Quorum-Sensing Signal DSF Inhibits the Proliferation of Intestinal Pathogenic Bacteria and Alleviates Inflammatory Response to Suppress DSS-Induced Colitis in Zebrafish
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Preparation of DSF
2.3. Zebrafish Maintenance and Embryo Handling
2.4. Construction of DSS-Induced Inflammatory Bowel Disease Zebrafish Model
2.5. Histological Analysis
2.5.1. Alcian Blue Staining
2.5.2. Hematoxylin–Eosin Staining
2.6. Inflammatory Response Analysis
2.6.1. Analysis of Macrophage Accumulation by Neutral Red Staining
2.6.2. Analysis of Neutrophilic Infiltration by Live Imaging
2.7. Microbial DNA Extraction and 16S rRNA Gene Sequencing
2.8. Real-Time Quantitative PCR (qRT-PCR)
2.9. Infecting Zebrafish Embryos with A. hydrophila or S. aureus
2.10. Quantitative Analysis of A. hydrophila or S. aureus in the Gut of Zebrafish
2.11. Statistical Analysis
3. Results
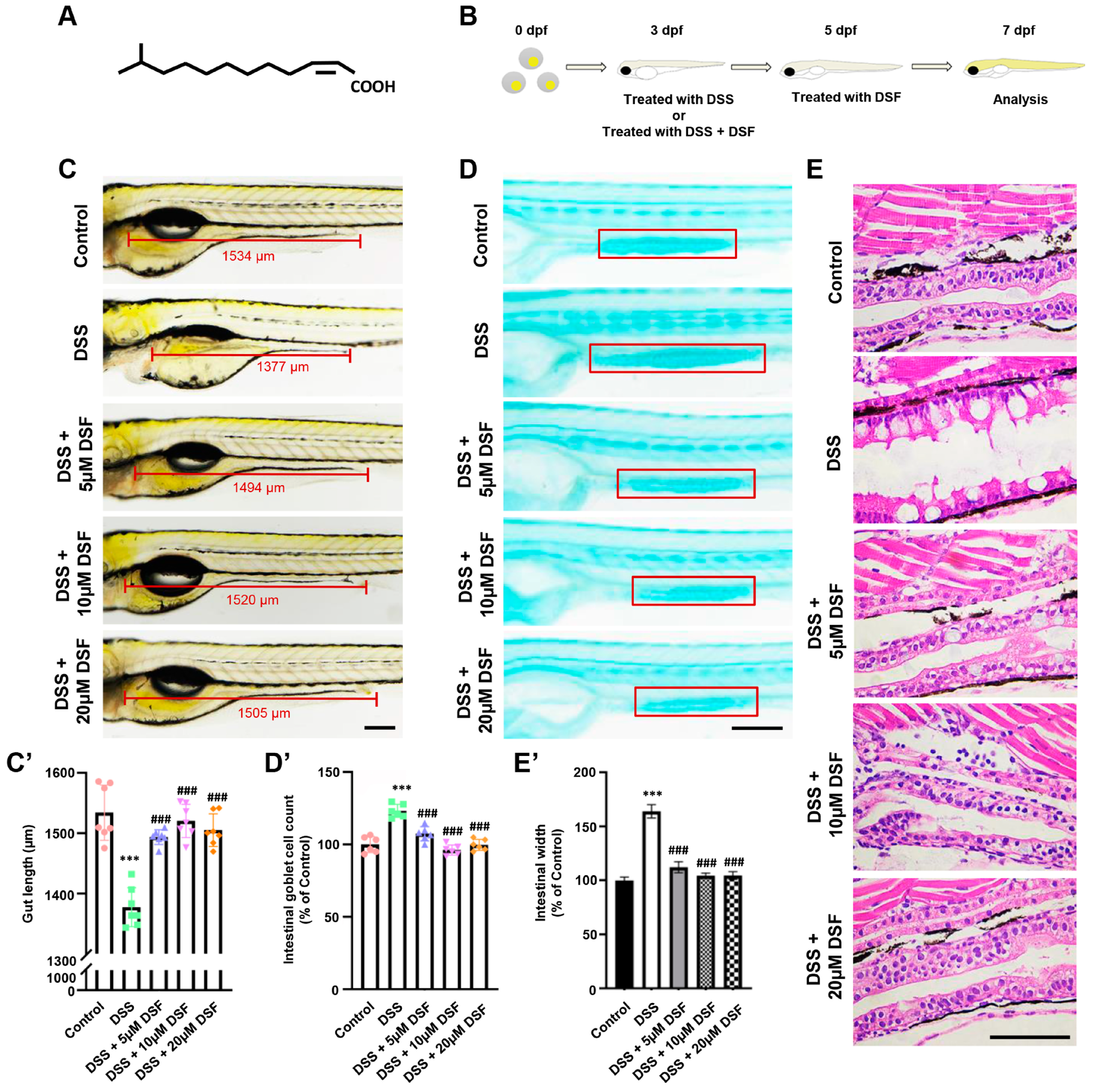
3.1. DSF Alleviates DSS-Induced Intestinal Damage in Zebrafish
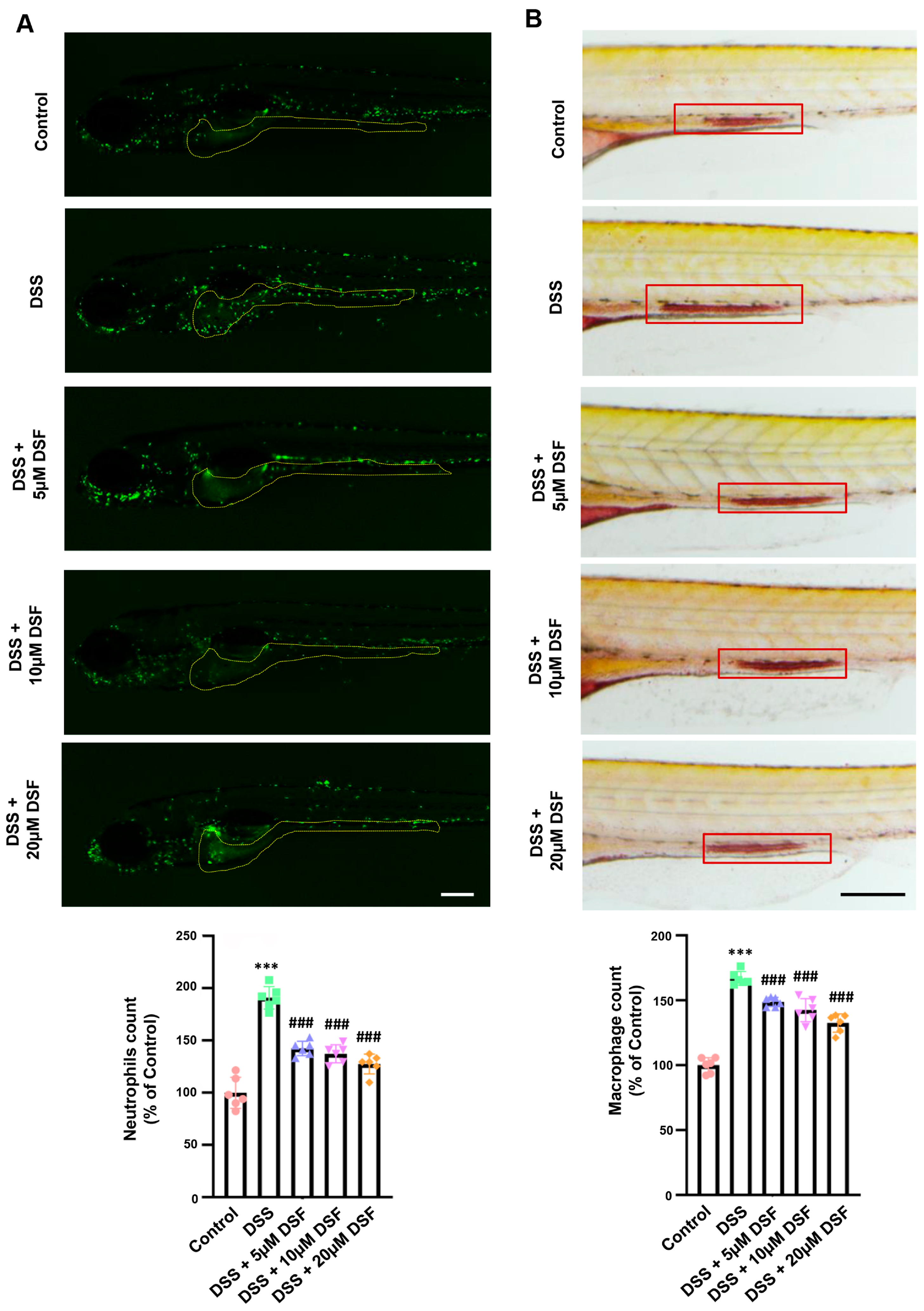
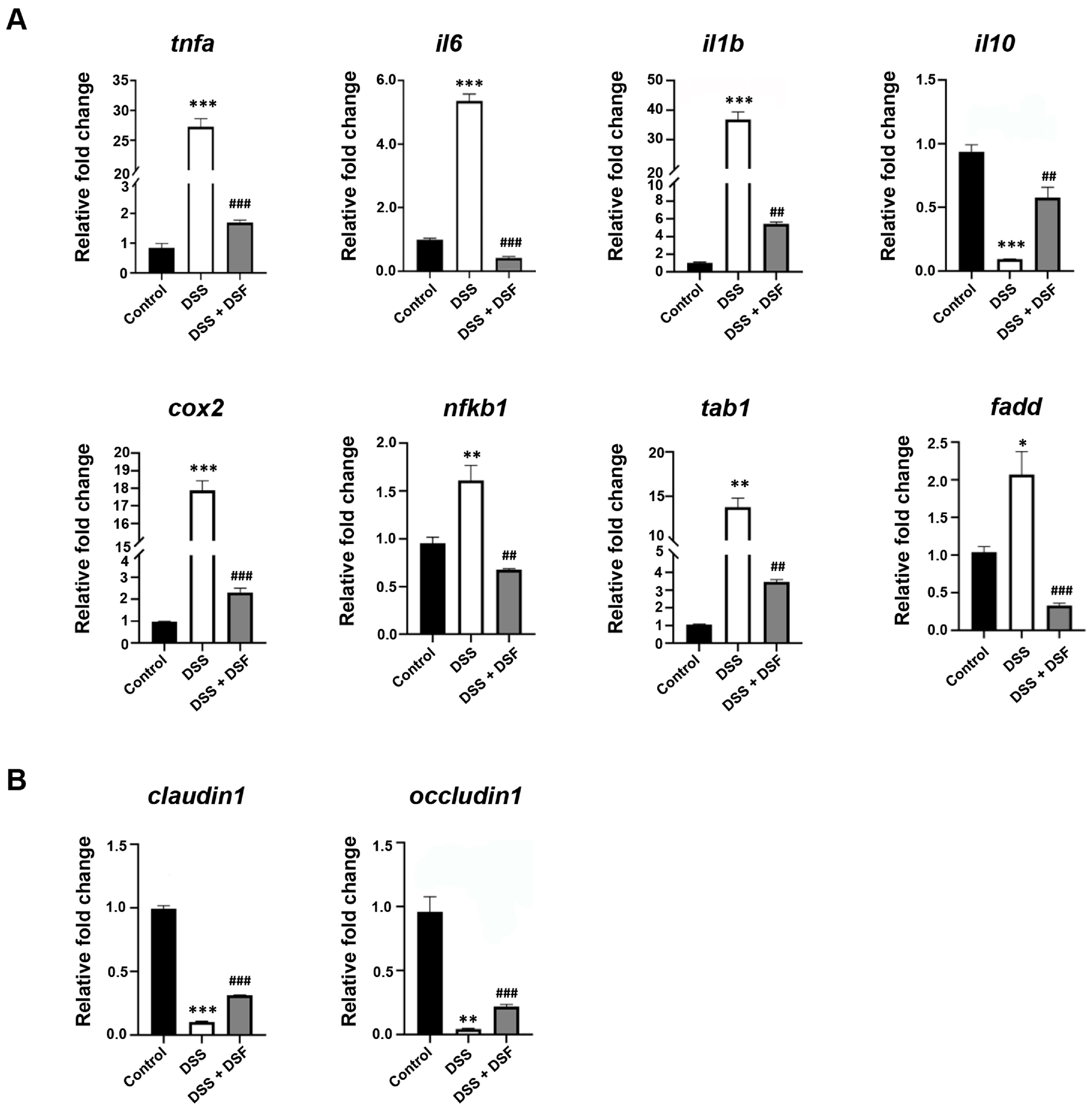
3.2. DSF Inhibits DSS-Induced Inflammation in Zebrafish
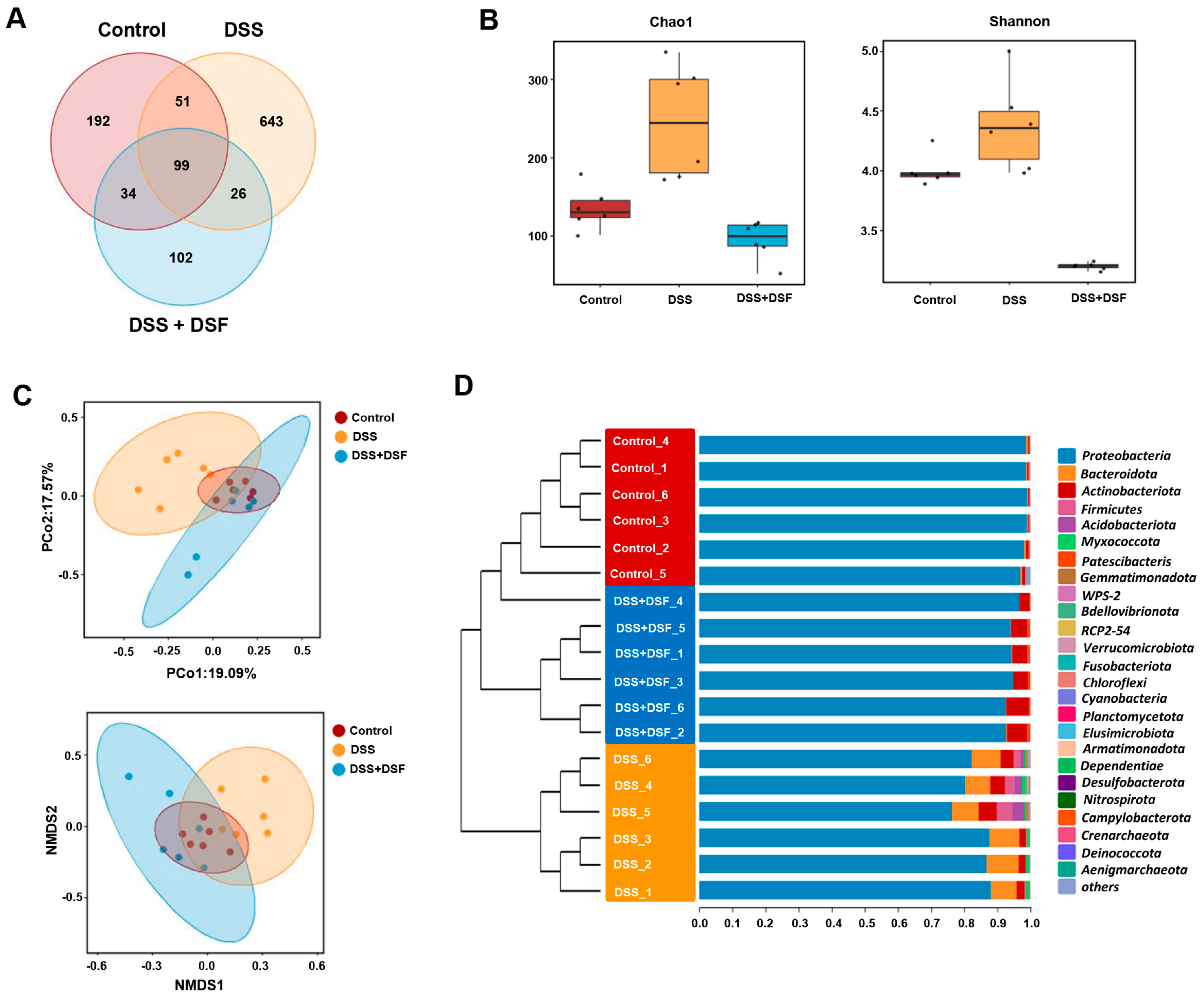
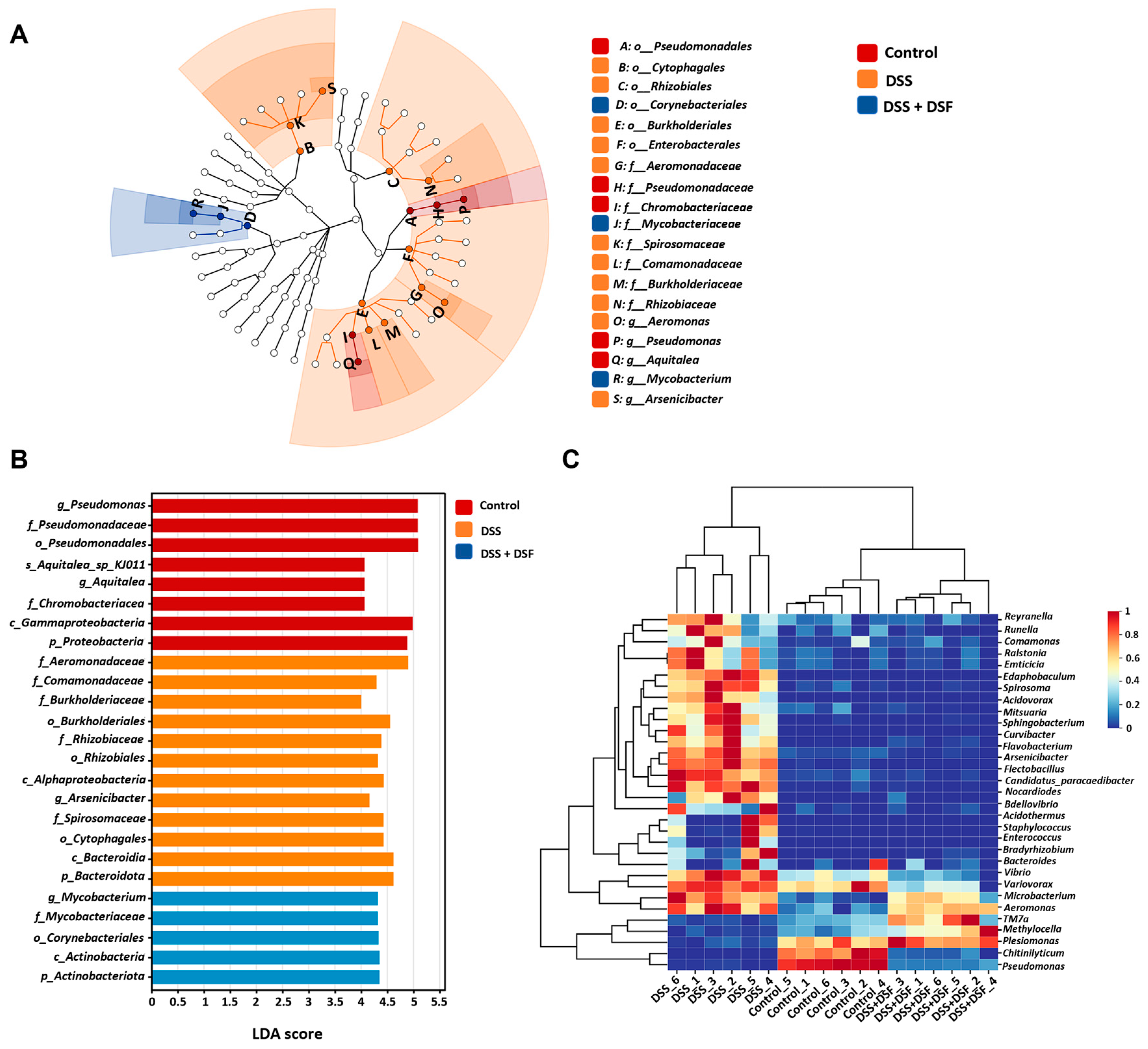
3.3. DSF Modulates the Intestinal Flora Structure in DSS-Induced Zebrafish Colitis
3.4. LEfSe Analysis of Intestinal Microbiota
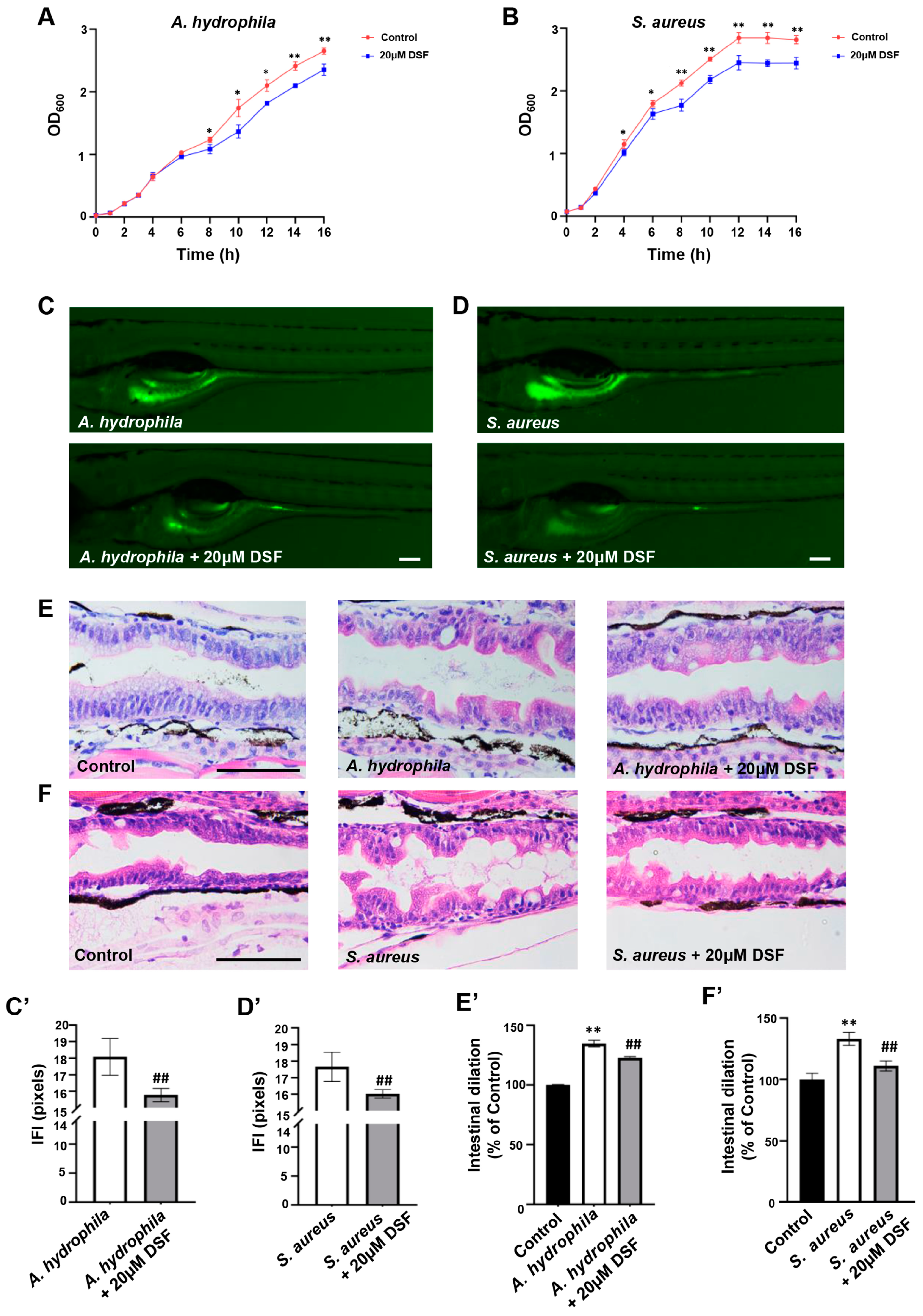
3.5. Inhibitory Effect of DSF on the Proliferation and Infection of A. hydrophila and S. aureus in Zebrafish
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, Q.; Cinzia, C.; Ciccacci, C. A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019, 2019, 7247238. [Google Scholar] [CrossRef] [PubMed]
