The Impact of Haplotypes of the FTO Gene, Lifestyle, and Dietary Patterns on BMI and Metabolic Syndrome in Polish Young Adult Men
Abstract
1. Introduction
2. Materials and Methods
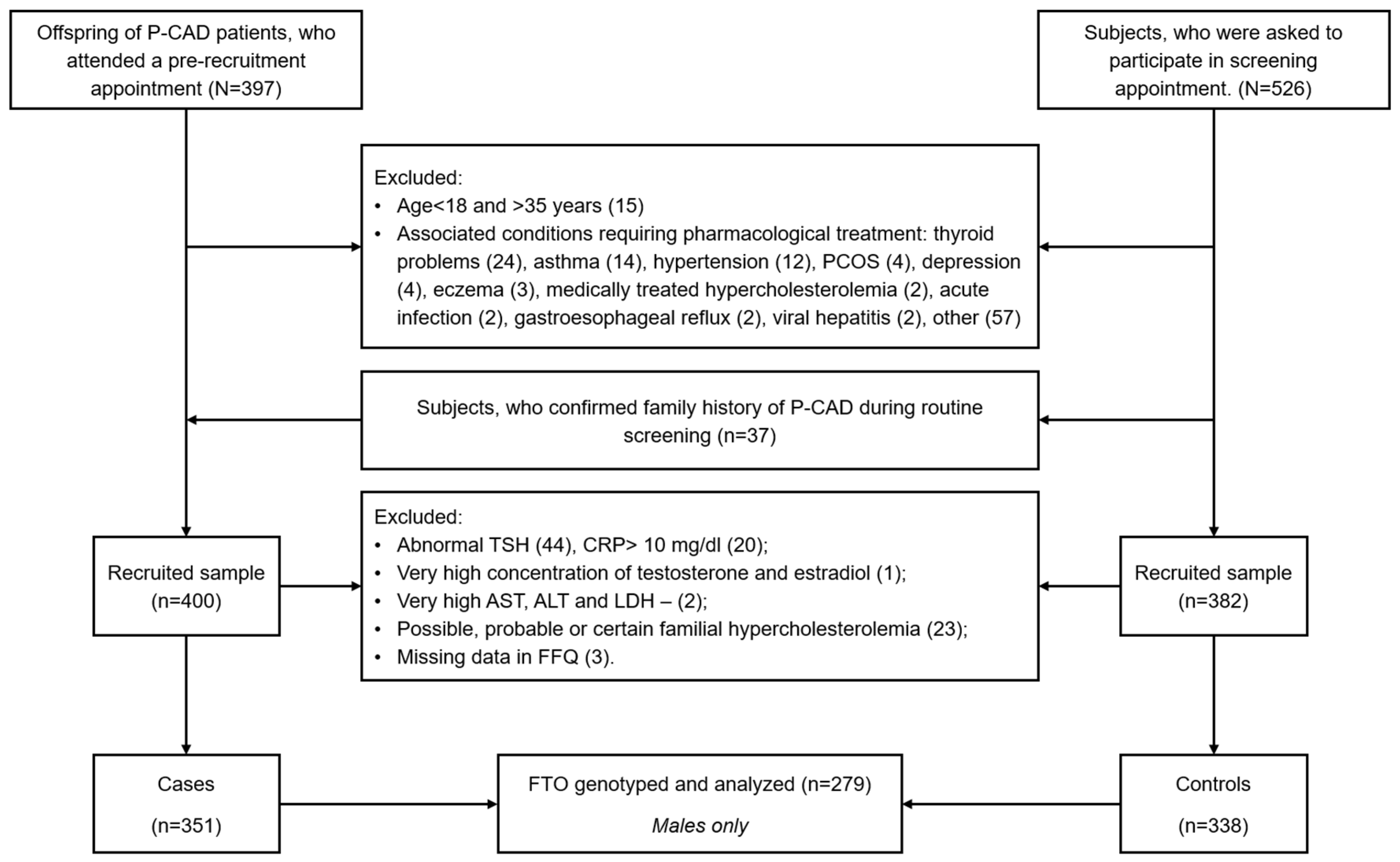
2.1. Study Group
- Waist circumference ≥80 cm in women and ≥94 cm in men;
- Fasting blood triglycerides > 1.7 mmol/L or treatment of hypertriglyceridemia;
- HDL-C < 1.3 mmol/L in women and <1.0 mmol/L in men or treatment of low HDL-c levels;
- Systolic blood pressure ≥ 130 mmHg or diastolic blood pressure ≥ 85 mm Hg or treatment for hypertension;
- Fasting blood glucose levels ≥ 5.6 mmol/L or treatment of DM.
