Impact of Maternal Mediterranean-Type Diet Adherence on Microbiota Composition and Epigenetic Programming of Offspring
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation
2.2.1. Illumina Epicarray-Based DNA Methylation Analysis
2.2.2. 16s rRNA Sequencing
2.3. Data Analysis
3. Results
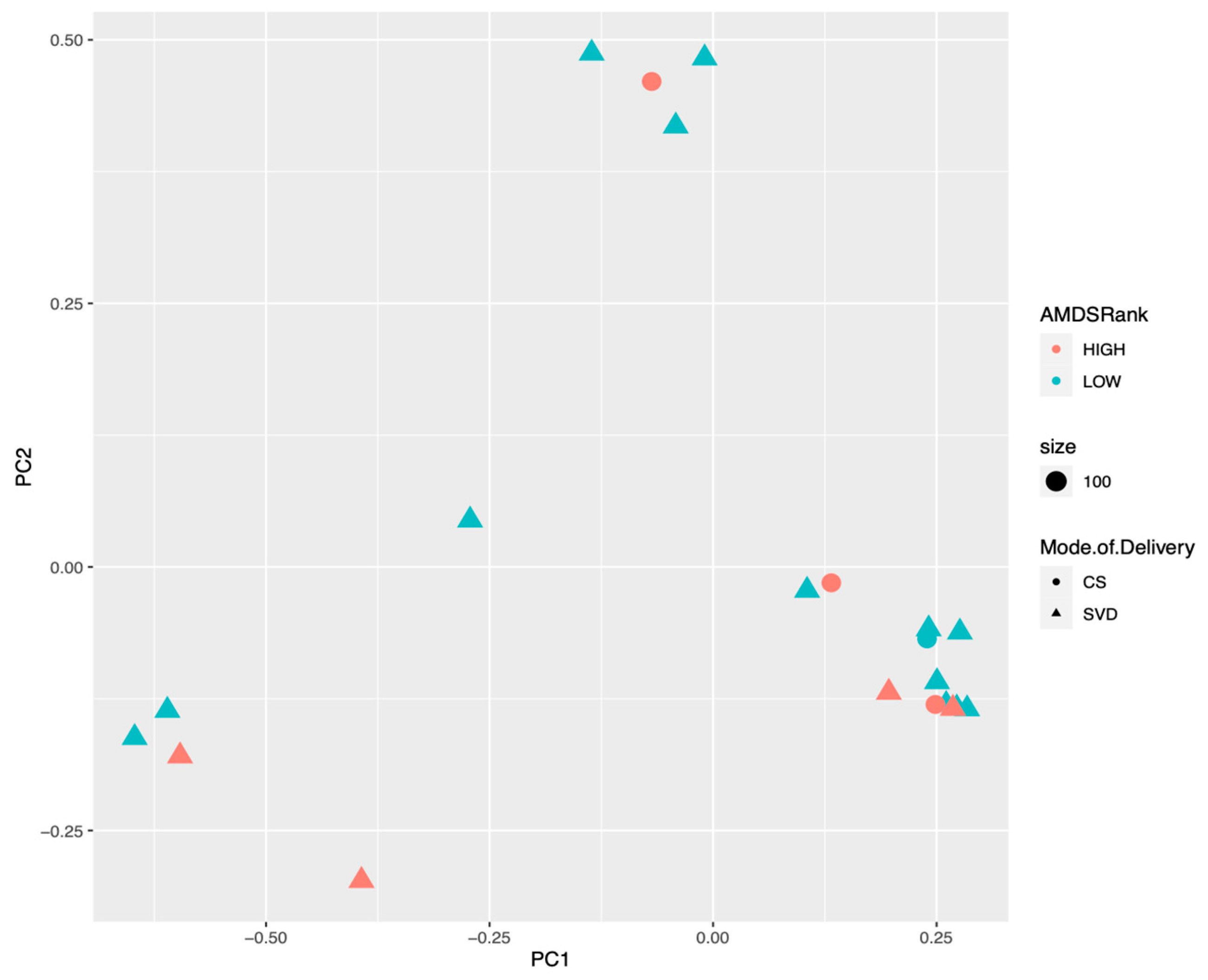
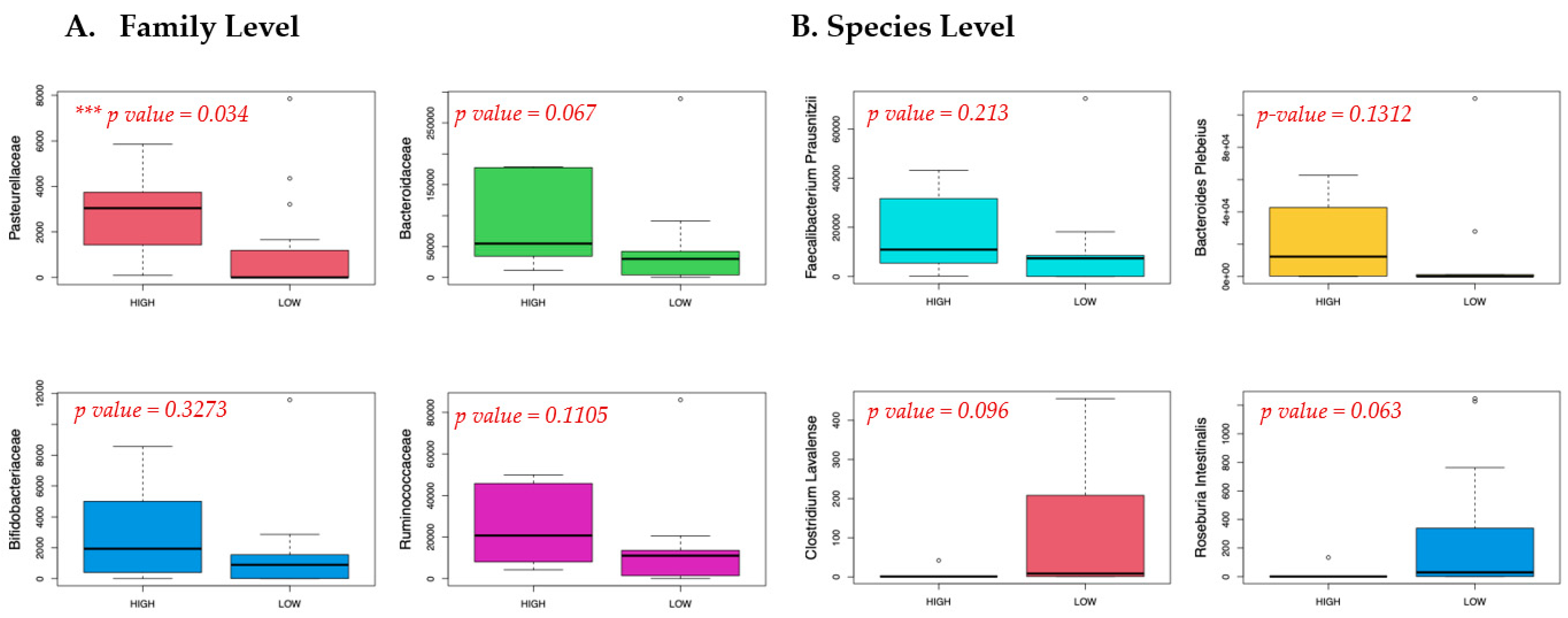
3.1. Meconium Microbiome Results
3.2. Methylation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gohir, W.; Kennedy, K.M.; Wallace, J.G.; Saoi, M.; Bellissimo, C.J.; Britz-McKibbin, P.; Petrik, J.J.; Surette, M.G.; Sloboda, D.M. High-fat diet intake modulates maternal intestinal adaptations to pregnancy and results in placental hypoxia, as well as altered fetal gut barrier proteins and immune markers. J. Physiol. 2019, 597, 3029–3051. [Google Scholar] [CrossRef] [PubMed]
- Gonseth, S.; Roy, R.; Houseman, E.A.; de Smith, A.J.; Zhou, M.; Lee, S.-T.; Nusslé, S.; Singer, A.W.; Wrensch, M.R.; Metayer, C.; et al. Periconceptional folate consumption is associated with neonatal DNA methylation modifications in neural crest regulatory and cancer development genes. Epigenetics 2015, 10, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Lehnen, H.; Zechner, U.; Haaf, T. Epigenetics of gestational diabetes mellitus and offspring health: The time for action is in early stages of life. Mol. Hum. Reprod. 2013, 19, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Barres, S.; Vrijheid, M.; Manzano-Salgado, C.B.; Valvi, D.; Martínez, D.; Iñiguez, C.; Jimenez-Zabala, A.; Riaño-Galán, I.; Navarrete-Muñoz, E.M.; Santa-Marina, L.; et al. The Association of Mediterranean Diet during Pregnancy with Longitudinal Body Mass Index Trajectories and Cardiometabolic Risk in Early Childhood. J. Pediatr. 2019, 206, 119–127.e6. [Google Scholar] [CrossRef] [PubMed]
- Carlisle, E.M.; Poroyko, V.; Caplan, M.S.; Alverdy, J.; Morowitz, M.J.; Liu, D. Murine gut microbiota and transcriptome are diet dependent. Ann. Surg. 2013, 257, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Bhagavata Srinivasan, S.P.; Raipuria, M.; Bahari, H.; Kaakoush, N.O.; Morris, M.J. Impacts of Diet and Exercise on Maternal Gut Microbiota Are Transferred to Offspring. Front. Endocrinol. 2018, 9, 716. [Google Scholar] [CrossRef] [PubMed]
- Wilczyńska, P.; Skarżyńska, E.; Lisowska-Myjak, B. Meconium microbiome as a new source of information about long-term health and disease: Questions and answers. J. Matern. Fetal Neonatal Med. 2019, 32, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Nomura, Y.; Bashir, A.; Fernandez-Hernandez, H.; Itzkowitz, S.; Pei, Z.; Stone, J.; Loudon, H.; Peter, I. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE 2013, 8, e78257. [Google Scholar] [CrossRef]
