The Mechanism Underlying the Hypoglycemic Effect of Epimedin C on Mice with Type 2 Diabetes Mellitus Based on Proteomic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
2.2. Animal Experiments
2.2.1. Experimental Design and Drug Administration
2.2.2. Biochemical and Physiological Analyses
Oral Glucose Tolerance Test
Assays of Insulin, Hepatic Glycogen, SOD, GSH-Px, MDA, HDL-C, and LDL-C and the Homeostasis Model Assessment of Insulin Resistance (HOMA-IR)
2.2.3. Histological Assessment
2.3. Proteomic and Bioinformatic Analyses
2.4. Statistical Analysis
3. Results
3.1. Physiological and Biochemical Analysis
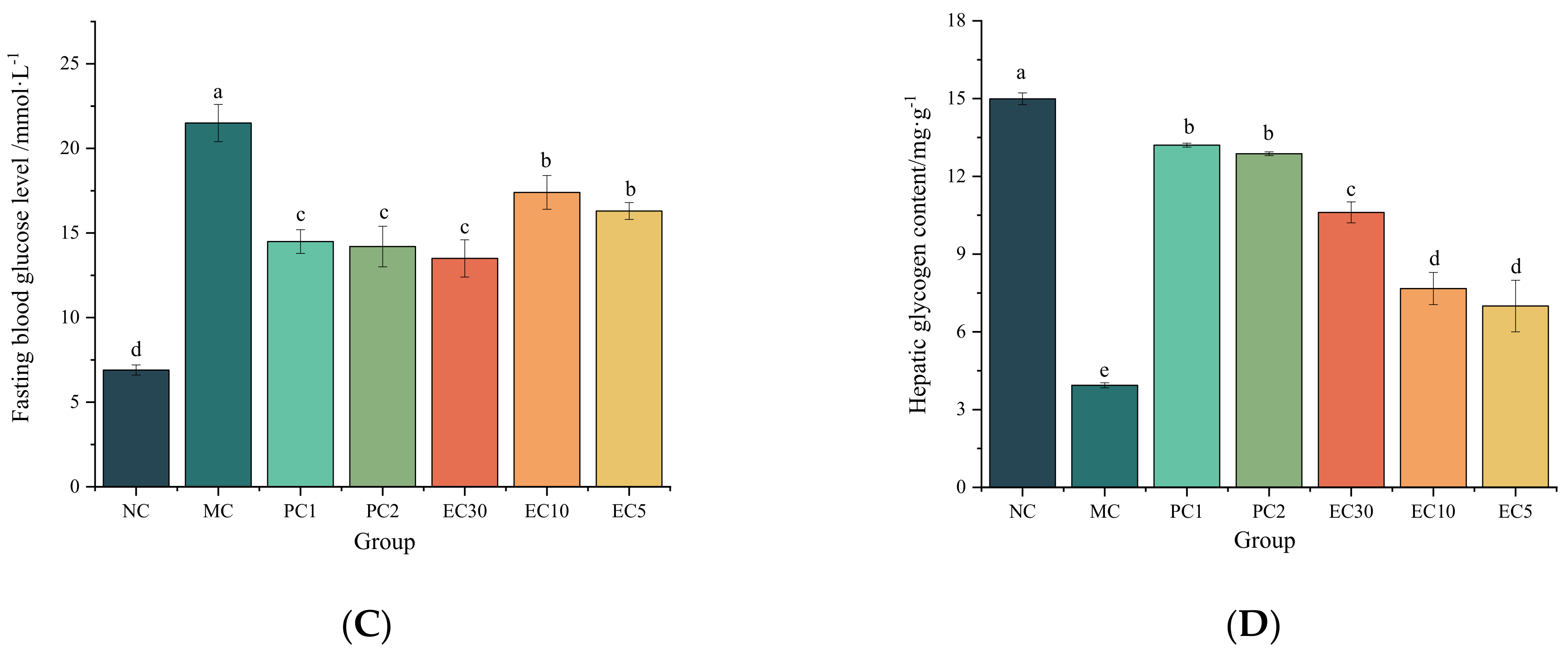
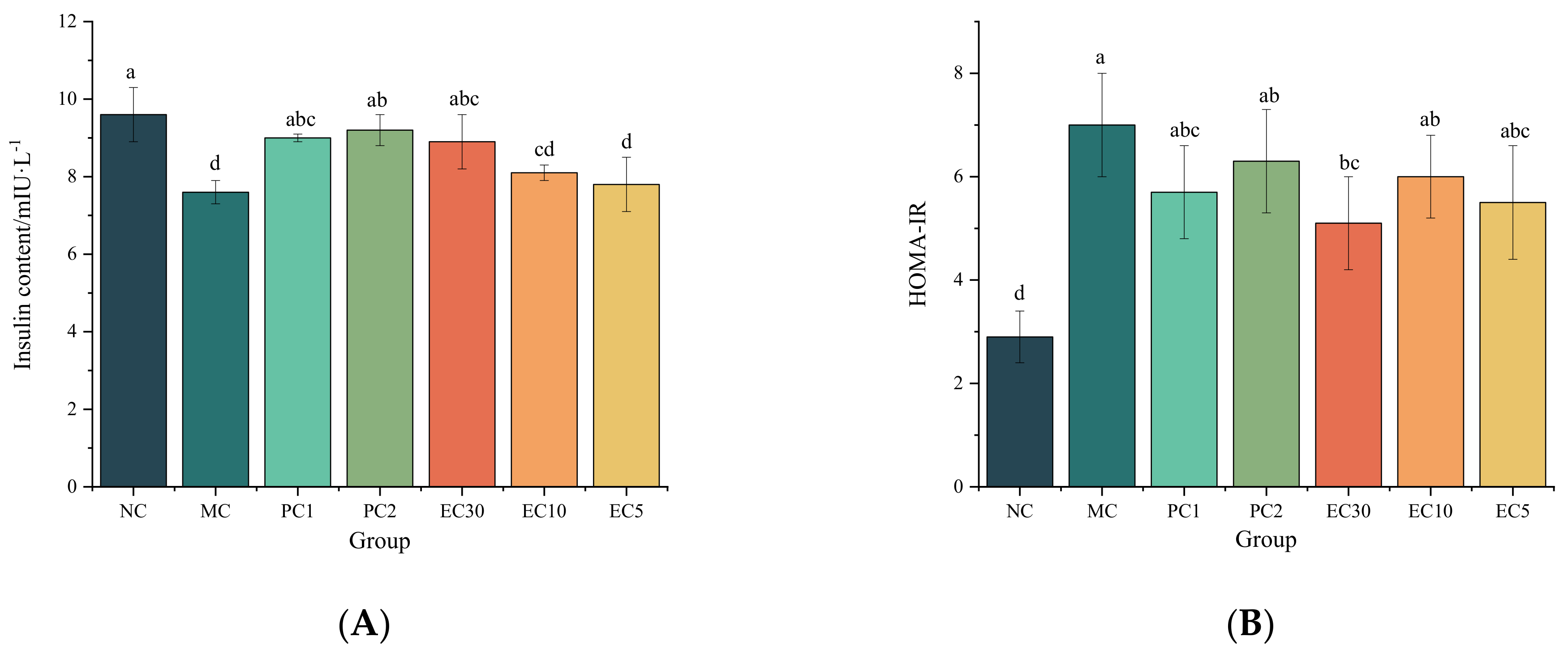
3.1.1. Effects of Epimedin C on the FBG, Hepatic Glycogen, Insulin, HOMA-IR, and Oral Glucose Tolerance of T2DM Mice
3.1.2. Effects of Epimedin C on Oxidative Stress
3.1.3. Effects of Epimedin C on Blood Lipid Levels
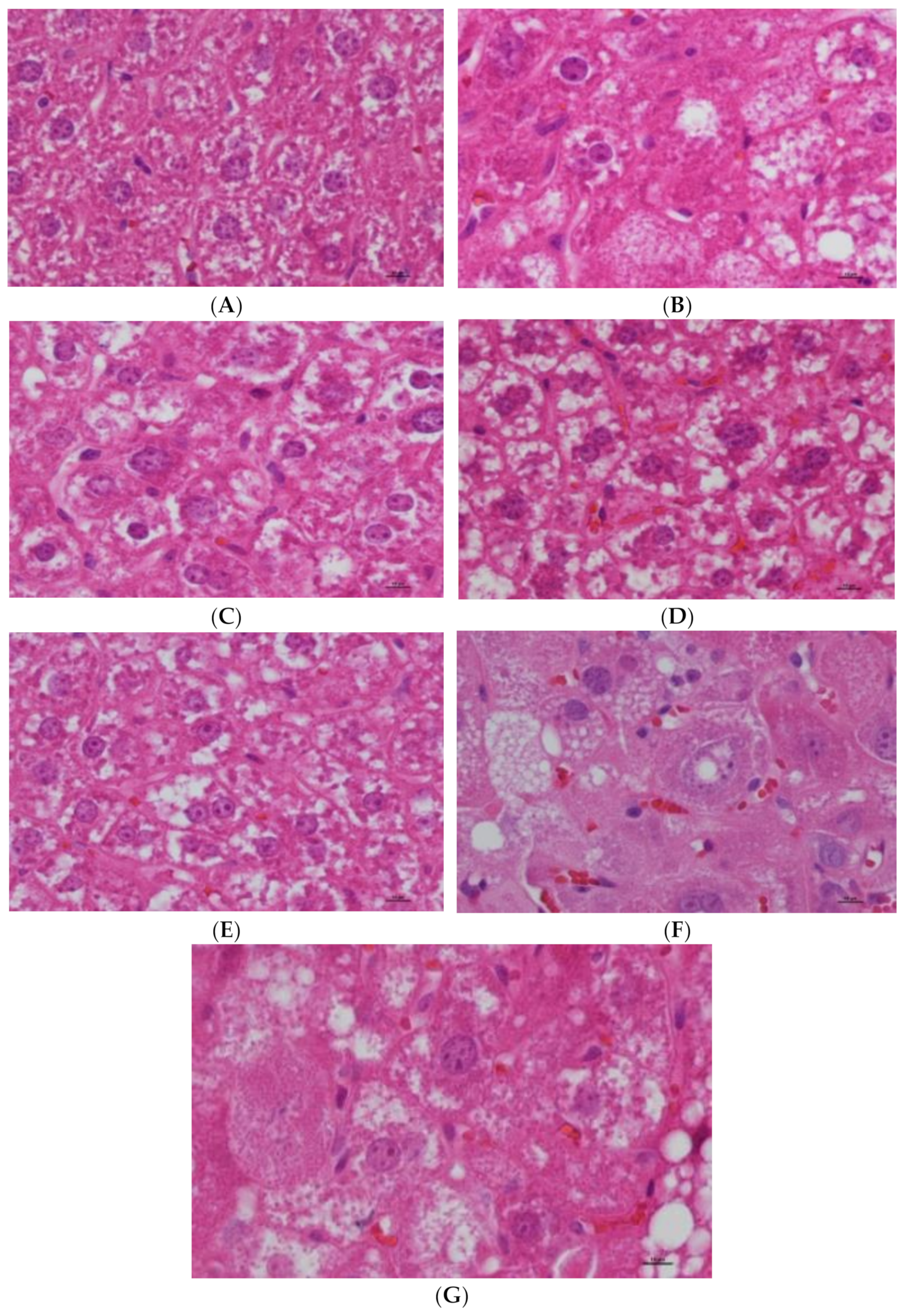