2.2. Measurements of Anthropometric Parameters
2.3. Laboratory Tests
2.4. Diet Quality and Lifestyle
2.5. Genetic Analysis
2.6. Statistical Analysis
3. Results
3.1. Study Group Characteristics
3.2. FTO SNP
3.3. Dietary Patterns, Dietary Quality, and Physical Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Obesity. Available online: https://www.who.int/health-topics/obesity (accessed on 29 September 2023).
- Guh, D.P.; Zhang, W.; Bansback, N.; Amarsi, Z.; Birmingham, C.L.; Anis, A.H. The Incidence of Co-Morbidities Related to Obesity and Overweight: A Systematic Review and Meta-Analysis. BMC Public Health 2009, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolski, P.; Prejbisz, A.; Kuryłowicz, A.; Baska, A.; Burchardt, P.; Chlebus, K.; Dzida, G.; Jankowski, P.; Jaroszewicz, J.; Jaworski, P.; et al. Metabolic Syndrome—A New Definition and Management guidelinesA Joint Position Paper by the Polish Society of Hypertension, Polish Society for the Treatment of Obesity, Polish Lipid Association, Polish Association for Study of Liver, Polish Society of Family Medicine, Polish Society of Lifestyle Medicine, Division of Prevention and Epidemiology Polish Cardiac Society, “Club 30” Polish Cardiac Society, and Division of Metabolic and Bariatric Surgery Society of Polish Surgeons. Arch. Med. Sci. 2022, 18, 1133–1156. [Google Scholar] [CrossRef]
- Do Vale Moreira, N.C.; Hussain, A.; Bhowmik, B.; Mdala, I.; Siddiquee, T.; Fernandes, V.O.; Montenegro Júnior, R.M.; Meyer, H.E. Prevalence of Metabolic Syndrome by Different Definitions, and Its Association with Type 2 Diabetes, Pre-Diabetes, and Cardiovascular Disease Risk in Brazil. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Alberti, K.G.M.M.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.-C.; James, W.P.T.; Loria, C.M.; Smith, S.C. Harmonizing the Metabolic Syndrome: A Joint Interim Statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009, 120, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Osadnik, K.; Osadnik, T.; Gierlotka, M.; Windak, A.; Tomasik, T.; Mastej, M.; Kuras, A.; Jóźwiak, K.; Penson, P.E.; Lip, G.Y.H.; et al. Metabolic Syndrome Is Associated with Similar Long-Term Prognosis in Those Living with and without Obesity: An Analysis of 45 615 Patients from the Nationwide LIPIDOGRAM 2004–2015 Studies. Eur. J. Prev. Cardiol. 2023, 30, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Racette, S.B.; Deusinger, S.S.; Deusinger, R.H. Obesity: Overview of Prevalence, Etiology, and Treatment. Phys. Ther. 2003, 83, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Elks, C.E.; Den Hoed, M.; Zhao, J.H.; Sharp, S.J.; Wareham, N.J.; Loos, R.J.F.; Ong, K.K. Variability in the Heritability of Body Mass Index: A Systematic Review and Meta-Regression. Front. Endocrin. 2012, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Maes, H.H.M.; Neale, M.C.; Eaves, L.J. Genetic and Environmental Factors in Relative Body Weight and Human Adiposity. Behav. Genet. 1997, 27, 325–351. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The Genetics of Obesity: From Discovery to Biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef]