- Hu, J.; Ly, J.; Zhang, W.; Huang, Y.; Glover, V.; Peter, I.; Hurd, Y.L.; Nomura, Y. Microbiota of newborn meconium is associated with maternal anxiety experienced during pregnancy. Dev. Psychobiol. 2019, 61, 640–649. [Google Scholar] [CrossRef]
- Gosalbes, M.J.; Llop, S.; Valles, Y.; Moya, A.; Ballester, F.; Francino, M.P. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 2013, 43, 198–211. [Google Scholar] [CrossRef]
- Tapiainen, T.; Paalanne, N.; Tejesvi, M.V.; Koivusaari, P.; Korpela, K.; Pokka, T.; Salo, J.; Kaukola, T.; Pirttilä, A.M.; Uhari, M.; et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr. Res. 2018, 84, 371–379. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.T.; McCullough, M.L.; Newby, P.K.; Manson, J.E.; Meigs, J.B.; Rifai, N.; Willett, W.C.; Hu, F.B. Diet-quality scores and plasma concentrations of markers of inflammation and endothelial dysfunction. Am. J. Clin. Nutr. 2005, 82, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The Mediterranean diets: What is so special about the diet of Greece? The scientific evidence. J. Nutr. 2001, 131, 3065S–3073S. [Google Scholar] [CrossRef] [PubMed]
- Trichopoulou, A.; Costacou, T.; Bamia, C.; Trichopoulos, D. Adherence to a Mediterranean diet and survival in a Greek population. N. Engl. J. Med. 2003, 348, 2599–2608. [Google Scholar] [CrossRef] [PubMed]
- Amati, F.; Hassounah, S.; Swaka, A. The Impact of Mediterranean Dietary Patterns During Pregnancy on Maternal and Offspring Health. Nutrients 2019, 11, 1098. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Laghi, L.; Gobbetti, M.; Ercolini, D. Unusual sub-genus associations of faecal Prevotella and Bacteroides with specific dietary patterns. Microbiome 2016, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, M.; Martín, R.; Torres-Maravilla, E.; Chadi, S.; González-Dávila, P.; Sokol, H.; Langella, P.; Chain, F.; Bermúdez-Humarán, L.G. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 2020, 12, 1–16. [Google Scholar] [CrossRef]
- Telle-Hansen, V.H.; Holven, K.B.; Ulven, S.M. Impact of a Healthy Dietary Pattern on Gut Microbiota and Systemic Inflammation in Humans. Nutrients 2018, 10, 1783. [Google Scholar] [CrossRef]
- Barrett, H.L.; Gomez-Arango, L.F.; Wilkinson, S.A.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Nitert, M.D. A Vegetarian Diet Is a Major Determinant of Gut Microbiota Composition in Early Pregnancy. Nutrients 2018, 10, 890. [Google Scholar] [CrossRef]
