Modulation of Gut Microbial Community and Metabolism by Bacillus licheniformis HD173 Promotes the Growth of Nursery Piglets Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Dietary Treatments
2.2. Sample Collections
2.3. Enzyme-Linked Immunosorbent Assay
2.4. DNA Extraction, 16S rRNA Gene Amplicon and Sequencing
2.5. Quantitative Analysis of Short-Chain Fatty Acids (SCFAs)
2.6. Untargeted Metabolomics Analysis
2.7. Statistical Analysis
3. Results
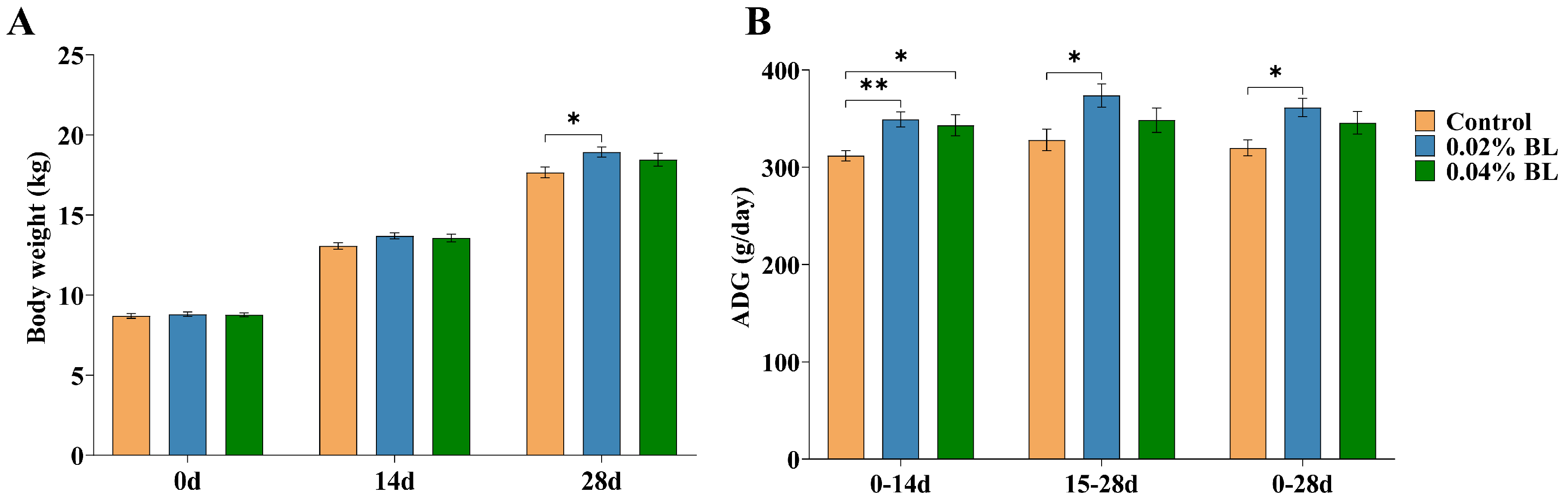
3.1. The Effects of B. licheniformis HD173 on the Growth Performance of Nursery Piglets
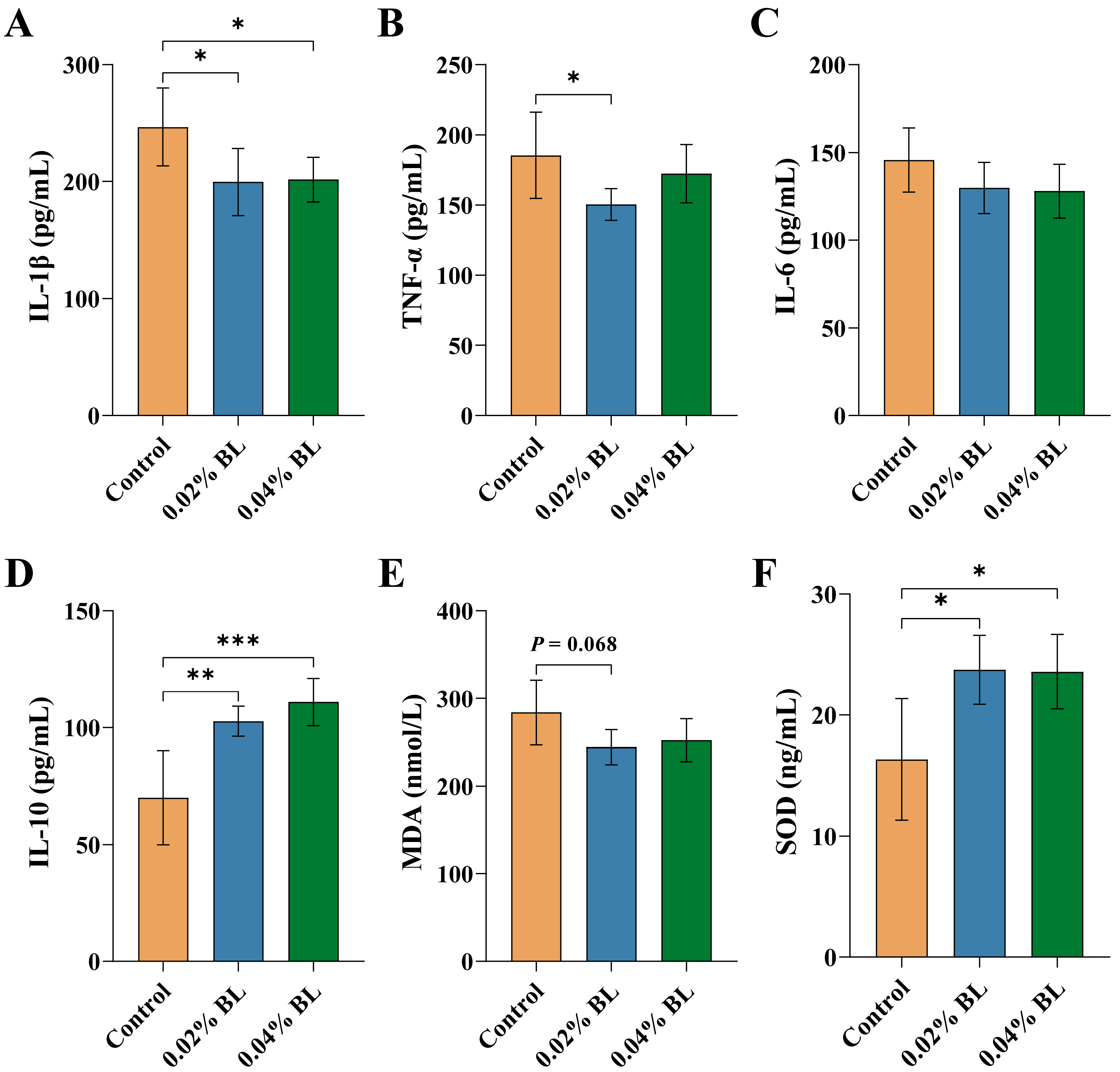
3.2. The Effects of B. licheniformis HD173 on Level of Serum Inflammatory Cytokines and Oxidative Stress Indicators
3.3. The Effect of B. licheniformis HD173 on the Gut Microbiota of Nursery Piglets
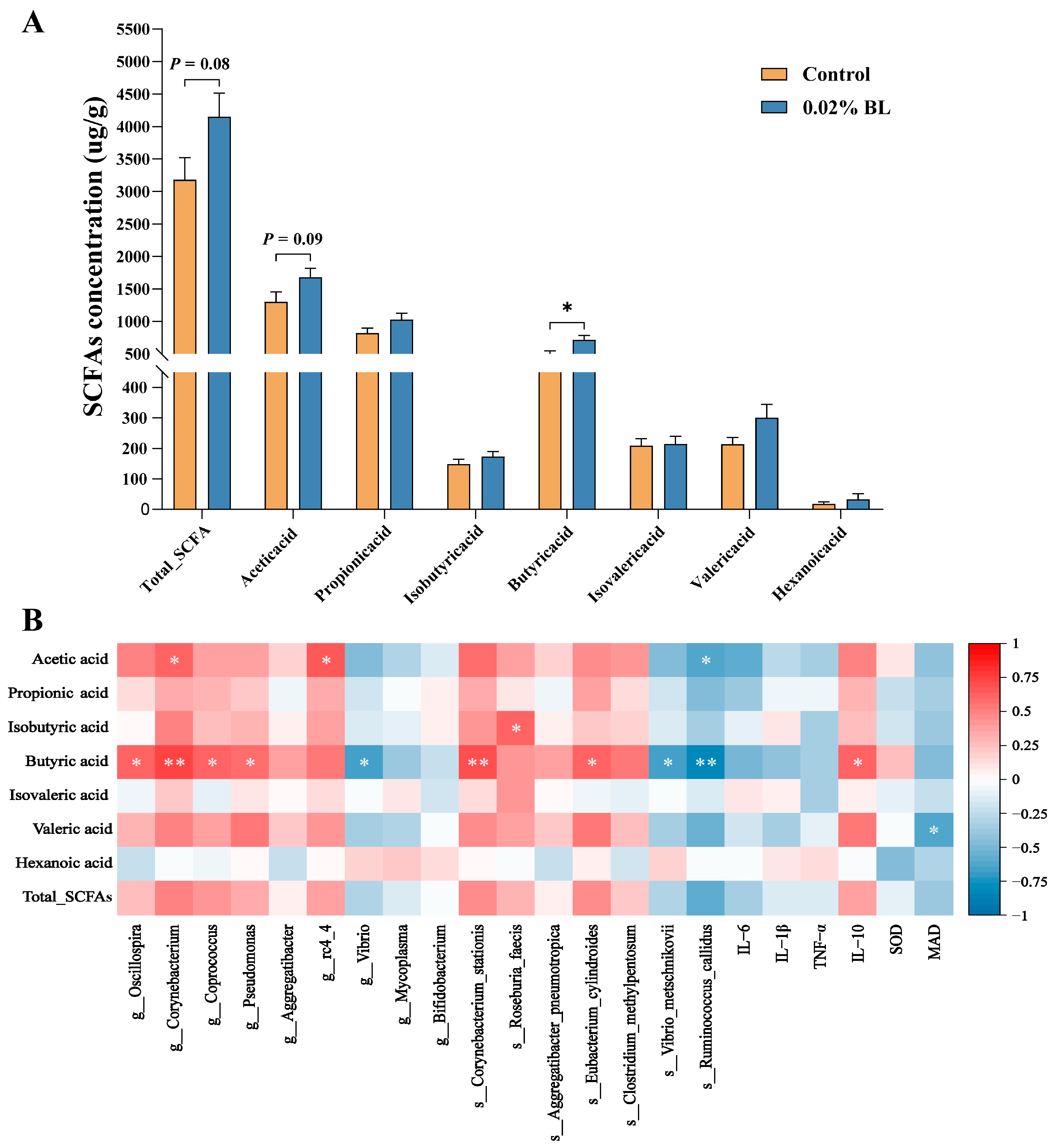
3.4. The Effects of B. licheniformis HD173 on the Production of SCFAs
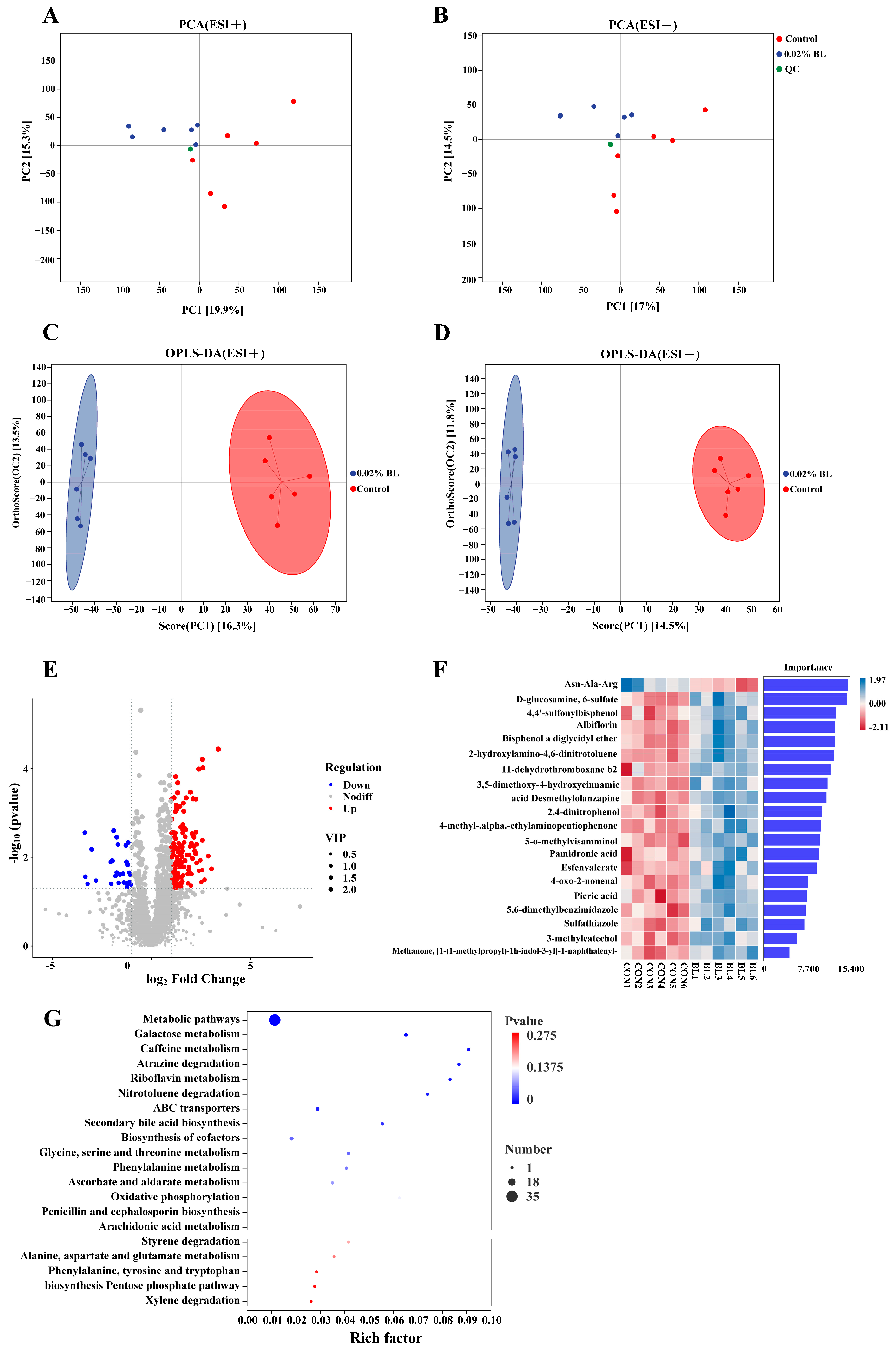
3.5. The Effects of B. licheniformis HD173 on the Gut Metabolites of Nursery Piglets
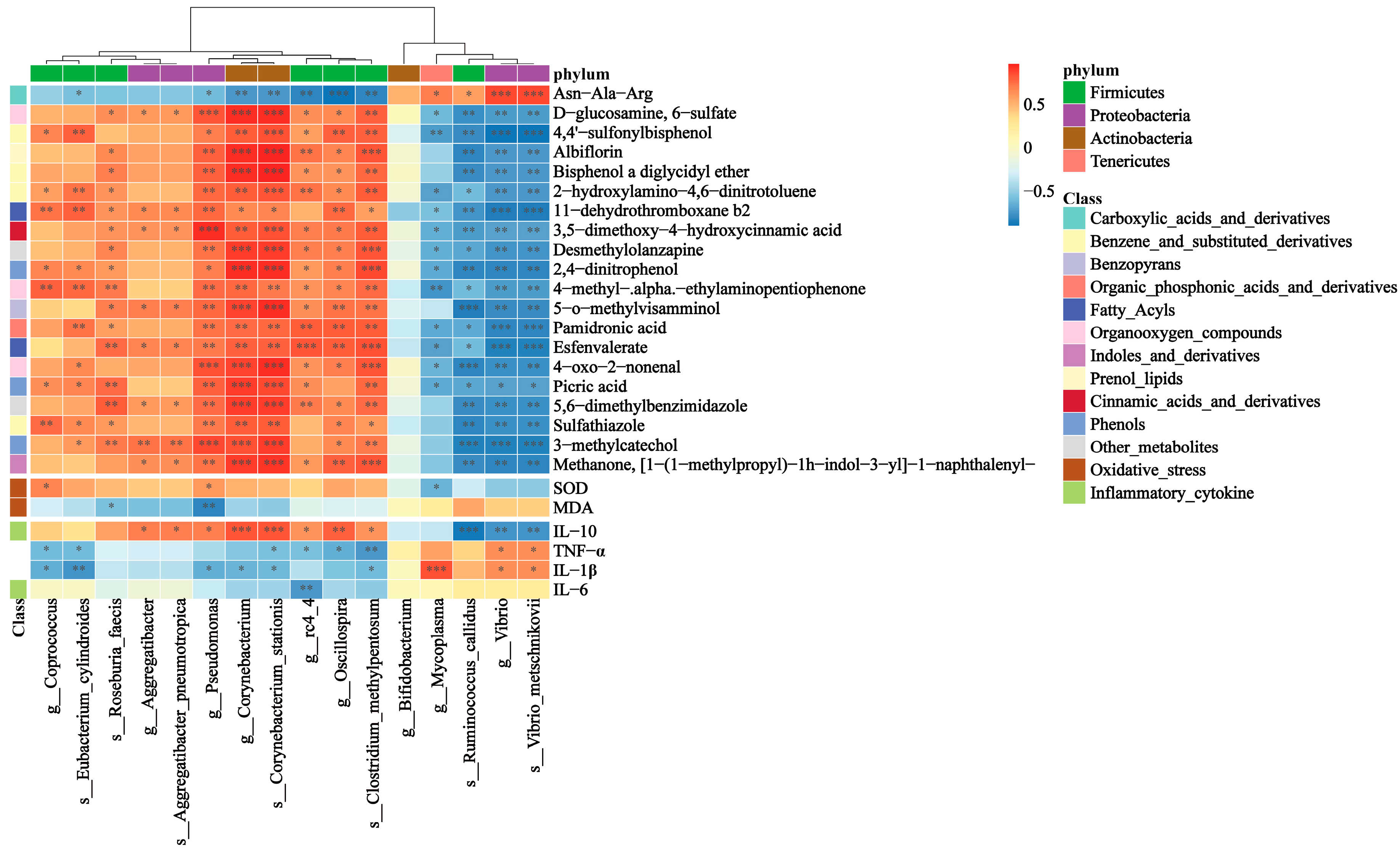
3.6. Correlations between Gut Microbiota and Inflammatory Cytokines, Oxidative Stress Indicators, Gut Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Ahmad, W.; Din, A.U.; Khan, T.M.; Rehman, M.U.; Hassan, A.; Aziz, T.; Alharbi, M.; Wu, J. Lacticaseibacillus paracasei BNCC345679 revolutionizes DSS-induced colitis and modulates gut microbiota. Front. Microbiol. 2024, 15, 1343891. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Naveed, M.; Makhdoom, S.I.; Ali, U.; Mughal, M.S.; Sarwar, A.; Khan, A.A.; Zhennai, Y.; Sameeh, M.Y.; Dablool, A.S.; et al. Genome Investigation and Functional Annotation of Lactiplantibacillus plantarum YW11 Revealing Streptin and Ruminococcin-A as Potent Nutritive Bacteriocins against Gut Symbiotic Pathogens. Molecules 2023, 28, 491. [Google Scholar] [CrossRef] [PubMed]
- Kuziel, G.A.; Rakoff-Nahoum, S. The gut microbiome. Curr. Biol. 2022, 32, R257–R264. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, J.; Lin, Z.; Liu, C.; Zhang, Y.; Zhang, S.; Zhou, M.; Zhao, J.; Liu, H.; Ma, X. Clostridium butyricum alleviates weaned stress of piglets by improving intestinal immune function and gut microbiota. Food Chem. 2023, 405, 135014. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Bola, S.H.; Gang, W.; Silvia, G.-P.; Alexander, O.M.; Ricardo, N.R.; Andrés, R.M.-R.; Kathleen, S.; Min, W.; Christophe, B.; Diane, M. The gut microbiota promotes distal tissue regeneration via RORγ+ regulatory T cell emissaries. Immunity 2023, 56, 829–846.e828. [Google Scholar] [CrossRef] [PubMed]
- Violi, F.; Cammisotto, V.; Bartimoccia, S.; Pignatelli, P.; Carnevale, R.; Nocella, C. Gut-derived low-grade endotoxaemia, atherothrombosis and cardiovascular disease. Nat. Rev. Cardiol. 2023, 20, 24–37. [Google Scholar] [CrossRef]
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut Microbiota Dysbiosis in Postweaning Piglets: Understanding the Keys to Health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [CrossRef]
- Fletcher, J.R.; Pike, C.M.; Parsons, R.J.; Rivera, A.J.; Foley, M.H.; McLaren, M.R.; Montgomery, S.A.; Theriot, C.M. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat. Commun. 2021, 12, 462. [Google Scholar] [CrossRef]