- Huvenne, H.; Dubern, B.; Clément, K.; Poitou, C. Rare Genetic Forms of Obesity: Clinical Approach and Current Treatments in 2016. Obes Facts 2016, 9, 158–173. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Larder, R.; Cheung, M.K.M.; Tung, Y.C.L.; Yeo, G.S.H.; Coll, A.P. Where to Go with FTO? Trends Endocrinol. Metab. 2011, 22, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Berulava, T.; Horsthemke, B. The Obesity-Associated SNPs in Intron 1 of the FTO Gene Affect Primary Transcript Levels. Eur. J. Hum. Genet. 2010, 18, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Karra, E.; O’Daly, O.G.; Choudhury, A.I.; Yousseif, A.; Millership, S.; Neary, M.T.; Scott, W.R.; Chandarana, K.; Manning, S.; Hess, M.E.; et al. A Link between FTO, Ghrelin, and Impaired Brain Food-Cue Responsivity. J. Clin. Investig. 2013, 123, 3539–3551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.; Ma, J.; Guo, F.; Cao, Q.; Zhang, Y.; Zhou, B.; Chai, J.; Zhao, W.; Zhao, R. The Demethylase Activity of FTO (Fat Mass and Obesity Associated Protein) Is Required for Preadipocyte Differentiation. PLoS ONE 2015, 10, e0133788. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, M.S.; Moshitch-Moshkovitz, S.; Rechavi, G. FTO: Linking m6A Demethylation to Adipogenesis. Cell Res. 2015, 25, 3–4. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-Dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Merkestein, M.; Laber, S.; McMurray, F.; Andrew, D.; Sachse, G.; Sanderson, J.; Li, M.; Usher, S.; Sellayah, D.; Ashcroft, F.M.; et al. FTO Influences Adipogenesis by Regulating Mitotic Clonal Expansion. Nat. Commun. 2015, 6, 6792. [Google Scholar] [CrossRef]
- Azzam, S.K.; Alsafar, H.; Sajini, A.A. FTO m6A Demethylase in Obesity and Cancer: Implications and Underlying Molecular Mechanisms. IJMS 2022, 23, 3800. [Google Scholar] [CrossRef]
- Stratigopoulos, G.; Martin Carli, J.F.; O’Day, D.R.; Wang, L.; LeDuc, C.A.; Lanzano, P.; Chung, W.K.; Rosenbaum, M.; Egli, D.; Doherty, D.A.; et al. Hypomorphism for RPGRIP1L, a Ciliary Gene Vicinal to the FTO Locus, Causes Increased Adiposity in Mice. Cell Metab. 2014, 19, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.-H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Smemo, S.; Tena, J.J.; Kim, K.-H.; Gamazon, E.R.; Sakabe, N.J.; Gómez-Marín, C.; Aneas, I.; Credidio, F.L.; Sobreira, D.R.; Wasserman, N.F.; et al. Obesity-Associated Variants within FTO Form Long-Range Functional Connections with IRX3. Nature 2014, 507, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Doaei, S.; Kalantari, N.; Mohammadi, N.K.; Tabesh, G.A.; Gholamalizadeh, M. Macronutrients and the FTO Gene Expression in Hypothalamus; a Systematic Review of Experimental Studies. Indian Heart J. 2017, 69, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Mehrdad, M.; Doaei, S.; Gholamalizadeh, M.; Eftekhari, M.H. The Association between FTO Genotype with Macronutrients and Calorie Intake in Overweight Adults. Lipids Health Dis. 2020, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, P.K.; Fredriksson, R.; Olszewska, A.M.; Stephansson, O.; Alsiö, J.; Radomska, K.J.; Levine, A.S.; Schiöth, H.B. Hypothalamic FTO Is Associated with the Regulation of Energy Intake Not Feeding Reward. BMC Neurosci. 