- Fan, H.-Y.; Tung, Y.-T.; Yang, Y.-C.S.H.; Hsu, J.B.; Lee, C.-Y.; Chang, T.-H.; Su, E.C.-Y.; Hsieh, R.-H.; Chen, Y.-C. Maternal Vegetable and Fruit Consumption during Pregnancy and Its Effects on Infant Gut Microbiome. Nutrients 2021, 13, 1559. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.M.; Antony, K.M.; Ma, J.; Prince, A.L.; Showalter, L.; Moller, M.; Aagaard, K.M. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes 2022, 14, 2022407. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Epigenetic Mechanisms Link Maternal Diets and Gut Microbiome to Obesity in the Offspring. Front. Genet. 2018, 9, 342. [Google Scholar] [CrossRef] [PubMed]
- Ziętek, M.; Celewicz, Z.; Szczuko, M. Short-Chain Fatty Acids, Maternal Microbiota and Metabolism in Pregnancy. Nutrients 2021, 13, 1244. [Google Scholar] [CrossRef]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.; Salvador, C.; Skibola, C.; Tollefsbol, T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 2015, 7, 112. [Google Scholar] [CrossRef]
- Lorite Mingot, D.; Gesteiro, E.; Bastida, S.; Sánchez-Muniz, F.J. Epigenetic effects of the pregnancy Mediterranean diet adherence on the offspring metabolic syndrome markers. J. Physiol. Biochem. 2017, 73, 495–510. [Google Scholar] [CrossRef]
- Harmon, B.E.; Boushey, C.J.; Shvetsov, Y.B.; Ettienne, R.; Reedy, J.; Wilkens, L.R.; Le Marchand, L.; Henthederson, B.E.; Kolonel, L.N. Associations of key diet-quality indexes with mortality in the Multiethnic Cohort: The Dietary Patterns Methods Project. Am. J. Clin. Nutr. 2015, 101, 587–597. [Google Scholar] [CrossRef]
- Gonzalez-Nahm, S.; Mendez, M.; Robinson, W.; Murphy, S.K.; Hoyo, C.; Hogan, V.; Rowley, D. Low maternal adherence to a Mediterranean diet is associated with increase in methylation at the MEG3-IG differentially methylated region in female infants. Environ. Epigenet. 2017, 3, dvx007. [Google Scholar] [CrossRef] [PubMed]
- House, J.S.; Mendez, M.; Maguire, R.L.; Gonzalez-Nahm, S.; Huang, Z.; Daniels, J.; Murphy, S.K.; Fuemmeler, B.F.; Wright, F.A.; Hoyo, C. Periconceptional Maternal Mediterranean Diet Is Associated With Favorable Offspring Behaviors and Altered CpG Methylation of Imprinted Genes. Front. Cell Dev. Biol. 2018, 6, 107. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Barone, G.; Mazzoleni, P.; Catalfo, A.; De Guidi, G.; Iemmolo, M.G.; Crimi, N.; Agodi, A. Mediterranean Diet and Particulate Matter Exposure Are Associated With LINE-1 Methylation: Results From a Cross-Sectional Study in Women. Front. Genet. 2018, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.B.; Benny, P.; Riel, J.; Boushey, C.; Perez, R.; Khadka, V.; Qin, Y.; Maunakea, A.K.; Lee, M.J. Adherence to Mediterranean diet impacts gastrointestinal microbial diversity throughout pregnancy. BMC Pregnancy Childbirth 2021, 21, 558. [Google Scholar] [CrossRef] [PubMed]