- Stokkers, P.C.F.; Hommes, D.W. New cytokine therapeutics for inflammatory bowel disease. Cytokine 2004, 28, 167–173. [Google Scholar] [CrossRef]
- Wędrychowicz, A.; Zając, A.; Tomasik, P. Advances in nutritional therapy in inflammatory bowel diseases: Review. World J. Gastroenterol. 2016, 22, 1045–1066. [Google Scholar] [CrossRef]
- Cai, Z.; Wang, S.; Li, J. Treatment of Inflammatory Bowel Disease: A Comprehensive Review. Front. Med. 2021, 8, 765474. [Google Scholar] [CrossRef] [PubMed]
- Ordas, I.; Eckmann, L.; Talamini, M.; Baumgart, D.C.; Sandborn, W.J. Ulcerative colitis. Lancet 2012, 380, 1606–1619. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Li, Y.Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, P.; Vollmer-Conna, U.; Hadzi-Pavlovic, D.; Grimm, M.C. A Review of Inflammatory Bowel Disease: A Model of Microbial, Immune and Neuropsychological Integration. Public Health Rev. 2021, 42, 1603990. [Google Scholar] [CrossRef]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef]
- Iacob, S.; Iacob, D.G. Infectious Threats, the Intestinal Barrier, and Its Trojan Horse: Dysbiosis. Front. Microbiol. 2019, 10, 1676. [Google Scholar] [CrossRef]
- Stecher, B.; Hardt, W.D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011, 14, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Hou, K.; Valencak, T.G.; Luo, X.M.; Liu, J.; Wang, H. AI-2/LuxS Quorum Sensing System Promotes Biofilm Formation of Lactobacillus rhamnosus GG and Enhances the Resistance to Enterotoxigenic Escherichia coli in Germ-Free Zebrafish. Microbiol. Spectr. 2022, 10, e00610-22. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Gupta, J.; Kumar, S.; Kumar, A. Gut biofilm forming bacteria in inflammatory bowel disease. Microb. Pathog. 2017, 112, 5–14. [Google Scholar] [CrossRef]
