The Crosstalk between Gut Microbiota and White Adipose Tissue Mitochondria in Obesity
Abstract
1. Introduction
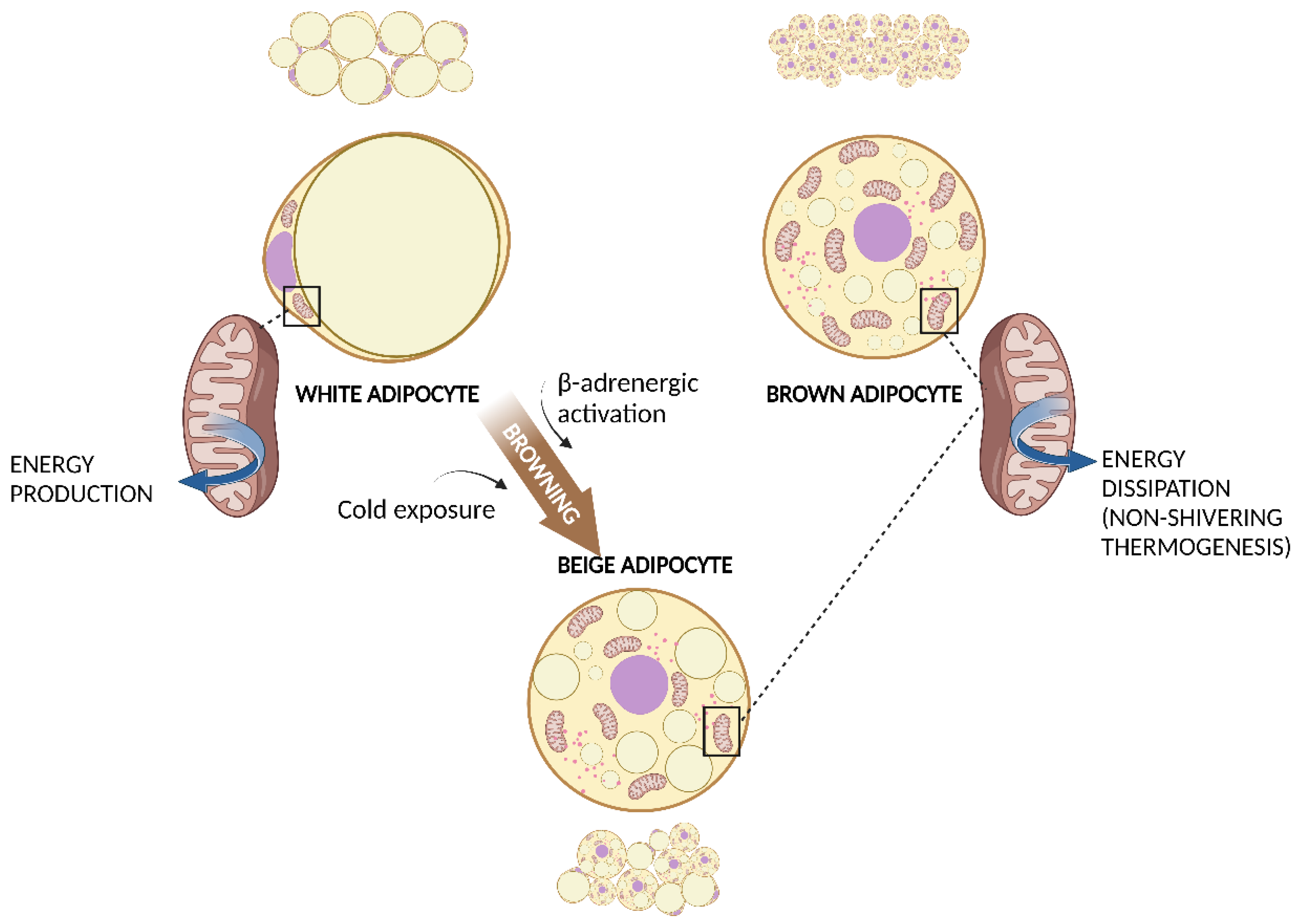
2. White and Brown Adipose Tissue
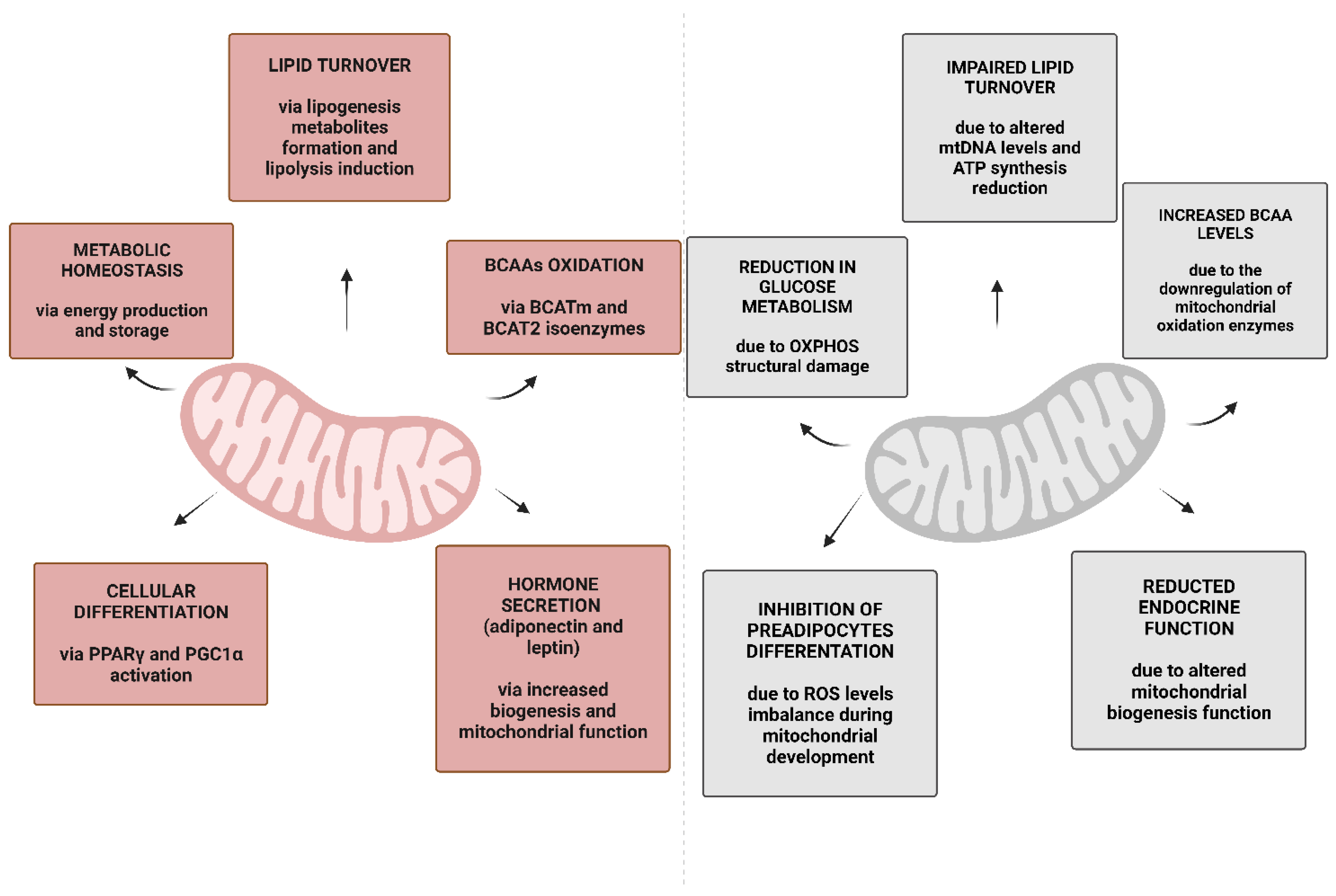