2009, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Dina, C.; Meyre, D.; Gallina, S.; Durand, E.; Körner, A.; Jacobson, P.; Carlsson, L.M.S.; Kiess, W.; Vatin, V.; Lecoeur, C.; et al. Variation in FTO Contributes to Childhood Obesity and Severe Adult Obesity. Nat. Genet. 2007, 39, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Kolackov, K.; Łaczmański, Ł.; Lwow, F.; Ramsey, D.; Zdrojowy-Wełna, A.; Tupikowska, M.; Bednarek-Tupikowska, G. The Frequencies of Haplotypes of FTO Gene Variants and Their Association with the Distribution of Body Fat in Non-Obese Poles. Adv. Clin. Exp. Med. 2016, 25, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Jarczak, J.; Grochowalski, Ł.; Marciniak, B.; Lach, J.; Słomka, M.; Sobalska-Kwapis, M.; Lorkiewicz, W.; Pułaski, Ł.; Strapagiel, D. Mitochondrial DNA Variability of the Polish Population. Eur. J. Hum. Genet. 2019, 27, 1304–1314. [Google Scholar] [CrossRef]
- Osadnik, T.; Osadnik, K.; Pawlas, N.; Strzelczyk, J.; Kasperczyk, J.; Poloński, L.; Gąsior, M. Metabolic and Genetic Profiling of Young Adults with and without a Family History of Premature Coronary Heart Disease (MAGNETIC). Study Design and Methodology. Arch. Med. Sci. 2019, 15, 590–597. [Google Scholar] [CrossRef]
- Niedzwiedzka, E.; Wadolowska, L.; Kowalkowska, J. Reproducibility of A Non-Quantitative Food Frequency Questionnaire (62-Item FFQ-6) and PCA-Driven Dietary Pattern Identification in 13–21-Year-Old Females. Nutrients 2019, 11, 2183. [Google Scholar] [CrossRef] [PubMed]
- Kowalkowska, J.; Wadolowska, L.; Czarnocinska, J.; Czlapka-Matyasik, M.; Galinski, G.; Jezewska-Zychowicz, M.; Bronkowska, M.; Dlugosz, A.; Loboda, D.; Wyka, J. Reproducibility of a Questionnaire for Dietary Habits, Lifestyle and Nutrition Knowledge Assessment (KomPAN) in Polish Adolescents and Adults. Nutrients 2018, 10, 1845. [Google Scholar] [CrossRef] [PubMed]
- Posit. Available online: https://www.posit.co/ (accessed on 29 September 2023).
- Wigginton, J.E.; Cutler, D.J.; Abecasis, G.R. A Note on Exact Tests of Hardy-Weinberg Equilibrium. Am. J. Hum. Genet. 2005, 76, 887–893. [Google Scholar] [CrossRef] [PubMed]
- González, J.R.; Armengol, L.; Solé, X.; Guinó, E.; Mercader, J.M.; Estivill, X.; Moreno, V. SNPassoc: An R Package to Perform Whole Genome Association Studies. Bioinformatics 2007, 23, 654–655. [Google Scholar] [CrossRef] [PubMed]
- Osadnik, T.; Pawlas, N.; Lonnie, M.; Osadnik, K.; Lejawa, M.; Wądołowska, L.; Bujak, K.; Fronczek, M.; Reguła, R.; Gawlita, M.; et al. Family History of Premature Coronary Artery Disease (P-CAD)—A Non-Modifiable Risk Factor? Dietary Patterns of Young Healthy Offspring of P-CAD Patients: A Case-Control Study (MAGNETIC Project). Nutrients 2018, 10, 1488. [Google Scholar] [CrossRef] [PubMed]
- Osadnik, K.; Osadnik, T.; Lonnie, M.; Lejawa, M.; Reguła, R.; Fronczek, M.; Gawlita, M.; Wądołowska, L.; Gąsior, M.; Pawlas, N. Metabolically Healthy Obese and Metabolic Syndrome of the Lean: The Importance of Diet Quality. Analysis of MAGNETIC Cohort. Nutr. J. 2020, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.; Wills, A.K.; Wong, A.; Elks, C.E.; Wareham, N.J.; Loos, R.J.F.; Kuh, D.; Ong, K.K. Life Course Variations in the Associations between FTO and MC4R Gene Variants and Body Size. Hum. Mol. Genet. 2009, 19, 545–552. [Google Scholar] [CrossRef]