- Illiano, P.; Brambilla, R.; Parolini, C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020, 287, 833–855. [Google Scholar] [CrossRef]
- Hossain, M.I.; Sadekuzzaman, M.; Ha, S.D. Probiotics as potential alternative biocontrol agents in the agriculture and food industries: A review. Food Res. Int. 2017, 100, 63–73. [Google Scholar] [CrossRef]
- Hafiz Arbab, S.; Heping, Z. Trends in Probiotic(s)-Fermented milks and their in vivo functionality: A review. Trends Food Sci. Technol. 2021, 110, 55–65. [Google Scholar] [CrossRef]
- Ma, T.; Shen, X.; Shi, X.; Sakandar, H.A.; Quan, K.; Li, Y.; Jin, H.; Kwok, L.-Y.; Zhang, H.; Sun, Z. Targeting gut microbiota and metabolism as the major probiotic mechanism—An evidence-based review. Trends Food Sci. Technol. 2023, 138, 178–198. [Google Scholar] [CrossRef]
- Sun, W.; Chen, W.; Meng, K.; Cai, L.; Li, G.; Li, X.; Jiang, X. Dietary Supplementation with Probiotic Bacillus licheniformis S6 Improves Intestinal Integrity via Modulating Intestinal Barrier Function and Microbial Diversity in Weaned Piglets. Biology 2023, 12, 238. [Google Scholar] [CrossRef]
- Muras, A.; Romero, M.; Mayer, C.; Otero, A. Biotechnological applications of Bacillus licheniformis. Crit. Rev. Biotechnol. 2021, 41, 609–627. [Google Scholar] [CrossRef]
- Yu, X.; Dai, Z.; Cao, G.; Cui, Z.; Zhang, R.; Xu, Y.; Wu, Y.; Yang, C. Protective effects of Bacillus licheniformis on growth performance, gut barrier functions, immunity and serum metabolome in lipopolysaccharide-challenged weaned piglets. Front. Immunol. 2023, 14, 1140564. [Google Scholar] [CrossRef]