- Kolonel, L.N.; Henderson, B.E.; Hankin, J.H.; Nomura, A.M.Y.; Wilkens, L.R.; Pike, M.C.; Stram, D.O.; Monroe, K.R.; Earle, M.E.; Nagamine, F.S. A multiethnic cohort in Hawaii and Los Angeles: Baseline characteristics. Am. J. Epidemiol. 2000, 151, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Shvetsov, Y.B.; Harmon, B.E.; Ettienne, R.; Wilkens, L.R.; Le Marchand, L.; Kolonel, L.N.; Boushey, C.J. The influence of energy standardisation on the alternate Mediterranean diet score and its association with mortality in the Multiethnic Cohort. Br. J. Nutr. 2016, 116, 1592–1601. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.M.; Yaktine, A.L. The National Academies Collection: Reports funded by National Institutes of Health. In Weight Gain During Pregnancy: Reexamining the Guidelines; Institutes of Medicine National Research Council Committee to Reexamine Gestational Weight Gain in Pregnancy, Ed.; National Academies Press (US): Washington, DC, USA, 2009. [Google Scholar]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef]
- Gesteiro, E.; Bastida, S.; Bernal, B.R.; Sánchez-Muniz, F.J. Adherence to Mediterranean diet during pregnancy and serum lipid, lipoprotein and homocysteine concentrations at birth. Eur. J. Nutr. 2015, 54, 1191–1199. [Google Scholar] [CrossRef]
- Gesteiro, E.; Rodríguez Bernal, B.; Bastida, S.; Sánchez-Muniz, F.J. Maternal diets with low healthy eating index or Mediterranean diet adherence scores are associated with high cord-blood insulin levels and insulin resistance markers at birth. Eur. J. Clin. Nutr. 2012, 66, 1008–1015. [Google Scholar] [CrossRef]
- Slomski, A. Mediterranean Diet During Pregnancy. JAMA 2019, 322, 1134. [Google Scholar] [CrossRef]
- Al Wattar, B.H.; Dodds, J.; Placzek, A.; Spyreli, E.; Higgins, S.; Moore, A.; Hooper, R.; Beresford, L.; Roseboom, T.J.; Bes-Rastrollo, M.; et al. Mediterranean diet based intervention in pregnancy to improve maternal and fetal outcomes: Methodological challenges and lessons learned from the multicentre ESTEEM study. Contemp. Clin. Trials Commun. 2017, 6, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Al Wattar, B.H.; Dodds, J.; Placzek, A.; Beresford, L.; Spyreli, E.; Moore, A.; Carreras, F.J.G.; Austin, F.; Murugesu, N.; Roseboom, T.J.; et al. Mediterranean-style diet in pregnant women with metabolic risk factors (ESTEEM): A pragmatic multicentre randomised trial. PLoS Med. 2019, 16, e1002857. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, N.G.; Assaf-Balut, C.; Jiménez Varas, I.; Del Valle, L.; Durán, A.; Fuentes, M.; Del Prado, N.; Bordiú, E.; Valerio, J.J.; Herraiz, M.A.; et al. Effectiveness of Following Mediterranean Diet Recommendations in the Real World in the Incidence of Gestational Diabetes Mellitus (GDM) and Adverse Maternal-Foetal Outcomes: A Prospective, Universal, Interventional Study with a Single GrouThe St Carlos Study. Nutrients 2019, 11, 1210. [Google Scholar] [PubMed]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Quevrain, E.; Maubert, M.