- Wrzosek, M.; Zakrzewska, A.; Ruczko, L.; Jabłonowska-Lietz, B.; Nowicka, G. Association between Rs9930506 Polymorphism of the Fat Mass & Obesity-Associated (FTO) Gene & Onset of Obesity in Polish Adults. Indian J. Med. Res. 2016, 143, 281. [Google Scholar] [CrossRef] [PubMed]
- Piwonska, A.M.; Cicha-Mikolajczyk, A.; Sobczyk-Kopciol, A.; Piwonski, J.; Drygas, W.; Kwasniewska, M.; Pajak, A.; Zdrojewski, T.; Tykarski, A.; Kozakiewicz, K.; et al. Independent Association of FTO Rs9939609 Polymorphism with Overweight and Obesity in Polish Adults. Results from the Representative Population-Based WOBASZ Study. J. Physiol. Pharmacol. 2022, 73, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Luczynski, W.; Zalewski, G.; Bossowski, A. The Association of the FTO Rs9939609 Polymorphism with Obesity and Metabolic Risk Factors for Cardiovascular Diseases in Polish Children. J. Physiol. Pharmacol. 2012, 63, 241–248. [Google Scholar]
- Sobalska-Kwapis, M.; Suchanecka, A.; Słomka, M.; Siewierska-Górska, A.; Kępka, E.; Strapagiel, D. Genetic Association of FTO/IRX Region with Obesity and Overweight in the Polish Population. PLoS ONE 2017, 12, e0180295. [Google Scholar] [CrossRef] [PubMed]
- Zdrojowy-Wełna, A.; Bednarek-Tupikowska, G.; Zatońska, K.; Kolackov, K.; Jokiel-Rokita, A.; Bolanowski, M. The Association between FTO Gene Polymorphism Rs9939609 and Obesity Is Sex-Specific in the Population of PURE Study in Poland. Adv. Clin. Exp. Med. 2020, 29, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Zdrojowy-Wełna, A.; Ramsey, D.; Kolačkov, K.; Słoka, N.; Zatońska, K.; Szuba, A.; Bolanowski, M. The FTO Gene Is Not Associated with Weight Gain during Six Years of Observation in the Population of the PURE Study in Poland. Endokrynol. Pol. 2020, 71, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Guclu-Geyik, F.; Onat, A.; Yuzbasıogulları, A.B.; Coban, N.; Can, G.; Lehtimäki, T.; Erginel-Unaltuna, N. Risk of Obesity and Metabolic Syndrome Associated with FTO Gene Variants Discloses Clinically Relevant Gender Difference among Turks. Mol. Biol. Rep. 2016, 43, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Karns, R.; Viali, S.; Tuitele, J.; Sun, G.; Cheng, H.; Weeks, D.E.; McGarvey, S.T.; Deka, R. Common Variants in FTO Are Not Significantly Associated with Obesity-Related Phenotypes among Samoans of Polynesia: FTO and Obesity among Samoans. Ann. Hum. Genet. 2012, 76, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Leite, L.C.G.; Dos Santos, M.C.; Duarte, N.E.; Horimoto, A.R.V.R.; Crispim, F.; Vieira Filho, J.P.B.; Dal Fabbro, A.L.; Franco, L.J.; Moises, R.S. Association of Fat Mass and Obesity-Associated (FTO) Gene Rs9939609 with Obesity-Related Traits and Glucose Intolerance in an Indigenous Population, the Xavante. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102358. [Google Scholar] [CrossRef] [PubMed]
- Reuter, C.P.; Rosane De Moura Valim, A.; Gaya, A.R.; Borges, T.S.; Klinger, E.I.; Possuelo, L.G.; Franke, S.I.R.; Kmetzsch, L.; Vainstein, M.H.; Prá, D.; et al. FTO Polymorphism, Cardiorespiratory Fitness, and Obesity in B Razilian Youth. Am. J. Hum. Biol. 2016, 28, 381–386. [Google Scholar] [CrossRef]