3. Mitochondria in WAT and BAT
4. Obesity and Mitochondrial Metabolism in WAT and BAT
5. Connections between Gut Microbiota and AT Mitochondria in Obesity
5.1. Gut Microbiota
5.2. Gut Microbiota Changes in Obesity
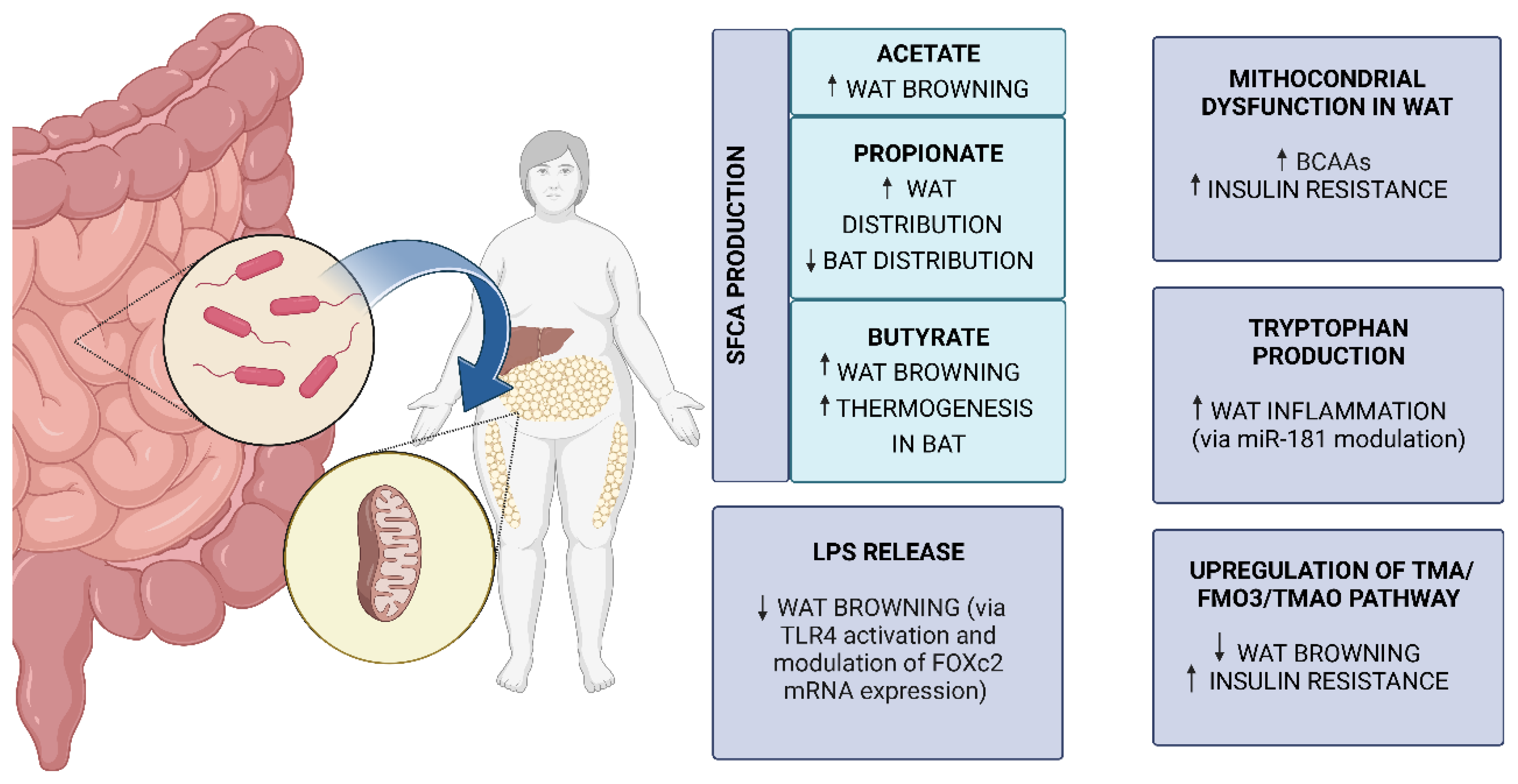
5.3. Molecular Mechanisms of Gut Microbiota and AT Mitochondria Crosstalk in Obesity
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 20 March 2022).