3.2. Effects of Epimedin C on Liver Histopathological Changes
3.3. Proteomic Analysis
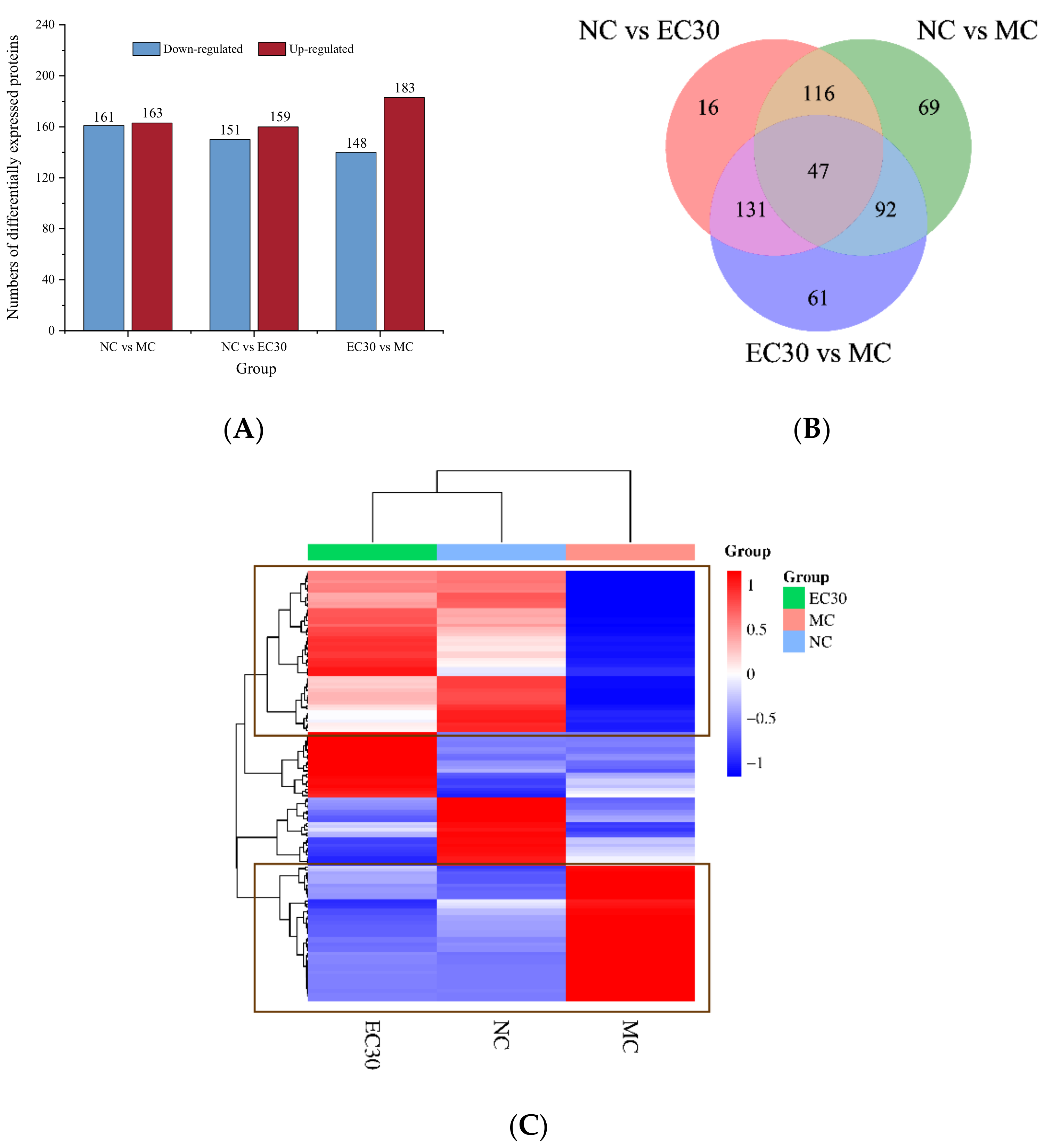
3.3.1. Identification of DEPs
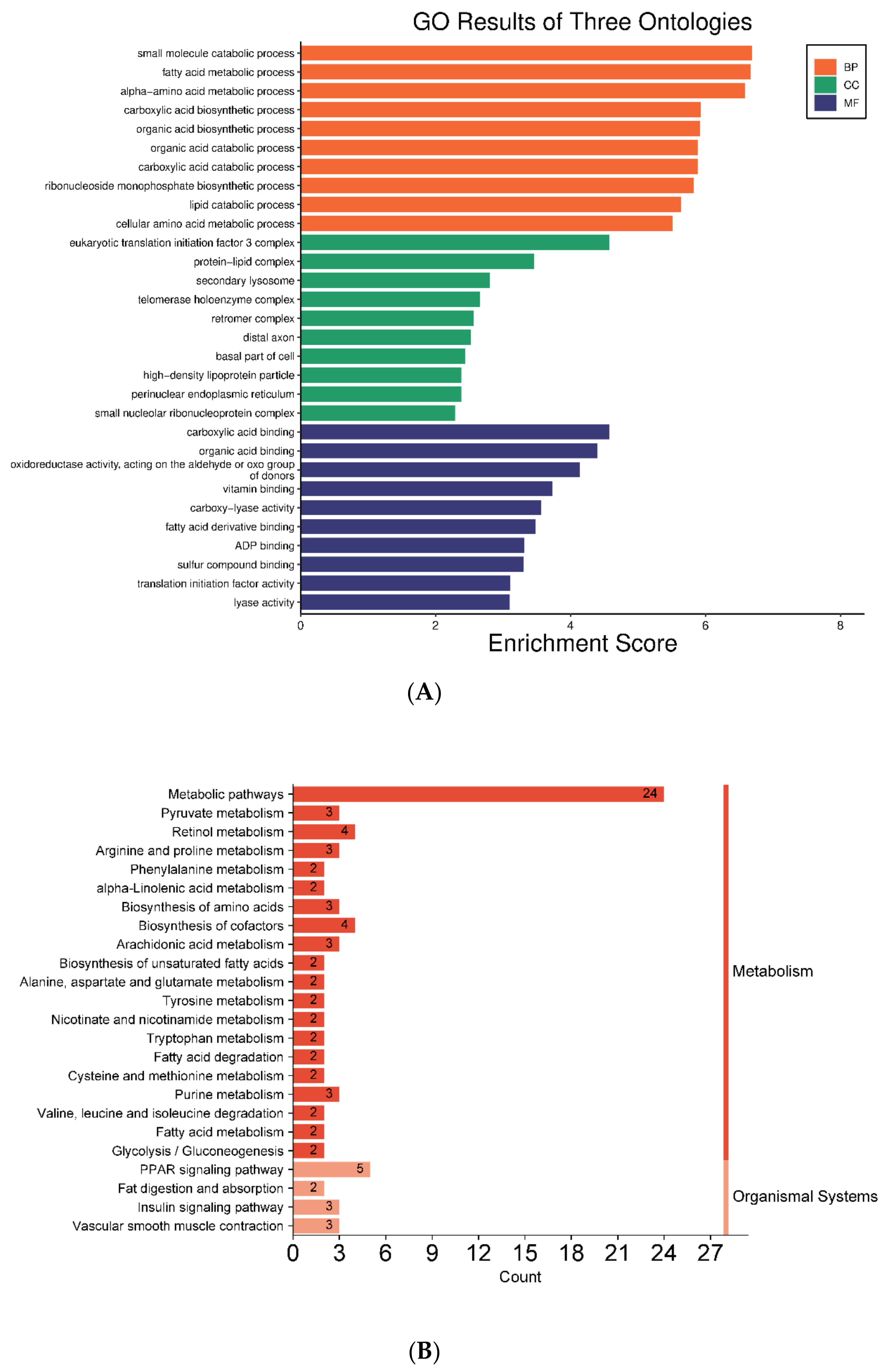
3.3.2. GO and KEGG Pathway Analyses
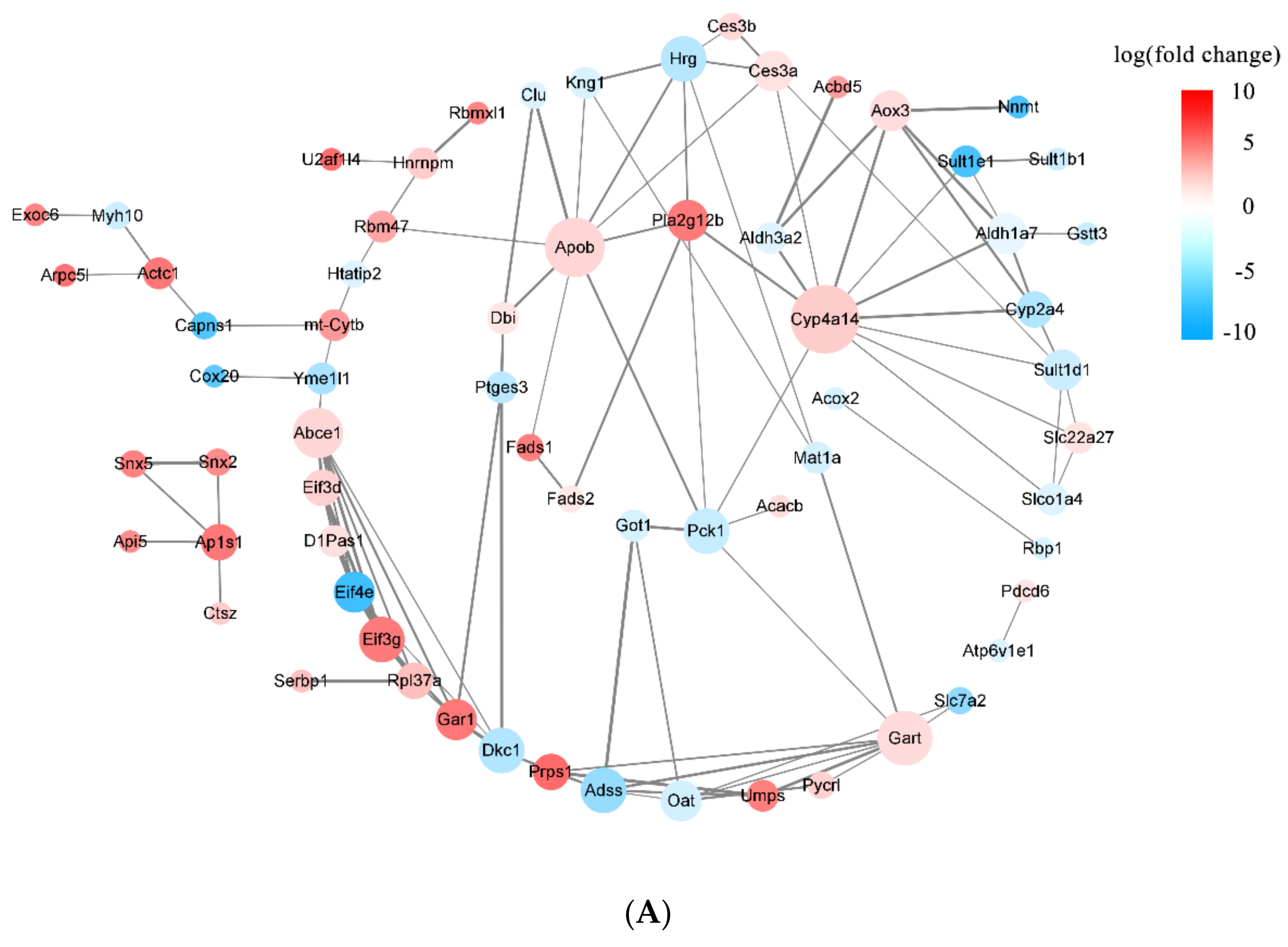
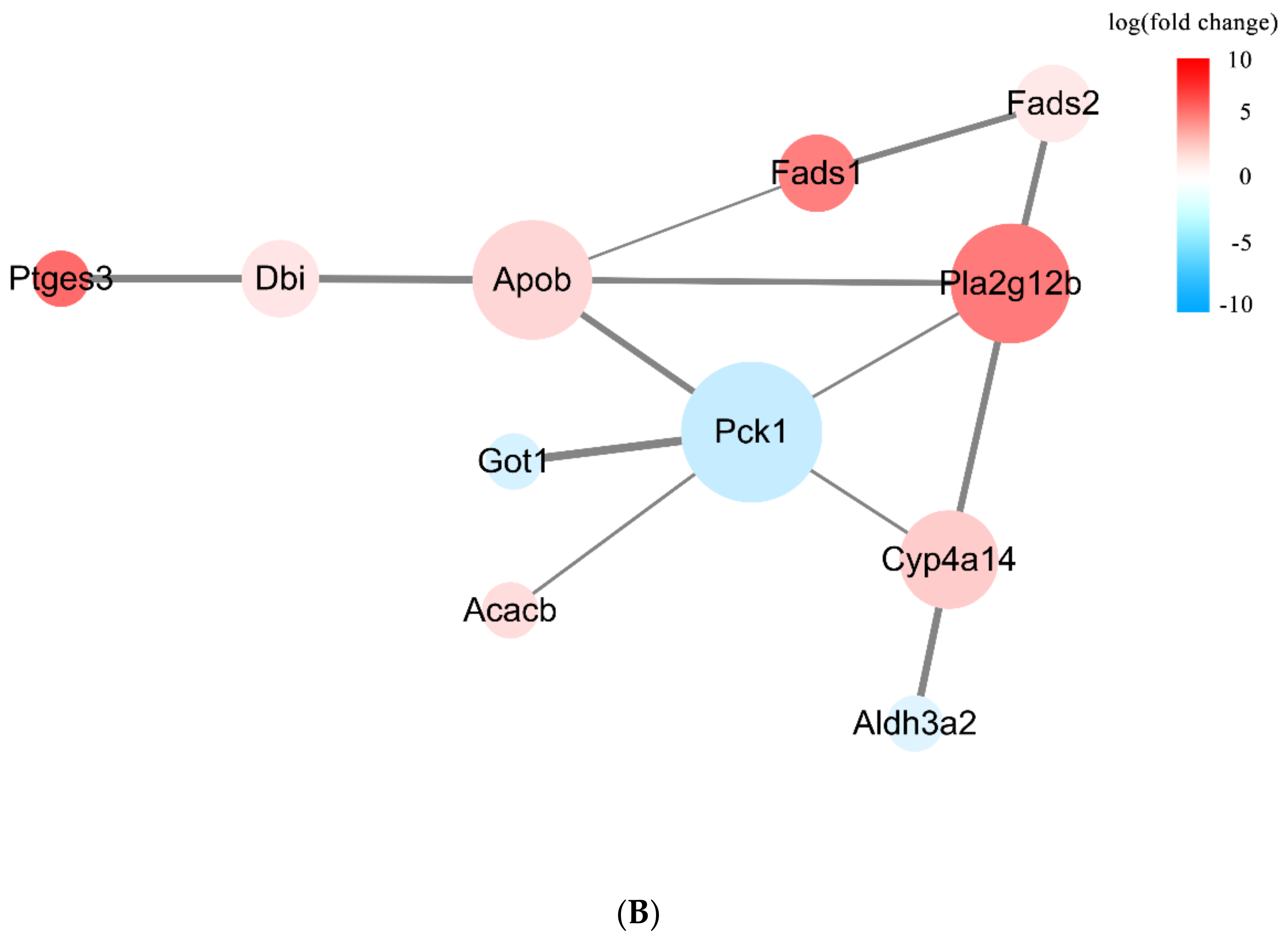
3.3.3. DEPs Network Interaction Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magliano, D.J.; Boyko, E.J.; IDF Diabetes Atlas 10th Edition Scientific Committee. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021. Available online: http://www.ncbi.nlm.nih.gov/books/NBK581934/ (accessed on 23 September 2022).
