In Vitro Circadian Clock Gene Expression Assessments in Mesenchymal Stem Cells from Human Infants: A Pilot Study
Abstract
:1. Introduction
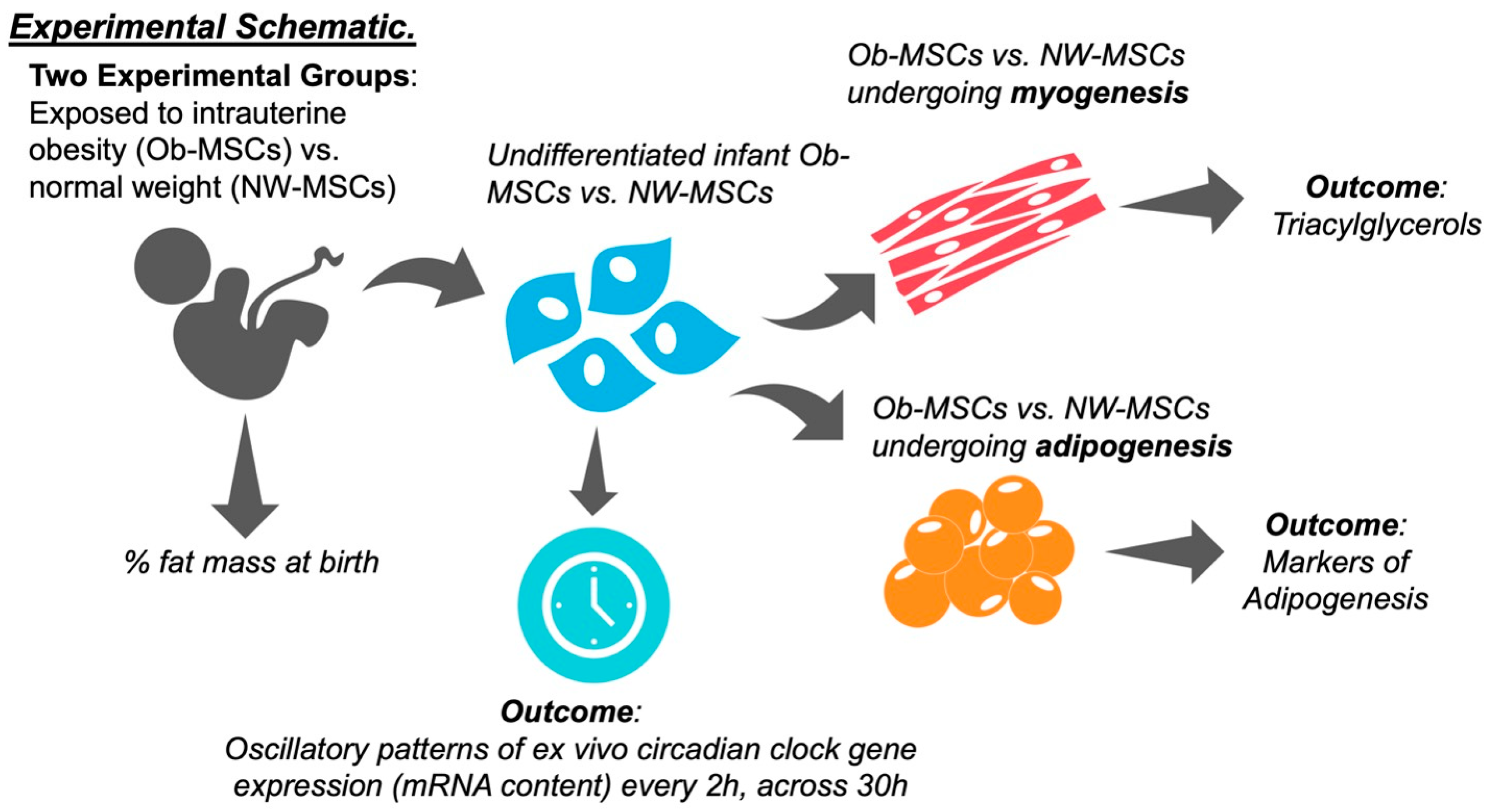
2. Materials and Methods
2.1. Mesenchymal Stem Cells
2.2. Participant Measures
2.3. Mesenchymal Stem Cell Culture
2.4. Rhythmic Gene Expression Patterns in Mesenchymal Stem Cells
2.5. RNA Isolation and qPCR
2.6. Mesenchymal Stem Cell Differentiation
2.7. Triacylglycerol Content
2.8. Protein Content
2.9. Statistical Analysis
3. Results
3.1. Participant Characteristics
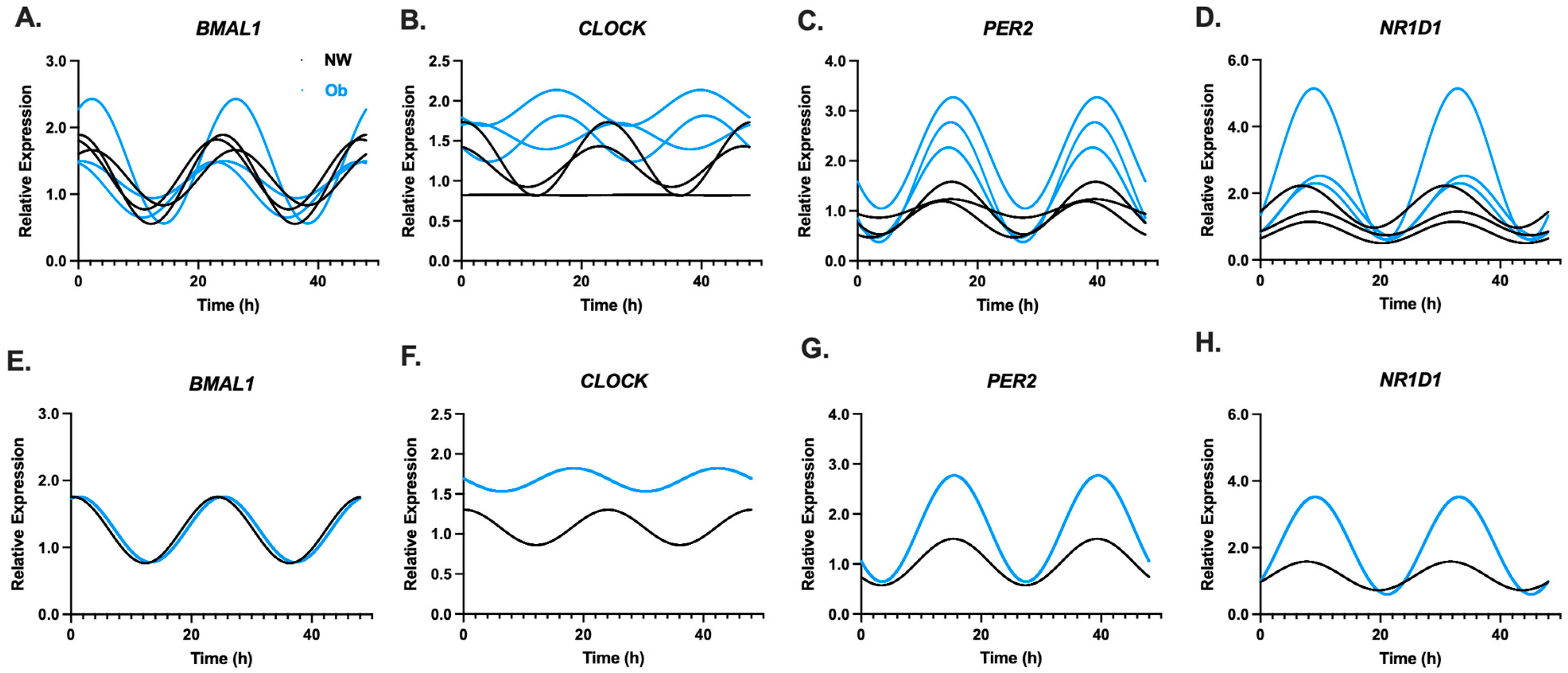
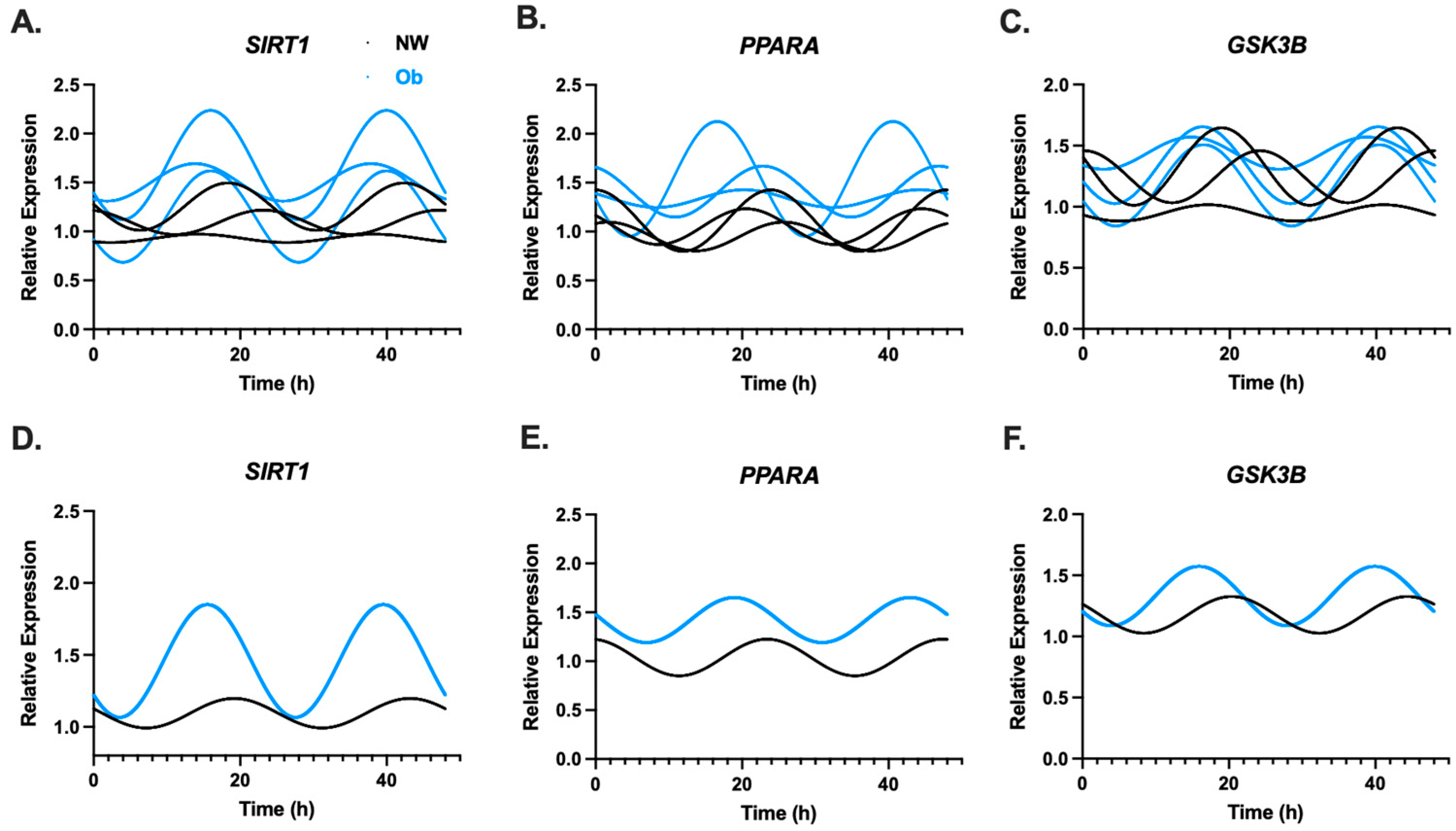
3.2. Infant MSCs Have Detectible Rhythmic Expression Patterns in Components of the Circadian Clock Network
3.3. PER2 Oscillatory Amplitude Is Higher in Ob-MSCs as Compared to NW-MSCs
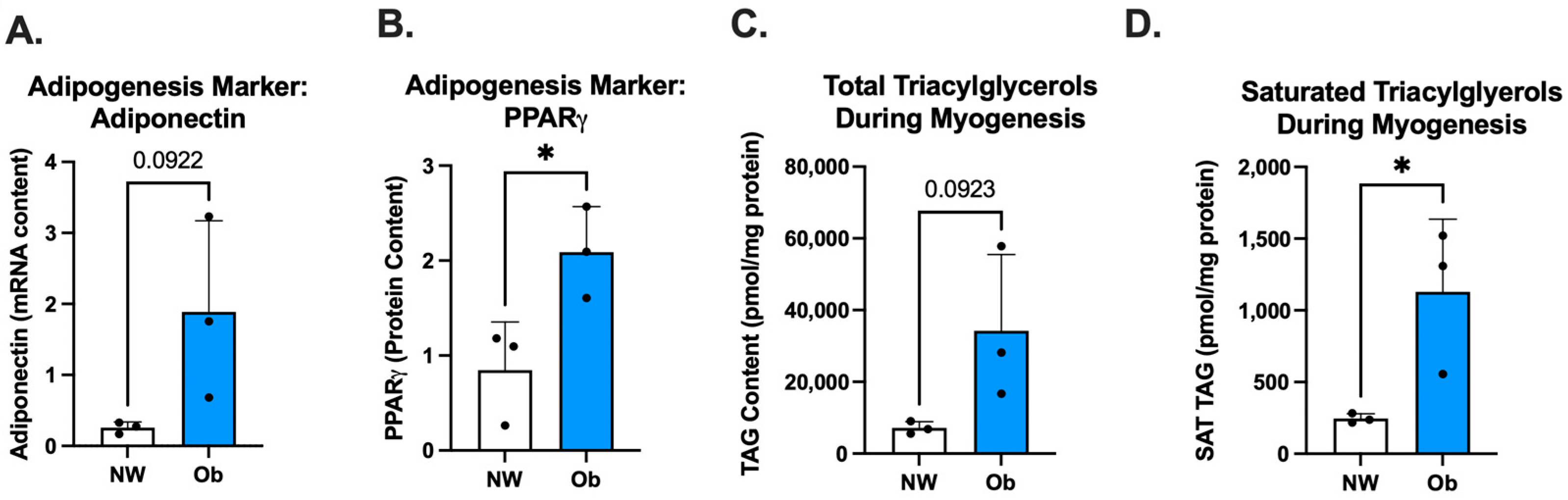
3.4. Ob-MSCs Have Greater Markers of Adipogenesis and Higher TAGs during Myogenesis
3.5. PER2 Amplitude May Be Linked to MSC Phenotype as Well as Maternal and Infant Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boney, C.M.; Verma, A.; Tucker, R.; Vohr, B.R. Metabolic syndrome in childhood: Association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 2005, 115, e290–e296. [Google Scholar] [CrossRef] [PubMed]
- Ismail-Beigi, F.; Catalano, P.M.; Hanson, R.W. Metabolic programming: Fetal origins of obesity and metabolic syndrome in the adult. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E439–E440. [Google Scholar] [CrossRef] [PubMed]