- Coquant, G.; Grill, J.; Seksik, P. Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 2020, 11, 1827. [Google Scholar] [CrossRef] [PubMed]
- Aguanno, D.; Coquant, G.; Postal, B.G.; Osinski, C.; Wieckowski, M.; Stockholm, D.; Grill, J.; Carrière, V.; Seksik, P.; Thenet, S. The intestinal quorum sensing 3-oxo-C12:2 Acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissue Barriers 2020, 8, 1832877. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Yang, T.; Ai, Q.; Shi, Y.; Ji, Y.; Sun, Q.; Tong, B.; Chen, J.; Wang, Z. Autoinducer-2 promotes the colonization of Lactobacillus rhamnosus GG to improve the intestinal barrier function in a neonatal mouse model of antibiotic-induced intestinal dysbiosis. J. Transl. Med. 2024, 22, 177. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Sun, Q.; Fu, C.; She, X.; Liu, X.; He, Y.; Ai, Q.; Li, L.; Wang, Z. Exogenous Autoinducer-2 Rescues Intestinal Dysbiosis and Intestinal Inflammation in a Neonatal Mouse Necrotizing Enterocolitis Model. Front. Cell. Infect. Microbiol. 2021, 11, 694395. [Google Scholar] [CrossRef]
- He, Y.W.; Zhang, L.H. Quorum sensing and virulence regulation in Xanthomonas campestris. Fems Microbiol. Rev. 2008, 32, 842–857. [Google Scholar] [CrossRef] [PubMed]
- Bosire, E.M.; Eade, C.R.; Schiltz, C.J.; Reid, A.J.; Troutman, J.; Chappie, J.S.; Altier, C.; Monack, D. Diffusible Signal Factors Act through AraC-Type Transcriptional Regulators as Chemical Cues to Repress Virulence of Enteric Pathogens. Infect. Immun. 2020, 88, e00220–e00226. [Google Scholar] [CrossRef]