- Yu, X.; Cui, Z.; Qin, S.; Zhang, R.; Wu, Y.; Liu, J.; Yang, C. Effects of Bacillus licheniformis on Growth Performance, Diarrhea Incidence, Antioxidant Capacity, Immune Function, and Fecal Microflora in Weaned Piglets. Animals 2022, 12, 1609. [Google Scholar] [CrossRef]
- Lan, R.; Kim, I.H. Effects of Bacillus licheniformis and Bacillus subtilis complex on growth performance and faecal noxious gas emissions in growing-finishing pigs. J. Sci. Food Agric. 2019, 99, 1554–1560. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Li, Y.L.; Yu, D.Y.; Rajput, I.R.; Li, W.F. Influence of dietary inclusion of Bacillus licheniformis on laying performance, egg quality, antioxidant enzyme activities, and intestinal barrier function of laying hens. Poult. Sci. 2013, 92, 2389–2395. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Ivanova, I.V.; Popov, I.; Weeks, R.; Chikindas, M.L. Bacillus spore-forming probiotics: Benefits with concerns? Crit. Rev. Microbiol. 2022, 48, 513–530. [Google Scholar] [CrossRef] [PubMed]
- Casula, G.; Cutting, S.M. Bacillus probiotics: Spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Mingmongkolchai, S.; Panbangred, W. Bacillus probiotics: An alternative to antibiotics for livestock production. J. Appl. Microbiol. 2018, 124, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Sorokulova, I.B. A comparative study of the biological properties of Biosporin and other commercial Bacillus-based preparations. Mikrobiol. Z. 1997, 59, 43–49. [Google Scholar] [PubMed]
- McFarlin, B.K.; Henning, A.L.; Bowman, E.M.; Gary, M.A.; Carbajal, K.M. Oral spore-based probiotic supplementation was associated with reduced incidence of post-prandial dietary endotoxin, triglycerides, and disease risk biomarkers. World J. Gastrointest. Pathophysiol. 2017, 8, 117–126. [Google Scholar] [CrossRef] [PubMed]