- Oken, E.; Taveras, E.M.; Kleinman, K.P.; Rich-Edwards, J.W.; Gillman, M.W. Gestational weight gain and child adiposity at age 3 years. Am. J. Obstet. Gynecol. 2007, 196, 322.e1–e8. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.H.T.; Ma, R.C.W.; Yuen, L.Y.; Ozaki, R.; Li, A.M.; Hou, Y.; Chan, M.H.M.; Ho, C.S.; Yang, X.; Chan, J.C.N.; et al. The impact of maternal gestational weight gain on cardiometabolic risk factors in children. Diabetologia 2018, 61, 2539–2548. [Google Scholar] [CrossRef] [PubMed]
- Stotland, N.E.; Cheng, Y.W.; Hopkins, L.M.; Caughey, A.B. Gestational weight gain and adverse neonatal outcome among term infants. Obstet. Gynecol. 2006, 108, 635–643. [Google Scholar] [CrossRef]
- Starling, A.P.; Brinton, J.T.; Glueck, D.H.; Shapiro, A.L.; Harrod, C.S.; Lynch, A.M.; Siega-Riz, A.M.; Dabelea, D. Associations of maternal BMI and gestational weight gain with neonatal adiposity in the Healthy Start study. Am. J. Clin. Nutr. 2015, 101, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Hillier, T.A.; Pedula, K.L.; Vesco, K.K.; Oshiro, C.E.; Ogasawara, K.K. Impact of Maternal Glucose and Gestational Weight Gain on Child Obesity over the First Decade of Life in Normal Birth Weight Infants. Matern. Child. Health J. 2016, 20, 1559–1568. [Google Scholar] [CrossRef]
- Whitaker, R.C. Predicting preschooler obesity at birth: The role of maternal obesity in early pregnancy. Pediatrics 2004, 114, e29–e36. [Google Scholar] [CrossRef]
- Catalano, P.M.; Farrell, K.; Thomas, A.; Huston-Presley, L.; Mencin, P.; de Mouzon, S.H.; Amini, S.B. Perinatal risk factors for childhood obesity and metabolic dysregulation. Am. J. Clin. Nutr. 2009, 90, 1303–1313. [Google Scholar] [CrossRef]
