2.895,60.534) = 18.164 | p = 1.887 × 1011 | p = 7.56 × 1011 | F(2.895,60.534) = 2.139 | p = 0.095 | ns |
| CU | F(1,2.85) = 0.006 | p = 0.937 | ns | F(2.85,78.972) = 17.603 | p = 5.79 × 1011 | p = 4.05 × 1010 | F(2.85,78.972) = 0.784 | p = 0.497 | n/a | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, R.S.; Bowers, S.J.; Vargas, F.; Hopkins, S.; Kelley, T.; Gonzalez, A.; Lowry, C.A.; Dorrestein, P.C.; Vitaterna, M.H.; Turek, F.W.; et al. A Prebiotic Diet Containing Galactooligosaccharides and Polydextrose Produces Dynamic and Reproducible Changes in the Gut Microbial Ecosystem in Male Rats. Nutrients 2024, 16, 1790. https://doi.org/10.3390/nu16111790
Thompson RS, Bowers SJ, Vargas F, Hopkins S, Kelley T, Gonzalez A, Lowry CA, Dorrestein PC, Vitaterna MH, Turek FW, et al. A Prebiotic Diet Containing Galactooligosaccharides and Polydextrose Produces Dynamic and Reproducible Changes in the Gut Microbial Ecosystem in Male Rats. Nutrients. 2024; 16(11):1790. https://doi.org/10.3390/nu16111790
Chicago/Turabian StyleThompson, Robert S., Samuel J. Bowers, Fernando Vargas, Shelby Hopkins, Tel Kelley, Antonio Gonzalez, Christopher A. Lowry, Pieter C. Dorrestein, Martha Hotz Vitaterna, Fred W. Turek, and et al. 2024. "A Prebiotic Diet Containing Galactooligosaccharides and Polydextrose Produces Dynamic and Reproducible Changes in the Gut Microbial Ecosystem in Male Rats" Nutrients 16, no. 11: 1790. https://doi.org/10.3390/nu16111790
APA StyleThompson, R. S., Bowers, S. J., Vargas, F., Hopkins, S., Kelley, T., Gonzalez, A., Lowry, C. A., Dorrestein, P. C., Vitaterna, M. H., Turek, F. W., Knight, R., Wright, K. P., Jr., & Fleshner, M. (2024). A Prebiotic Diet Containing Galactooligosaccharides and Polydextrose Produces Dynamic and Reproducible Changes in the Gut Microbial Ecosystem in Male Rats. Nutrients, 16(11), 1790. https://doi.org/10.3390/nu16111790