- de Boer, A.S.; Priest, F.; Diderichsen, B. On the industrial use of Bacillus licheniformis: A review. Appl. Microbiol. Biotechnol. 1994, 40, 595–598. [Google Scholar] [CrossRef]
- Noohi, N.; Papizadeh, M.; Rohani, M.; Talebi, M.; Pourshafie, M.R. Screening for probiotic characters in lactobacilli isolated from chickens revealed the intra-species diversity of Lactobacillus brevis. Anim. Nutr. 2021, 7, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Y.; Shen, Y.; Li, Q.; Lan, J.; Wu, Y.; Zhang, R.; Cao, G.; Yang, C. Effects of Bacillus subtilis and Bacillus licheniformis on growth performance, immunity, short chain fatty acid production, antioxidant capacity, and cecal microflora in broilers. Poult. Sci. 2021, 100, 101358. [Google Scholar] [CrossRef]
- Wang, X.; Tian, Z.; Azad, M.A.K.; Zhang, W.; Blachier, F.; Wang, Z.; Kong, X. Dietary supplementation with Bacillus mixture modifies the intestinal ecosystem of weaned piglets in an overall beneficial way. J. Appl. Microbiol. 2021, 130, 233–246. [Google Scholar] [CrossRef]
- Lin, K.H.; Yu, Y.H. Evaluation of Bacillus licheniformis-Fermented Feed Additive as an Antibiotic Substitute: Effect on the Growth Performance, Diarrhea Incidence, and Cecal Microbiota in Weaning Piglets. Animals 2020, 10, 1649. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, Y.H.; Zhang, W.; Wang, M.L.; Fan, W.Y.; Song, D.; Yang, G.Y.; Jensen, B.B.; Wang, J.F. Oral administration of a select mixture of Bacillus probiotics generates Tr1 cells in weaned F4ab/acR-pigs challenged with an F4+ ETEC/VTEC/EPEC strain. Vet. Res. 2015, 46, 95. [Google Scholar] [CrossRef] [PubMed]