- Chen, P.L.; Yang, W.S. Human Genetics of Diabetes Mellitus in Taiwan. Front. Biosci. 2009, 14, 4535–4545. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Artasensi, A.; Pedretti, A.; Vistoli, G.; Fumagalli, L. Type 2 Diabetes Mellitus: A Review of Multi-Target Drugs. Molecules 2020, 25, 1987. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, N.; Hashimoto, Y.; Hamaguchi, M.; Osaka, T.; Fukuda, T.; Yamazaki, M.; Fukui, M. Liver Stiffness Is Associated with Progression of Albuminuria in Adults with Type 2 Diabetes: Nonalcoholic Fatty Disease Cohort Study. Can. J. Diabetes 2020, 44, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Ceriello, A. Postprandial Hyperglycemia and Diabetes Complications: Is it Time to Treat? Diabetes 2005, 54, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Spínola, V.; Llorent-Martínez, E.J.; Castilho, P.C. Polyphenols of Myrica faya Inhibit Key Enzymes Linked to Type II Diabetes and Obesity and Formation of Advanced Glycation End-Products (In Vitro): Potential Role in the Prevention of Diabetic Complications. Food Res. Int. 2019, 116, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Delgado, F.; Katsiki, N.; Lopez-Miranda, J.; Perez-Martinez, P. Dietary Habits, Lipoprotein Metabolism and Cardiovascular Disease: From Individual Foods to Dietary Patterns. Crit. Rev. Food Sci. Nutr. 2021, 61, 1651–1669. [Google Scholar] [CrossRef] [PubMed]
- Etxeberria, U.; de la Garza, A.L.; Campión, J.; Martínez, J.A.; Milagro, F.I. Antidiabetic Effects of Natural Plant Extracts via Inhibition of Carbohydrate Hydrolysis Enzymes with Emphasis on Pancreatic Alpha Amylase. Expert Opin. Ther. Targets 2012, 16, 269–297. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, X.; Hu, M.; Zhang, D.; Yuan, S.; Li, P.; Feng, L. Medicinal and Edible Plants in the Treatment of Dyslipidemia: Advances and Prospects. Chin. Med. 2022, 17, 113. [Google Scholar] [CrossRef]
- Zhang, H.F.; Yang, X.H. Asian Medicine: Protect Rare Plants. Nature 2012, 482, 35. [Google Scholar] [CrossRef]
- The State Pharmacopoeia Commission of the People’s Republic of China. Pharmacopoeia of the People’s Republic of China; Chinese Medical Science Press: Beijing, China, 2020; Volume I. [Google Scholar]
- Zhang, H.F.; Yang, X.H.; Zhao, L.D.; Wang, Y. Ultrasonic-Assisted Extraction of Epimedin C from Fresh Leaves of Epimedium and Extraction Mechanism. Innov. Food Sci. Emerg. Technol. 2009, 10, 54–60. [Google Scholar] [CrossRef]
- Xu, A.; Yao, M. The Mechanism of Improving Insulin Resistance of Epimedium in Type 2 Diabetic Rats. Chin. J. Clin. Res. 2012, 25, 731–733. [Google Scholar]
- Song, L.; Wang, L.-W.; Zhou, Y.-T.; Liu, Y.; Bi, Y.-N.; Zhou, K. The Mechanism of Anti-Osteoporosis Effect of Epimedin C In Vivo. Lishizhen Med. Mater. Med. Res. 2021, 32, 555–558. [Google Scholar] [CrossRef]
- Han, D.; Zhuang, Y.; Meng, J. Research Progress on the Role of SGK in the Development of Diabetes Mellitus. Chin. J. Diabetes 2023, 31, 394–397. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, M.; Fan, M.; Pan, S.; Li, S.; Chen, M.; Wang, H. Metabolomic-Proteomic Combination Analysis Reveals the Targets and Molecular Pathways Associated with Hydrogen Sulfide Alleviating NAFLD. Life Sci. 2021, 264, 118629. [Google Scholar] [CrossRef]
- Rahman, M.O.; Ahmed, S.; Mazumder, T.; Salam, A.; Baral, P.K.; Rana, F.; Mitra, S.; Hossain, S.; Rahman, R.; Hussain, S. A Comparative Evaluation of Cardiac and Neurological Safety Status of Two Commonly Used Oral Hypoglycaemic Agents in T2-DM Swiss Albino Mice Model. Metab. Open 2022, 14, 100191. [Google Scholar] [CrossRef]
- Liu, W.; Lv, X.; Huang, W.; Yao, W.; Gao, X. Characterization and Hypoglycemic Effect of a Neutral Polysaccharide Extracted from the Residue of Codonopsis pilosula. Carbohydr. Polym. 2018, 197, 215–226. [Google Scholar] [CrossRef]