- Marino, L.; Castaldi, M.A.; Rosamilio, R.; Ragni, E.; Vitolo, R.; Fulgione, C.; Castaldi, S.G.; Serio, B.; Bianco, R.; Guida, M.; et al. Mesenchymal Stem Cells from the Wharton’s Jelly of the Human Umbilical Cord: Biological Properties and Therapeutic Potential. Int. J. Stem Cells 2019, 12, 218–226. [Google Scholar] [CrossRef]
- Boyle, K.E.; Patinkin, Z.W.; Shapiro, A.L.B.; Bader, C.; Vanderlinden, L.; Kechris, K.; Janssen, R.C.; Ford, R.J.; Smith, B.K.; Steinberg, G.R.; et al. Maternal obesity alters fatty acid oxidation, AMPK activity, and associated DNA methylation in mesenchymal stem cells from human infants. Mol. Metab. 2017, 6, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.E.; Patinkin, Z.W.; Shapiro, A.L.; Baker, P.R., 2nd; Dabelea, D.; Friedman, J.E. Mesenchymal Stem Cells From Infants Born to Obese Mothers Exhibit Greater Potential for Adipogenesis: The Healthy Start BabyBUMP Project. Diabetes 2016, 65, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Erickson, M.L.; Patinkin, Z.W.; Duensing, A.M.; Dabelea, D.; Redman, L.M.; Boyle, K.E. Maternal metabolic health drives mesenchymal stem cell metabolism and infant fat mass at birth. JCI Insight 2021, 6, e146606. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Yan, X.; Tong, J.F.; Zhao, J.; Zhu, M.J. Maternal obesity, inflammation, and fetal skeletal muscle development. Biol. Reprod. 2010, 82, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, C.; Wu, G.; Smith, S.B.; Dunlap, K.A.; Satterfield, M.C. Maternal Nutrient Restriction and Skeletal Muscle Development: Consequences for Postnatal Health. Adv. Exp. Med. Biol. 2020, 