- Chuanqi, C.; Jinchi, J.; Leilei, Y.; Yiwen, L.; Songli, Z.; Wei, Z.; Qun, W.; Jianxin, Z.; Qixiao, Z.; Fengwei, T.; et al. Bifidobacterium longum CCFM1077 Attenuates Hyperlipidemia by Modulating the Gut Microbiota Composition and Fecal Metabolites: A Randomized, Double-Blind, Placebo-Controlled Clinical Trial. Engineering 2023, 28, 193–205. [Google Scholar] [CrossRef]
- Wang, G.; Wang, X.; Ma, Y.; Cai, S.; Yang, L.; Fan, Y.; Zeng, X.; Qiao, S. Lactobacillus reuteri improves the development and maturation of fecal microbiota in piglets through mother-to-infant microbe and metabolite vertical transmission. Microbiome 2022, 10, 211. [Google Scholar] [CrossRef]
- Zhou, J.; Luo, J.; Yang, S.; Xiao, Q.; Wang, X.; Zhou, Z.; Xiao, Y.; Shi, D. Different Responses of Microbiota across Intestinal Tract to Enterococcus faecium HDRsEf1 and Their Correlation with Inflammation in Weaned Piglets. Microorganisms 2021, 9, 1767. [Google Scholar] [CrossRef]
- Kim, K.; He, Y.; Xiong, X.; Ehrlich, A.; Li, X.; Raybould, H.; Atwill, E.R.; Maga, E.A.; Jørgensen, J.; Liu, Y. Dietary supplementation of Bacillus subtilis influenced intestinal health of weaned pigs experimentally infected with a pathogenic E. coli. J. Anim. Sci. Biotechnol. 2019, 10, 52. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, K.; Lan, R.; Glenn Morris, J.; Xiao, Y.; Ye, J.; Zhang, L.; Luo, L.; Gao, H.; Bai, X.; et al. Vibrio metschnikovii as an emergent pathogen: Analyses of phylogeny and O-antigen and identification of possible virulence characteristics. Emerg. Microbes Infect. 2023, 12, 2252522. [Google Scholar] [CrossRef]