- Geidenstam, N.; Danielsson, A.P.H.; Spégel, P.; Ridderstråle, M. Changes in Glucose-Elicited Blood Metabolite Responses Following Weight Loss and Long Term Weight Maintenance in Obese Individuals with Impaired Glucose Tolerance. Diabetes Res. Clin. Pract. 2016, 113, 187–197. [Google Scholar] [CrossRef]
- Jack, B.U.; Ramharack, P.; Malherbe, C.; Gabuza, K.; Joubert, E.; Pheiffer, C. Cyclopia intermedia (Honeybush) Induces Uncoupling Protein 1 and Peroxisome Proliferator-Activated Receptor Alpha Expression in Obese Diabetic Female Db/Db Mice. Int. J. Mol. Sci. 2023, 24, 3868. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, H.; Li, L.; Zhou, X.; Liu, Y.; Lai, J. Proteomic Analysis of Protective Effects of Epimedium Flavonoids against Ethanol-Induced Toxicity in Retinoic Acid-Treated SH-SY5Y Cells. Molecules 2022, 27, 1026. [Google Scholar] [CrossRef]
- Liu, S.; Wang, F.; Chen, H.; Yang, Z.; Ning, Y.; Chang, C.; Yang, D. New Insights into Radio-Resistance Mechanism Revealed by (Phospho)Proteome Analysis of Deinococcus radiodurans after Heavy Ion Irradiation. Int. J. Mol. Sci. 2023, 24, 14817. [Google Scholar] [CrossRef]
- Li, Y.X.; Lu, Y.P.; Tang, D.; Hu, B.; Zhang, Z.Y.; Wu, H.W.; Fan, L.J.; Cai, K.W.; Tang, C.; Zhang, Y.Q.; et al. Anthocyanin Improves Kidney Function in Diabetic Kidney Disease by Regulating Amino Acid Metabolism. J. Transl. Med. 2022, 20, 510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, G.; Zhu, Y.; Zhang, D.; Xia, J.; Li, W. Effects of NAFLD Act on Glucose, insulin Metabolism and Liver Glycogen Synthesis in Rats. J. Southeast Univ. Med. Sci. Ed. 2020, 39, 420–424. [Google Scholar] [CrossRef]
- Winzell, M.S.; Ahrén, B. The High-Fat Diet-Fed Mouse: A Model for Studying Mechanisms and Treatment of Impaired Glucose Tolerance and Type 2 Diabetes. Diabetes 2004, 53, S215–S219. [Google Scholar] [CrossRef] [PubMed]
- McGarry, J.D. Banting Lecture 2001: Dysregulation of Fatty Acid Metabolism in the Etiology of Type 2 Diabetes. Diabetes 2002, 51, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.X.; Yang, X.H.; Zhang, H.F.; Feng, J.; Zhang, M.Y. Chemical Structure, Hypoglycemic Activity, and Mechanism of Action of Selenium Polysaccharides. Biol. Trace Elem. Res. 2022, 200, 4404–4418. [Google Scholar] [CrossRef] [PubMed]
- San Martin, A.; Foncea, R.; Laurindo, F.R.; Ebensperger, R.; Griendling, K.K.; Leighton, F. Nox1-Based NADPH Oxidase-Derived Superoxide Is Required for VSMC Activation by Advanced Glycation End-Products. Free Radic. Biol. Med. 2007, 42, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, S.; Maeda, S.; Matsui, T.; Ueda, S.; Fukami, K.; Okuda, S. Role of Advanced Glycation End Products (AGEs) and Oxidative Stress in Vascular Complications in Diabetes. Biochim. Biophys. Acta 2012, 1820, 663–671. [Google Scholar] [CrossRef]
- Stefan, N.; Cusi, K. A Global View of the Interplay between Non-Alcoholic Fatty Liver Disease and Diabetes. Lancet Diabetes Endocrinol. 2022, 10, 284–296. [Google Scholar] [CrossRef]
- Kuzmenko, D.I.; Udintsev, S.N.; Klimentyeva, T.K.; Serebrov, V.Y. Oxidative Stress in Adipose Tissue as a Primary Link in Pathogenesis of Insulin Resistance. Biomed. Khim. 2016, 62, 14–21. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef]
- Wang, L.; Song, B. The Research Perspective of Intestinal Cholesterol Absorption. Chem. Life 2014, 34, 318–328. [Google Scholar] [CrossRef]
- Subramaniam, K.; Babu, L.A.; Shah, N. A Case of Premature and Recurrent Myocardial Infarction Associated with ABCA.1 Gene Mutation. J. Postgrad. Med. 2021, 67, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, C.; Tang, C. Regulation Mechanism of Intracellular Cholesterol Level. Prog. Biochem. Biophys. 2022, 49, 292–302. [Google Scholar] [CrossRef]