1265, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Alfaradhi, M.Z.; Kusinski, L.C.; Fernandez-Twinn, D.S.; Pantaleão, L.C.; Carr, S.K.; Ferland-McCollough, D.; Yeo, G.S.; Bushell, M.; Ozanne, S.E. Maternal Obesity in Pregnancy Developmentally Programs Adipose Tissue Inflammation in Young, Lean Male Mice Offspring. Endocrinology 2016, 157, 4246–4256. [Google Scholar] [CrossRef] [PubMed]
- Borengasser, S.J.; Kang, P.; Faske, J.; Gomez-Acevedo, H.; Blackburn, M.L.; Badger, T.M.; Shankar, K. High fat diet and in utero exposure to maternal obesity disrupts circadian rhythm and leads to metabolic programming of liver in rat offspring. PLoS ONE 2014, 9, e84209. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15, R271–R277. [Google Scholar] [CrossRef]
- Bass, J.; Lazar, M.A. Circadian time signatures of fitness and disease. Science 2016, 354, 994–999. [Google Scholar] [CrossRef]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Harfmann, B.D.; Schroder, E.A.; Kachman, M.T.; Hodge, B.A.; Zhang, X.; Esser, K.A. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet. Muscle 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.; Timmers, S.; Moonen-Kornips, E.; Duez, H.; Staels, B.; Hesselink, M.K.; Schrauwen, P. Synchronized human skeletal myotubes of lean, obese and type 2 diabetic patients maintain circadian oscillation of clock genes. Sci. Rep. 2016, 6, 35047. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Besing, R.C.; Paul, J.R.; Hablitz, L.M.; Rogers, C.O.; Johnson, R.L.; Young, M.E.; Gamble, K.L. Circadian rhythmicity of active GSK3 isoforms modulates molecular clock gene rhythms in the suprachiasmatic