- Gómez Rufo, D.; García Sánchez, E.; García Sánchez, J.E.; García Moro, M. [Clinical implications of the genus Mycoplasma]. Rev. Esp. Quimioter. 2021, 34, 169–184. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Tom, K.; Uri, G. Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Gophna, U.; Konikoff, T.; Nielsen, H.B. Oscillospira and related bacteria—From metagenomic species to metabolic features. Environ. Microbiol. 2017, 19, 835–841. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Guilloteau, P.; Martin, L.; Eeckhaut, V.; Ducatelle, R.; Zabielski, R.; Van Immerseel, F. From the gut to the peripheral tissues: The multiple effects of butyrate. Nutr. Res. Rev. 2010, 23, 366–384. [Google Scholar] [CrossRef]
- Jacobi, S.K.; Odle, J. Nutritional factors influencing intestinal health of the neonate. Adv. Nutr. 2012, 3, 687–696. [Google Scholar] [CrossRef]
- de Clercq, N.C.; Groen, A.K.; Romijn, J.A.; Nieuwdorp, M. Gut Microbiota in Obesity and Undernutrition. Adv. Nutr. 2016, 7, 1080–1089. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Su, L.; Tan, J.; Ding, T.; Yue, Y. Albiflorin alleviates DSS-induced ulcerative colitis in mice by reducing inflammation and oxidative stress. Iran. J. Basic. Med. Sci. 2023, 26, 48–56. [Google Scholar] [CrossRef]
- Yu, H.; Wang, Y.; He, Z.; Chen, R.; Dai, Y.; Tang, Y.; Chen, Y. Albiflorin ameliorates mesangial proliferative glomerulonephritis by PI3K/AKT/NF-κB pathway. Hum. Exp. Toxicol. 