- YANG, D.L.; ZHANG, Y.Q.; HU, Y.L.; WENG, L.X.; ZENG, G.S.; WANG, L.H. Protective Effects of cis-2-Dodecenoic Acid in an Experimental Mouse Model of Vaginal Candidiasis. Biomed. Environ. Sci. 2018, 31, 816–828. [Google Scholar]
- Zhu, H.; Wang, Z.; Wang, W.; Lu, Y.; He, Y.W.; Tian, J. Bacterial Quorum-Sensing Signal DSF Inhibits LPS-Induced Inflammations by Suppressing Toll-like Receptor Signaling and Preventing Lysosome-Mediated Apoptosis in Zebrafish. Int. J. Mol. Sci. 2022, 23, 7110. [Google Scholar] [CrossRef] [PubMed]
- He, Y.W.; Wu, J.; Cha, J.S.; Zhang, L.H. Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol. 2010, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Lyu, Z.; Wang, Z.; Kou, Y.; Liu, C.; Li, S.; Hu, M.; Zhu, H.; Wang, W.; Zhang, C.; et al. Lipin 1 deficiency causes adult-onset myasthenia with motor neuron dysfunction in humans and neuromuscular junction defects in zebrafish. Theranostics 2021, 11, 2788–2805. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ai, F.; Ji, C.; Tu, P.; Gao, Y.; Wu, Y.; Yan, F.; Yu, T. A Rapid Screening Method of Candidate Probiotics for Inflammatory Bowel Diseases and the Anti-inflammatory Effect of the Selected Strain Bacillus smithii XY1. Front. Microbiol. 2021, 12, 760385. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.; Morrison, J.; Hsu, N.Y.; Labrias, P.R.; Nayar, S.; Chen, E.; Villaverde, N.; Facey, J.A.; Boschetti, G.; Giri, M.; et al. Zebrafish modeling of intestinal injury, bacterial exposures and medications defines epithelial in vivo responses relevant to human inflammatory bowel disease. Dis. Model. Mech. 2019, 12, dmm037432. [Google Scholar] [CrossRef] [PubMed]