nucleus. J. Biol. Rhythms 2015, 30, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Sardon Puig, L.; Pillon, N.J.; Naslund, E.; Krook, A.; Zierath, J.R. Influence of obesity, weight loss, and free fatty acids on skeletal muscle clock gene expression. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E1–E10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yan, Q.; Yang, H.; Ren, A.; He, Z.; Tan, Z. Maternal intake restriction programs the energy metabolism, clock circadian regulator and mTOR signals in the skeletal muscles of goat offspring probably via the protein kinase A-cAMP-responsive element-binding proteins pathway. Anim. Nutr. 2021, 7, 1303–1314. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Chaves, A.B.; Zheng, D.; Johnson, J.A.; Bergman, B.C.; Patinkin, Z.W.; Zaegel, V.; Biagioni, E.M.; Krassovskaia, P.; Broskey, N.T.; May, L.E.; et al. Infant Mesenchymal Stem Cell Insulin Action Is Associated with Maternal Plasma Free Fatty Acids, Independent of Obesity Status: The Healthy Start Study. Diabetes 2022, 71, 1649–1659. [Google Scholar] [CrossRef]
- Wu, X.; Zvonic, S.; Floyd, Z.E.; Kilroy, G.; Goh, B.C.; Hernandez, T.L.; Eckel, R.H.; Mynatt, R.L.; Gimble, J.M. Induction of circadian gene expression in human subcutaneous adipose-derived stem cells. Obesity 2007, 15, 2560–2570. [Google Scholar] [CrossRef]
- Bonaconsa, M.; Malpeli, G.; Montaruli, A.; Carandente, F.; Grassi-Zucconi, G.; Bentivoglio, M. Differential modulation of clock gene expression in the suprachiasmatic nucleus, liver and heart of aged mice. Exp. Gerontol. 