- Yang, M.; Sui, D.; Chen, W.; Yang, M.; Yu, D. Study on Hypoglycemic Effect and Mechanism of Total Flavonoids of Propolis in STZ Diabetic Rats. J. Chin. Med. Mater. 2014, 37, 1623–1626. [Google Scholar] [CrossRef]
- Yuan, Q.; Yu, Y.; Wang, Y.; Xu, C. The Study of Effection and Mechanism of Tangkening Granule on Rats Blood Glucose and Blood Fat. Chin. J. Mod. Appl. Pharm. 2009, 26, 184–187. [Google Scholar] [CrossRef]
- Xiao, C.; Wu, Q.; Xie, Y.; Tan, J.; Ding, Y.; Bai, L. Hypoglycemic Mechanisms of Ganoderma lucidum Polysaccharides F31 in Db/Db Mice via RNA-Seq and iTRAQ. Food Funct. 2018, 9, 6495–6507. [Google Scholar] [CrossRef]
- Beale, E.G.; Hammer, R.E.; Antoine, B.; Forest, C. Disregulated Glyceroneogenesis: PCK1 as a Candidate Diabetes and Obesity Gene. Trends Endocrinol. Metab. 2004, 15, 129–135. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, Y.; Zhang, G.; Deng, H.; Wang, X.; Tuo, L.; Chen, C.; Pan, X.; Wu, K.; Fan, J.; et al. Deficiency of Gluconeogenic Enzyme PCK1 Promotes Metabolic-Associated Fatty Liver Disease through PI3K/AKT/PDGF Axis Activation in Male Mice. Nat. Commun. 2023, 14, 1402. [Google Scholar] [CrossRef]
- Guan, M.; Qu, L.; Tan, W.; Chen, L.; Wong, C.W. Hepatocyte Nuclear Factor-4 Alpha Regulates Liver Triglyceride Metabolism in Part through Secreted Phospholipase A2 GXIIB. Hepatology 2011, 53, 458–466. [Google Scholar] [CrossRef]
- Fisher, E.; Lake, E.; McLeod, R.S. Apolipoprotein B100 Quality Control and the Regulation of Hepatic Very Low Density Lipoprotein Secretion. J. Biomed. Res. 2014, 28, 178–193. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, M.; Fu, X.; Liu, R.; Sun, C.; Pan, H.; Wong, C.W.; Guan, M. Activation of Farnesoid X Receptor Promotes Triglycerides Lowering by Suppressing Phospholipase A2 G12B Expression. Mol. Cell Endocrinol. 2016, 436, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, X.A.; Wang, D.W. The Roles of CYP450 Epoxygenases and Metabolites, Epoxyeicosatrienoic Acids, in Cardiovascular and Malignant Diseases. Adv. Drug Deliv. Rev. 2011, 63, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.; Zhou, Y.; Su, W.; Ruan, X.; Wang, B.; Zheng, F.; Warner, M.; Gustafsson, J.-Å.; Guan, Y. Ablation of Cytochrome P450 Omega-Hydroxylase 4A14 Gene Attenuates Hepatic Steatosis and Fibrosis. Proc. Natl. Acad. Sci. USA 2017, 114, 3181–3185. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, L.; Li, D.; Wu, Q.; Zhang, Y.; Gao, M.; Ji, A.; Jiang, Q.; Chen, R.; Zhang, R.; et al. Real Ambient Particulate Matter-Induced Lipid Metabolism Disorder: Roles of Peroxisome Proliferators-Activated Receptor Alpha. Ecotoxicol. Environ. Saf. 2022, 231, 113173. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Liu, Z.; Yang, X.; Feng, J.; Gins, M.S.; Yan, T.; Han, L.; Zhang, H. The Mechanism Underlying the Hypoglycemic Effect of Epimedin C on Mice with Type 2 Diabetes Mellitus Based on Proteomic Analysis. Nutrients 2024, 16, 25. https://doi.org/10.3390/nu16010025
Zhou X, Liu Z, Yang X, Feng J, Gins MS, Yan T, Han L, Zhang H. The Mechanism Underlying the Hypoglycemic Effect of Epimedin C on Mice with Type 2 Diabetes Mellitus Based on Proteomic Analysis. Nutrients. 2024; 16(1):25. https://doi.org/10.3390/nu16010025
Chicago/Turabian StyleZhou, Xuexue, Ziqi Liu, Xiaohua Yang, Jing Feng, Murat Sabirovich Gins, Tingyu Yan, Lei Han, and Huafeng Zhang. 2024. "The Mechanism Underlying the Hypoglycemic Effect of Epimedin C on Mice with Type 2 Diabetes Mellitus Based on Proteomic Analysis" Nutrients 16, no. 1: 25. https://doi.org/10.3390/nu16010025
APA StyleZhou, X., Liu, Z., Yang, X., Feng, J., Gins, M. S., Yan, T., Han, L., & Zhang, H. (2024). The Mechanism Underlying the Hypoglycemic Effect of Epimedin C on Mice with Type 2 Diabetes Mellitus Based on Proteomic Analysis. Nutrients, 16(1), 25. https://doi.org/10.3390/nu16010025