- Oehlers, S.H.; Flores, M.V.; Hall, C.J.; Okuda, K.S.; Sison, J.O.; Crosier, K.E.; Crosier, P.S. Chemically Induced Intestinal Damage Models in Zebrafish Larvae. Zebrafish 2013, 10, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Minchin, P.R. An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 1987, 69, 89–107. [Google Scholar] [CrossRef]
- Kruskal, J.B. Nonmetric multidimensional scaling: A numerical method. Psychometrika 1964, 29, 115–129. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Weingart, G.; Tickle, T.L.; Huttenhower, C.; Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. Peerj 2015, 3, e1029. [Google Scholar] [CrossRef]
- Tian, J.; Shao, J.; Liu, C.; Hou, H.Y.; Chou, C.W.; Shboul, M.; Li, G.Q.; El-Khateeb, M.; Samarah, O.Q.; Kou, Y.; et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of Jagged-Notch signaling in fin and limb development. Cell. Mol. Life Sci. 2019, 76, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Murugan, R.; Subramaniyan, S.; Priya, S.; Ragavendran, C.; Arasu, M.V.; Al-Dhabi, N.A.; Choi, K.C.; Guru, A.; Arockiaraj, J. Bacterial clearance and anti-inflammatory effect of Withaferin A against human pathogen of Staphylococcus aureus in infected zebrafish. Aquat. Toxicol. 2023, 260, 106578. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Liu, C.; Dai, M.; Wang, Q.; Li, C.; Hung, W. Bifidobacterium lactis BL-99 modulates intestinal inflammation and functions in zebrafish models. PLoS ONE 2022, 17, e0262942. [Google Scholar] [CrossRef]