2014, 55, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, G.A.; Jordan, R.C.; Wood, P.A.; Li, Q.; Lincoln, D.W.; Sothern, R.B.; Hrushesky, W.J.; Ben-David, Y. Circadian expression of clock genes in human oral mucosa and skin: Association with specific cell-cycle phases. Am. J. Pathol. 2001, 158, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H.; Granneman, J.G.; Piomelli, D.; Leff, T.; Sassone-Corsi, P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Chen, Y.; Wang, S.; Ouyang, J.; Hu, L.; Yang, Q.; Wang, M.; Zhang, B.; Loor, J.J. Circadian Gene PER2 Silencing Downregulates PPARG and SREBF1 and Suppresses Lipid Synthesis in Bovine Mammary Epithelial Cells. Biology 2021, 10, 1226. [Google Scholar] [CrossRef]
- Barbour, L.A.; Hernandez, T.L. Maternal Lipids and Fetal Overgrowth: Making Fat from Fat. Clin. Ther. 2018, 40, 1638–1647. [Google Scholar] [CrossRef]

| Normal Weight n = 3 | Obese n = 3 | p | |
|---|---|---|---|
| Maternal Characteristics | |||
| Age (yrs) | 26.3 ± 6.4 | 33.7 ± 9.7 | 0.335 |
| Pre-pregnancy BMI (kg/m2) | 21.2 ± 1.7 | 32.3 ± 2.0 | 0.002 * |
| Race/ethnicity (self-reported), N | 0.519 | ||
| Hispanic | 1 | 1 | |
| White, non-Hispanic | 1 | 2 | |
| Black, non-Hispanic | 0 | 0 | |
| All others combined | 1 | 0 | |
| Fasting Plasma Glucose (mg/dL) | 75.0 ± 3.6 | 74.3 ± 8.0 | 0.902 |
| Fasting Plasma Insulin (mU/dL) | 9.0 ± 2.0 | 13.7 ± 2.1 | 0.049 * |
| Infant Characteristics | |||
| Sex (M/F) | 2/1 | 2/1 | -- |
| Birthweight (kg) | 3.2 ± 0.09 | 3.4 ± 0.3 | 0.244 |
| Fat Free Mass (g) | 2816.0 ± 154.3 | 2906.1 ± 260.7 | 0.632 |
| Fat Mass (g) | 191.7 ± 54.0 | 395.1 ± 136.0 | 0.074 |
| Fat Mass (%) | 6.4 ± 2.0 | 11.9 ± 3.4 | 0.074 |