-A.; Sokol, H.; Devreese, B.; Seksik, P. The presence of the anti-inflammatory protein MAM, from Faecalibacterium prausnitzii, in the intestinal ecosystem. Gut 2016, 65, 882. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liang, R.; Zhang, W.; Tian, K.; Li, J.; Chen, X.; Yu, T.; Chen, Q. Faecalibacterium prausnitzii-derived microbial anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J. Diabetes 2020, 12, 224–236. [Google Scholar] [CrossRef]
- Werlang, I.C.R.; Mueller, N.T.; Pizoni, A.; Wisintainer, H.; Matte, U.; Costa, S.H.d.A.M.; Ramos, J.G.L.; Goldani, M.Z.; Dominguez-Bello, M.G.; Goldani, H.A.S. Associations of birth mode with cord blood cytokines, white blood cells, and newborn intestinal bifidobacteria. PLoS ONE 2018, 13, e0205962. [Google Scholar] [CrossRef]
- Shi, Y.C.; Guo, H.; Chen, J.; Sun, G.; Ren, R.-R.; Guo, M.-Z.; Peng, L.-H.; Yang, Y.-S. Initial meconium microbiome in Chinese neonates delivered naturally or by cesarean section. Sci. Rep. 2018, 8, 3255. [Google Scholar] [CrossRef]
- Weng, T.H.; Huang, K.-Y.; Jhong, J.-H.; Kao, H.-J.; Chen, C.-H.; Chen, Y.-C.; Weng, S.-L. Microbiome analysis of maternal and neonatal microbial communities associated with the different delivery modes based on 16S rRNA gene amplicon sequencing. Taiwan. J. Obstet. Gynecol. 2023, 62, 687–696. [Google Scholar] [CrossRef]
- Martin, R.; Makino, H.; Cetinyurek Yavuz, A.; Ben-Amor, K.; Roelofs, M.; Ishikawa, E.; Kubota, H.; Swinkels, S.; Sakai, T.; Oishi, K.; et al. Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS ONE 2016, 11, e0158498. [Google Scholar] [CrossRef]
- Bianchi, M.; Alisi, A.; Fabrizi, M.; Vallone, C.; Ravà, L.; Giannico, R.; Vernocchi, P.; Signore, F.; Manco, M. Maternal Intake of n-3 Polyunsaturated Fatty Acids During Pregnancy Is Associated With Differential Methylation Profiles in Cord Blood White Cells. Front. Genet. 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, S.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; AS Langie, S.; Koppen, G.; Devlieger, R.; Godderis, L. Dietary and supplemental maternal methyl-group donor intake and cord blood DNA methylation. Epigenetics 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, A.A.; Sexton-Oates, A.; O’brien, E.C.; Alberdi, G.; Fransquet, P.; Saffery, R.; McAuliffe, F.M. A Low Glycaemic Index Diet in Pregnancy Induces DNA Methylation Variation in Blood of Newborns: Results from the ROLO Randomised Controlled Trial. Nutrients 2018, 10, 455. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Barraza-Villarreal, A.; Biessy, C.; Duarte-Salles, T.; Sly, P.D.; Ramakrishnan, U.; Rivera, J.; Herceg, Z.; Romieu, I. Dietary supplementation with polyunsaturated fatty acid during pregnancy modulates DNA methylation at IGF2/H19 imprinted genes and growth of infants. Physiol. Genom. 2014, 46, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Küpers, L.K.; Fernández-Barrés, S.; Nounu, A.; Friedman, C.; Fore, R.; Mancano, G.; Dabelea, D.; Rifas-Shiman, S.L.; Mulder, R.H.; Oken, E.; et al. Maternal Mediterranean diet in pregnancy and newborn DNA methylation: A meta-analysis in the PACE Consortium. Epigenetics 2022, 17, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.D.; Davis, M.N.; Barqué, A.; Thahab, A.H.; Ricard-Blum, S.; Naba, A. Computational and experimental characterization of the novel ECM glycoprotein SNED1 and prediction of its interactome. Biochem. J. 2021, 478, 1413–1434. [Google Scholar] [CrossRef] [PubMed]
- Rzehak, P.; Covic, M.; Saffery, R.; Reischl, E.; Wahl, S.; Grote, V.; Weber, M.; Xhonneux, A.; Langhendries, J.-P.; Ferre, N.; et al. DNA-Methylation and Body Composition in Preschool Children: Epigenome-Wide-Analysis in the European Childhood Obesity Project (CHOP)-Study. Sci. Rep. 2017, 7, 14349. [Google Scholar] [CrossRef]
- Wagner, G.; Fenzl, A.; Lindroos-Christensen, J.; Einwallner, E.; Husa, J.; Witzeneder, N.; Rauscher, S.; Gröger, M.; Derdak, S.; Mohr, T.; et al. LMO3 reprograms visceral adipocyte metabolism during obesity. J. Mol. Med. 2021, 99, 1151–1171. [Google Scholar] [CrossRef]
- Lindroos, J.; Husa, J.; Mitterer, G.; Haschemi, A.; Rauscher, S.; Haas, R.; Gröger, M.; Loewe, R.; Kohrgruber, N.; Schrögendorfer, K.F.; et al. Human but not mouse adipogenesis is critically dependent on LMO3. Cell. Metab. 2013, 18, 62–74. [Google Scholar] [CrossRef]
- Louwies, T.; Johnson, A.C.; Orock, A.; Yuan, T.; Meerveld, B.G.-V. The microbiota-gut-brain axis: An emerging role for the epigenome. Exp. Biol. Med. 2019, 45, 138–145. [Google Scholar] [CrossRef]
- Alam, R.; Abdolmaleky, H.M.; Zhou, J.R. Microbiome, inflammation, epigenetic alterations, and mental diseases. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017, 174, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrova, K.; Romero-Mosquera, B.; Hernandez, V. Diet, Gut Microbiome and Epigenetics: Emerging Links with Inflammatory Bowel Diseases and Prospects for Management and Prevention. Nutrients 2017, 9, 962. [Google Scholar] [CrossRef] [PubMed]
- Becerra, C.Y.; Wells, R.K.; Kunihiro, B.P.; Lee, R.H.; Umeda, L.; Allan, N.P.; Rubas, N.C.; McCracken, T.A.; Nunokawa, C.K.L.; Lee, M.-H.; et al. Examining the immunoepigenetic-gut microbiome axis in the context of self-esteem among Native Hawaiians and other Pacific Islanders. Front. Genet. 2023, 14, 1125217. [Google Scholar] [CrossRef] [PubMed]