- Jacobsson, J.A.; Risérus, U.; Axelsson, T.; Lannfelt, L.; Schiöth, H.B.; Fredriksson, R. The Common FTOvariant Rs9939609 Is Not Associated with BMI in a Longitudinal Study on a Cohort of Swedish Men Born 1920-1924. BMC Med. Genet. 2009, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Harbron, J.; Van der Merwe, L.; Zaahl, M.G.; Kotze, M.J.; Senekal, M. Fat Mass and Obesity-Associated (FTO) Gene Polymorphisms Are Associated with Physical Activity, Food Intake, Eating Behaviors, Psychological Health, and Modeled Change in Body Mass Index in Overweight/Obese Caucasian Adults. Nutrients 2014, 6, 3130–3152. [Google Scholar] [CrossRef]
- Goh, Y.; Choi, J.-H. Genetic Variation Rs1121980 in the Fat Mass and Obesity-Associated Gene (FTO) Is Associated with Dietary Intake in Koreans. Food Nutr. Res. 2022, 66, 8059. [Google Scholar] [CrossRef]
- Jacobsson, J.A.; Almén, M.S.; Benedict, C.; Hedberg, L.A.; Michaëlsson, K.; Brooks, S.; Kullberg, J.; Axelsson, T.; Johansson, L.; Ahlström, H.; et al. Detailed Analysis of Variants in FTO in Association with Body Composition in a Cohort of 70-Year-Olds Suggests a Weakened Effect among Elderly. PLoS ONE 2011, 6, e20158. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, J.; Zhang, Z.; Ren, J.; Li, Y.; Wang, J.; Cao, Y.; Rong, F.; Zhao, R.; Huang, X.; et al. Association of FTO Polymorphisms with Obesity and Metabolic Parameters in Han Chinese Adolescents. PLoS ONE 2014, 9, e98984. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, N.; Keshavarz Mohammadi, N.; Izadi, P.; Doaei, S.; Gholamalizadeh, M.; Eini-Zinab, H.; Salonurmi, T.; Rafieifar, S.; Janipoor, R.; Azizi Tabesh, G. A Haplotype of Three SNPs in FTO Had a Strong Association with Body Composition and BMI in Iranian Male Adolescents. PLoS ONE 2018, 13, e0195589. [Google Scholar] [CrossRef] [PubMed]
- Freathy, R.M.; Timpson, N.J.; Lawlor, D.A.; Pouta, A.; Ben-Shlomo, Y.; Ruokonen, A.; Ebrahim, S.; Shields, B.; Zeggini, E.; Weedon, M.N.; et al. Common Variation in the FTO Gene Alters Diabetes-Related Metabolic Traits to the Extent Expected Given Its Effect on BMI. Diabetes 2008, 57, 1419–1426. [Google Scholar] [CrossRef]
- Doney, A.S.F.; Dannfald, J.; Kimber, C.H.; Donnelly, L.A.; Pearson, E.; Morris, A.D.; Palmer, C.N.A. The FTO Gene Is Associated With an Atherogenic Lipid Profile and Myocardial Infarction in Patients With Type 2 Diabetes: A Genetics of Diabetes Audit and Research Study in Tayside Scotland (Go-DARTS) Study. Circ. Cardiovasc. Genet. 2009, 2, 255–259. [Google Scholar] [CrossRef]
| Variable | N * | Number of Patients (%) or Value (SD) † |
|---|---|---|
| Age | 279 | 28.92 (4.28) |
| Family History of P-CAD (%) | 279 | 178/279 (64%) |
| Family History of T2DM (%) | 99/279 (35%) | |
| Current smoking (vs. past smoker or non-smoker) | 279 | 72/279 (26%) |
| Physical activity level | 279 | |
| Low | 73/279 (26%) | |
| Moderate | 108/279 (39%) | |
| High | 98/279 (35%) | |
| SBP [mmHg] | 270 | 132 (15) |
| DBP [mmHg] | 270 | 81 (11) |
| BMI [kg/m2] | 279 | 26.14 (4.38) |
| VAI | 279 | 1.58 (2.65) |
| WC [m] | 279 | 0.89 (0.08) |
| WHTR | 279 | 0.50 (0.06) |
| TC [mmol/L] | 279 | 5.12 (1.11) |
| HDL-C [mmol/L] | 279 | 1.42 (0.37) |
| LDL-C [mmol/L] | 279 | 3.26 (0.96) |
| TG [mmol/L] | 279 | 1.42 (1.51) |
| Lp(a) [nmol/L] | 279 | 42.49 (65.64) |
| apoA1 [g/L] | 276 | 1.56 (0.25) |
| apoB [g/L] | 277 | 1.02 (0.60) |