- Sidhu, M.; van der Poorten, D. The gut microbiome. Aust Fam Physician 2017, 46, 206–211. [Google Scholar]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef]
- Fala, A.K.; Alvarez-Ordonez, A.; Filloux, A.; Gahan, C.; Cotter, P.D. Quorum sensing in human gut and food microbiomes: Significance and potential for therapeutic targeting. Front. Microbiol. 2022, 13, 1002185. [Google Scholar] [CrossRef]
- Barbara, G.; Barbaro, M.R.; Fuschi, D.; Palombo, M.; Falangone, F.; Cremon, C.; Marasco, G.; Stanghellini, V. Inflammatory and Microbiota-Related Regulation of the Intestinal Epithelial Barrier. Front. Nutr. 2021, 8, 718356. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.C. Mucus and mucins in diseases of the intestinal and respiratory tracts. J. Intern. Med. 2019, 285, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Van Itallie, C.M. Physiology and function of the tight junction. Cold Spring Harbor Perspect. Biol. 2009, 1, a002584. [Google Scholar] [CrossRef] [PubMed]
- Gajendran, M.; Loganathan, P.; Jimenez, G.; Catinella, A.P.; Ng, N.; Umapathy, C.; Ziade, N.; Hashash, J.G. A comprehensive review and update on ulcerative colitis. Dis.-A-Mon. 2019, 65, 100851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yao, X.; Ma, M.; Ding, Y.; Zhang, H.; He, X.; Song, Z. Protective Effect of L-Theanine against DSS-Induced Colitis by Regulating the Lipid Metabolism and Reducing Inflammation via the NF-κB Signaling Pathway. J. Agric. Food. Chem. 2021, 69, 14192–14203. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liu, H.; Xu, L.; Yu, T.; Zhao, X.; Yao, S.; Zhao, Q.; Barnes, S.; Cohn, S.M.; Dann, S.M.; et al. GPR120 Inhibits Colitis Through Regulation of CD4+ T Cell Interleukin 10 Production. Gastroenterology 2022, 162, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.J.; Wang, Q.W.; Hu, Y.Y.; He, J.M.; Ge, Q.W.; Qi, Y.D.; Chen, L.Y.; Zhang, Y.; Fan, L.N.; Lin, Y.F.; et al. Lactobacillus johnsonii alleviates colitis by TLR1/2-STAT3 mediated CD206(+) macrophages(IL-10) activation. Gut Microbes 2022, 14, 2145843. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Sun, Y.; Shao, X.; Zhou, Y.; Yu, Y.; Kuai, X.; Zhou, C. Leaky Gut in IBD: Intestinal Barrier-Gut Microbiota Interaction. J. Microbiol. Biotechnol. 2022, 32, 825–834. [Google Scholar] [CrossRef]
- Chen, Y.; Cui, W.; Li, X.; Yang, H. Interaction Between Commensal Bacteria, Immune Response and the Intestinal Barrier in Inflammatory Bowel Disease. Front. Immunol. 2021, 12, 761981. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.T.; Alves, S.J.; Neves, S.; Falcao, D.; Costa, P.; Lago, P.; Pedroto, I.; Salgado, M. Positioning Aeromonas Infection in Inflammatory Bowel Disease: A Retrospective Analysis. Ge Port. J. Gastroenterol. 2023, 30, 20–28. [Google Scholar]
- Azimirad, M.; Krutova, M.; Balaii, H.; Kodori, M.; Shahrokh, S.; Azizi, O.; Yadegar, A.; Aghdaei, H.A.; Zali, M.R. Coexistence of Clostridioides difficile and Staphylococcus aureus in gut of Iranian outpatients with underlying inflammatory bowel disease. Anaerobe 2020, 61, 102113. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.; Kapila, R.; Kapila, S. A novel gut inflammatory rat model by laparotomic injection of peptidoglycan from Staphylococcus aureus. Arch. Microbiol. 2022, 204, 684. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Russell, S.L.; Finlay, B.B. Shifting the balance: Antibiotic effects on host–microbiota mutualism. Nat. Rev. Microbiol. 2011, 9, 233–243. [Google Scholar] [CrossRef]