2023, 42, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Feng, L.; Yao, L. Albiflorin alleviates sepsis-induced acute liver injury through mTOR/p70S6K pathway. Curr. Mol. Med. 2024, 24, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Fitton, A.; McTavish, D. Pamidronate. A review of its pharmacological properties and therapeutic efficacy in resorptive bone disease. Drugs 1991, 41, 289–318. [Google Scholar] [CrossRef] [PubMed]
- Norman, S.J.; Reeves, D.J.; Saum, L.M. Use of Pamidronate for Hypercalcemia of Malignancy in Renal Dysfunction. J. Pharm. Pract. 2021, 34, 553–557. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Tian, C.; Feng, S.; Cheng, W.; Tao, S.; Li, C.; Xiao, Y.; Wei, H. Modulation of Gut Microbial Community and Metabolism by Bacillus licheniformis HD173 Promotes the Growth of Nursery Piglets Model. Nutrients 2024, 16, 1497. https://doi.org/10.3390/nu16101497
Li J, Tian C, Feng S, Cheng W, Tao S, Li C, Xiao Y, Wei H. Modulation of Gut Microbial Community and Metabolism by Bacillus licheniformis HD173 Promotes the Growth of Nursery Piglets Model. Nutrients. 2024; 16(10):1497. https://doi.org/10.3390/nu16101497
Chicago/Turabian StyleLi, Jiaxuan, Cheng Tian, Shuaifei Feng, Wei Cheng, Shiyu Tao, Changchun Li, Yuncai Xiao, and Hong Wei. 2024. "Modulation of Gut Microbial Community and Metabolism by Bacillus licheniformis HD173 Promotes the Growth of Nursery Piglets Model" Nutrients 16, no. 10: 1497. https://doi.org/10.3390/nu16101497
APA StyleLi, J., Tian, C., Feng, S., Cheng, W., Tao, S., Li, C., Xiao, Y., & Wei, H. (2024). Modulation of Gut Microbial Community and Metabolism by Bacillus licheniformis HD173 Promotes the Growth of Nursery Piglets Model. Nutrients, 16(10), 1497. https://doi.org/10.3390/nu16101497