- Eijsbouts, C.; Zheng, T.; Kennedy, N.A.; Bonfiglio, F.; Anderson, C.A.; Moutsianas, L.; Holliday, J.; Shi, J.; Shringarpure, S.; Agee, M.; et al. Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat. Genet. 2021, 53, 1543–1552. [Google Scholar] [CrossRef]
- Miro-Blanch, J.; Yanes, O. Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism. Front. Genet. 2019, 10, 638. [Google Scholar] [CrossRef]
- Cortese, R.; Lu, L.; Yu, Y.; Ruden, D.; Claud, E.C. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 2016, 11, 205–215. [Google Scholar] [CrossRef]


| Food Group | Foods Included | Criteria for 1 Point |
|---|---|---|
| Vegetables | All vegetables except potatoes | Greater than median intake (servings/d) |
| Legumes | Tofu, string beans, peas, beans | Greater than median intake (servings/d) |
| Fruit | All fruit and juices | Greater than median intake (servings/d) |
| Nuts | Nuts, peanut putter | Greater than median intake (servings/d) |
| Whole Grains | Whole-grain cereals, cooked cereals, crackers, dark breads, brown rice, other grains, wheat germ, bran, popcorn | Greater than median intake (servings/d) |
| Red and Processed Meats | Hot dogs, deli meat, bacon, hamburger, beef | Less than median intake (servings/d) |
| Fish | Fish and shrimp, breaded fish | Greater than median intake (servings/d) |
| Ratio of Monounsaturated to Saturated Fat | - | Greater than median intake (servings/d) |
| Ethanol | Wine, beer, light beer, liquor | <25 g/d |
| Low aMED Adherence (n = 21) | High aMED Adherence (n = 12) | p-Value | |
|---|---|---|---|
| Maternal Age Median (SE) | 28 (5.2) | 33 (5.5) | 0.123 |
| Ethnicity | 0.078 | ||
| Non-Hispanic White | 6 | 3 | |
| Filipino | 7 | 0 | |
| Native Hawaiian | 3 | 5 | |
| Japanese | 5 | 4 | |
| Parity | 0.452 | ||
| Nulliparous | 12/21 | 7/12 | |
| Primiparous | 8/21 | 3/12 | |
| Multiparous | 1/21 | 2/12 | |
| Pregnancy Complications | 0.064 | ||
| Gestational Diabetes | 1/21 | 2/12 | |
| Preeclampsia | 5/21 | 2/12 | |
| Preterm Labor | 1/21 | 0/12 | |
| Maternal Obesity 1 | 29.41% | 25% | 0.717 |
| Gestational Weight Gain (Mean, +/− SD) | 28.04 (9.89) | 26.83 (12.03) | 0.761 |
| Excess Gestational Weight Gain | 5 | 4 | 0.503 |
| Mode of Delivery | |||
| Vaginal (Spontaneous or Operative) | 17/21 | 7/12 | 0.071 |
| Cesarean Delivery | 4/21 | 5/12 | |
| Neonatal Birth Weight (g) | 3206.05 g (+/− 468.35) | 3412.67 g (+/− 495.57) | 0.251 |
| Gestational Age at Delivery (Median in weeks) | 39 weeks | 39 weeks | 1.000 |
| Total Kilocalories/day (mean, [+/− SD]) | 2297.9 (1267.18) | 1638.4 (612.21) | 0.238 |