| Gene Target | Normal Weight n = 3 | Obese n = 3 | ||
|---|---|---|---|---|
| R2 | p | R2 | p | |
| CLOCK | 0.117 | 0.136 | 0.019 | 0.716 |
| BMAL1 | 0.347 | 0.001 * | 0.222 | 0.021 * |
| PER2 | 0.367 | 0.001 * | 0.505 | 0.000 * |
| NR1D1 | 0.229 | 0.012 * | 0.444 | 0.000 * |
| GSK3B | 0.059 | 0.345 | 0.086 | 0.241 |
| PPARA | 0.124 | 0.099 | 0.069 | 0.318 |
| SIRT1 | 0.036 | 0.530 | 0.158 | 0.064 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erickson, M.L.; Dobias, D.; Keleher, M.R.; Dabelea, D.; Bergman, B.C.; Broussard, J.L.; Boyle, K.E. In Vitro Circadian Clock Gene Expression Assessments in Mesenchymal Stem Cells from Human Infants: A Pilot Study. Nutrients 2024, 16, 52. https://doi.org/10.3390/nu16010052
Erickson ML, Dobias D, Keleher MR, Dabelea D, Bergman BC, Broussard JL, Boyle KE. In Vitro Circadian Clock Gene Expression Assessments in Mesenchymal Stem Cells from Human Infants: A Pilot Study. Nutrients. 2024; 16(1):52. https://doi.org/10.3390/nu16010052
Chicago/Turabian StyleErickson, Melissa L., Devin Dobias, Madeline Rose Keleher, Dana Dabelea, Bryan C. Bergman, Josiane L. Broussard, and Kristen E. Boyle. 2024. "In Vitro Circadian Clock Gene Expression Assessments in Mesenchymal Stem Cells from Human Infants: A Pilot Study" Nutrients 16, no. 1: 52. https://doi.org/10.3390/nu16010052
APA StyleErickson, M. L., Dobias, D., Keleher, M. R., Dabelea, D., Bergman, B. C., Broussard, J. L., & Boyle, K. E. (2024). In Vitro Circadian Clock Gene Expression Assessments in Mesenchymal Stem Cells from Human Infants: A Pilot Study. Nutrients, 16(1), 52. https://doi.org/10.3390/nu16010052