| Glucose [mmol/L] | 279 | 5.15 (0.45) |
| HbA1c [%] | 278 | 5.06 (0.25) |
| hsCRP [mg/dL] | 279 | 1.33 (1.37) |
| Uric Acid [µmol/L] | 279 | 349.24 (63.96) |
| Fibrinogen [mg/dL] | 278 | 264.96 (56.89) |
| Variable | Protective Diplotype N = 65 1 | “Risk” Diplotype 1 N = 129 1 | “Risk” Diplotype 2 N = 55 1 | p-Value 2 |
|---|---|---|---|---|
| BMI [kg/m2] | 25.86 (4.59) | 26.46 (4.43) | 26.07 (4.39) | 0.53 |
| WC [m] | 0.89 (0.12) | 0.90 (0.11) | 0.90 (0.12) | 0.55 |
| TG [mmol/L] | 1.26 (0.79) | 1.39 (1.42) | 1.45 (1.16) | 0.72 |
| HDL-C [mmol/L] | 1.49 (0.40) | 1.37 (0.33) | 1.43 (0.39) | 0.33 |
| Glucose [mmol/L] | 5.08 (0.34) | 5.16 (0.47) | 5.21 (0.50) | 0.20 |
| SBP [mmHg] | 131.25 (12.41) | 130.81 (14.86) | 136.15 (15.33) | 0.060 |
| DBP [mmHg] | 81.19 (9.25) | 80.22 (11.33) | 82.56 (12.89) | 0.21 |
| Variable | N | Protective Diplotype, N = 65 1 | Risk Diplotype 1, N = 128 1 | Risk Diplotype 2, N = 55 1 | p-Value 2 |
|---|---|---|---|---|---|
| “Prudent” dietary pattern | 248 | 0.61 | |||
| Lowest adherence to DP | 23/65 (35%) | 44/128 (34%) | 19/55 (35%) | ||
| Moderate adherence to DP | 20/65 (31%) | 41/128 (32%) | 23/55 (42%) | ||
| Highest adherence to DP | 22/65 (34%) | 43/128 (34%) | 13/55 (24%) | ||
| “Western” dietary pattern | 248 | 0.30 | |||
| Lowest adherence to DP | 21/65 (32%) | 48/128 (38%) | 14/55 (25%) | ||
| Moderate adherence to DP | 21/65 (32%) | 46/128 (36%) | 18/55 (33%) | ||
| Highest adherence to DP | 23/65 (35%) | 34/128 (27%) | 23/55 (42%) | ||
| Physical activity | 248 | 0.68 | |||
| gentle | 17/65 (26%) | 33/128 (26%) | 14/55 (25%) | ||
| moderate | 25/65 (38%) | 55/128 (43%) | 18/55 (33%) | ||
| vigorous | 23/65 (35%) | 40/128 (31%) | 23/55 (42%) | ||
| nHDI (% points) | 246 | 5.57 (2.42) | 5.63 (2.18) | 5.53 (2.74) | 0.72 |
| pHDI (% points) | 247 | 6.87 (2.55) | 6.26 (2.08) | 7.18 (2.62) | 0.087 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Górczyńska-Kosiorz, S.; Lejawa, M.; Goławski, M.; Tomaszewska, A.; Fronczek, M.; Maksym, B.; Banach, M.; Osadnik, T. The Impact of Haplotypes of the FTO Gene, Lifestyle, and Dietary Patterns on BMI and Metabolic Syndrome in Polish Young Adult Men. Nutrients 2024, 16, 1615. https://doi.org/10.3390/nu16111615
Górczyńska-Kosiorz S, Lejawa M, Goławski M, Tomaszewska A, Fronczek M, Maksym B, Banach M, Osadnik T. The Impact of Haplotypes of the FTO Gene, Lifestyle, and Dietary Patterns on BMI and Metabolic Syndrome in Polish Young Adult Men. Nutrients. 2024; 16(11):1615. https://doi.org/10.3390/nu16111615
Chicago/Turabian StyleGórczyńska-Kosiorz, Sylwia, Mateusz Lejawa, Marcin Goławski, Agnieszka Tomaszewska, Martyna Fronczek, Beata Maksym, Maciej Banach, and Tadeusz Osadnik. 2024. "The Impact of Haplotypes of the FTO Gene, Lifestyle, and Dietary Patterns on BMI and Metabolic Syndrome in Polish Young Adult Men" Nutrients 16, no. 11: 1615. https://doi.org/10.3390/nu16111615
APA StyleGórczyńska-Kosiorz, S., Lejawa, M., Goławski, M., Tomaszewska, A., Fronczek, M., Maksym, B., Banach, M., & Osadnik, T. (2024). The Impact of Haplotypes of the FTO Gene, Lifestyle, and Dietary Patterns on BMI and Metabolic Syndrome in Polish Young Adult Men. Nutrients, 16(11), 1615. https://doi.org/10.3390/nu16111615