- Sun, Z.; Grimm, V.; Riedel, C.U. AI-2 to the rescue against antibiotic-induced intestinal dysbiosis? Trends Microbiol. 2015, 23, 327–328. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, R.; Yang, B.; Zhu, H.; Sun, Y.; Wu, H.; Wang, Z.; Lu, Y.; He, Y.-W.; Tian, J. Quorum-Sensing Signal DSF Inhibits the Proliferation of Intestinal Pathogenic Bacteria and Alleviates Inflammatory Response to Suppress DSS-Induced Colitis in Zebrafish. Nutrients 2024, 16, 1562. https://doi.org/10.3390/nu16111562
Yi R, Yang B, Zhu H, Sun Y, Wu H, Wang Z, Lu Y, He Y-W, Tian J. Quorum-Sensing Signal DSF Inhibits the Proliferation of Intestinal Pathogenic Bacteria and Alleviates Inflammatory Response to Suppress DSS-Induced Colitis in Zebrafish. Nutrients. 2024; 16(11):1562. https://doi.org/10.3390/nu16111562
Chicago/Turabian StyleYi, Ruiya, Bo Yang, Hongjie Zhu, Yu Sun, Hailan Wu, Zhihao Wang, Yongbo Lu, Ya-Wen He, and Jing Tian. 2024. "Quorum-Sensing Signal DSF Inhibits the Proliferation of Intestinal Pathogenic Bacteria and Alleviates Inflammatory Response to Suppress DSS-Induced Colitis in Zebrafish" Nutrients 16, no. 11: 1562. https://doi.org/10.3390/nu16111562
APA StyleYi, R., Yang, B., Zhu, H., Sun, Y., Wu, H., Wang, Z., Lu, Y., He, Y.-W., & Tian, J. (2024). Quorum-Sensing Signal DSF Inhibits the Proliferation of Intestinal Pathogenic Bacteria and Alleviates Inflammatory Response to Suppress DSS-Induced Colitis in Zebrafish. Nutrients, 16(11), 1562. https://doi.org/10.3390/nu16111562