| Macronutrients | |||
| % Energy from Carbohydrates | 46.33% | 51.20% | 0.139 |
| % Energy from Total Fat | 36.78% | 34.00% | 0.015 |
| % Energy from Protein | 16.88% | 14.78% | 0.415 |
| Micronutrients Mean (SD) | |||
| Fiber (g) | 14.10 (7.11) | 31.18 (20.04) | 0.003 |
| Vitamin D (International Units) | 126.00 (67.92) | 159.00 (105.46) | 0.275 |
| Vitamin B12 (mcg) | 5.12 (2.44) | 7.09 (4.72) | 0.120 |
| Monosaturated Fatty Acids (g) | 27.5 (11.75) | 32.16 (13.43) | 0.312 |
| Saturated Fatty Acids (g) | 24.28 (9.78) | 27.112 (10.12) | 0.440 |
| Monounsaturated:Saturated Fatty Acid Ratio | 1.13 (0.16) | 1.17 (0.17) | 0.508 |
| Polyunsaturated Fatty Acids (g) | 12.44 (5.27) | 18.93 (10.3) | 0.025 |
| Tissue | Chromosome | Base Pair Region | Overlapping Gene Symbol | FDR |
|---|---|---|---|---|
| Cord Blood— aMED Model | Chr1 | 67600546–67600963 | 5.68 × 10−7 | |
| Chr1 | 108023248–108023486 | 1.27 × 10−8 | ||
| Chr2 | 105853199–105853526 | NCK2, ENSG00000235522 | 2.53 × 10−7 | |
| Chr2 | 241076415–241076601 | SNED1, MTERF4 | 4.63 × 10−8 | |
| Chr3 | 133502622–133502917 | 1.73 × 10−7 | ||
| Chr4 | 74847709–74848016 | 6.89 × 10−9 | ||
| Chr6 | 31744523–31744628 | MSH5, MSH5-SAPCD1 | 1.14 × 10−4 | |
| Chr6 | 32063873–32064146 | TNXB | 7.17 × 10−5 | |
| Chr6 | 33084419–33085063 | HLA-DPB1 | 4.05 × 10−15 | |
| Chr7 | 30196738–30197130 | 1.04 × 10−10 | ||
| Cord Blood— Bacteroides | Chr6 | 31650734–31651158 | BAG6 | 1.03 × 10−11 |
| Placenta | Chr12 | 16758934–16759391 | MGST1, LMO3 | 2.64 × 10−9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasaki, T.; Kawamura, M.; Okuno, C.; Lau, K.; Riel, J.; Lee, M.-J.; Miller, C. Impact of Maternal Mediterranean-Type Diet Adherence on Microbiota Composition and Epigenetic Programming of Offspring. Nutrients 2024, 16, 47. https://doi.org/10.3390/nu16010047
Sasaki T, Kawamura M, Okuno C, Lau K, Riel J, Lee M-J, Miller C. Impact of Maternal Mediterranean-Type Diet Adherence on Microbiota Composition and Epigenetic Programming of Offspring. Nutrients. 2024; 16(1):47. https://doi.org/10.3390/nu16010047
Chicago/Turabian StyleSasaki, Tamlyn, Megan Kawamura, Chirstyn Okuno, Kayleen Lau, Jonathan Riel, Men-Jean Lee, and Corrie Miller. 2024. "Impact of Maternal Mediterranean-Type Diet Adherence on Microbiota Composition and Epigenetic Programming of Offspring" Nutrients 16, no. 1: 47. https://doi.org/10.3390/nu16010047
APA StyleSasaki, T., Kawamura, M., Okuno, C., Lau, K., Riel, J., Lee, M.-J., & Miller, C. (2024). Impact of Maternal Mediterranean-Type Diet Adherence on Microbiota Composition and Epigenetic Programming of Offspring. Nutrients, 16(1), 47. https://doi.org/10.3390/nu16010047