- Janssen, F.; Bardoutsos, A.; Vidra, N. Obesity Prevalence in the Long-Term Future in 18 European Countries and in the USA. Obes. Facts 2020, 13, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Gluvic, Z.; Zaric, B.; Resanovic, I.; Obradovic, M.; Mitrovic, A.; Radak, D.; Isenovic, E.R. Link between Metabolic Syndrome and Insulin Resistance. Curr. Vasc. Pharmacol. 2017, 15, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Lafontan, M. Adipose tissue and adipocyte dysregulation. Diabetes Metab. 2014, 40, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Tune, J.D.; Goodwill, A.G.; Sassoon, D.J.; Mather, K.J. Cardiovascular consequences of metabolic syndrome. Transl. Res. 2017, 183, 57–70. [Google Scholar] [CrossRef]
- Frühbeck, G.; Busetto, L.; Dicker, D.; Yumuk, V.; Goossens, G.H.; Hebebrand, J.; Halford, J.G.C.; Farpour-Lambert, N.J.; Blaak, E.E.; Woodward, E.; et al. The ABCD of Obesity: An EASO Position Statement on a Diagnostic Term with Clinical and Scientific Implications. Obes. Facts 2019, 12, 131–136. [Google Scholar] [CrossRef]
- Prasun, P. Mitochondrial dysfunction in metabolic syndrome. Biochim. Biophys. Acta. Mol. Basis. Dis. 2020, 1866, 165838. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Khan, M.T.; Nieuwdorp, M.; Bäckhed, F. Microbial modulation of insulin sensitivity. Cell Metab. 2014, 20, 753–760. [Google Scholar] [CrossRef]
- Colleluori, G.; Perugini, J.; Giordano, A.; Cinti, S. From Obesity to Diabetes: The Role of the Adipose Organ. Handb. Exp. Pharmacol. 2022, 274, 75–92. [Google Scholar] [CrossRef]
- Corvera, S. Cellular Heterogeneity in Adipose Tissues. Annu. Rev. Physiol. 2021, 83, 257–278. [Google Scholar] [CrossRef]
- Cinti, S. Adipose Organ Development and Remodeling. Compr. Physiol. 2018, 8, 1357–1431. [Google Scholar] [CrossRef]
- Smith, S.R.; Lovejoy, J.C.; Greenway, F.; Ryan, D.; deJonge, L.; de la Bretonne, J.; Volafova, J.; Bray, G.A. Contributions of total body fat, abdominal subcutaneous adipose tissue compartments, and visceral adipose tissue to the metabolic complications of obesity. Metabolism 2001, 50, 425–435. [Google Scholar] [CrossRef]
- Geer, E.B.; Shen, W. Gender differences in insulin resistance, body composition, and energy balance. Gend. Med. 2009, 6 (Suppl. S1), 60–75. [Google Scholar] [CrossRef] [PubMed]
- Caton, P.W.; Evans, E.A.; Philpott, M.P.; Hannen, R.F. Can the skin make you fat? A role for the skin in regulating adipose tissue function and whole-body glucose and lipid homeostasis. Curr. Opin. Pharmacol. 2017, 37, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Guglielmi, V.; Sbraccia, P. Obesity phenotypes: Depot-differences in adipose tissue and their clinical implications. Eat Weight Disord. 2018, 23, 3–14. [Google Scholar] [CrossRef]
- Zhang, C.; Rexrode, K.M.; van Dam, R.M.; Li, T.Y.; Hu, F.B. Abdominal obesity and the risk of all-cause, cardiovascular, and cancer mortality: Sixteen years of follow-up in US women. Circulation 2008, 117, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Sbraccia, P.; D’Adamo, M.; Guglielmi, V. Is type 2 diabetes an adiposity-based metabolic disease? From the origin of insulin resistance to the concept of dysfunctional adipose tissue. Eat Weight Disord. 2021, 26, 2429–2441. [Google Scholar] [CrossRef]
- Guglielmi, V.; Sbraccia, P. Epicardial adipose tissue: At the heart of the obesity complications. Acta Diabetol. 2017, 54, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Venteclef, N.; Guglielmi, V.; Balse, E.; Gaborit, B.; Cotillard, A.; Atassi, F.; Amour, J.; Leprince, P.; Dutour, A.; Clément, K.; et al. Human epicardial adipose tissue induces fibrosis of the atrial myocardium through the secretion of adipo-fibrokines. Eur. Heart J. 2015, 36, 795–805a. [Google Scholar] [CrossRef]
- Maliszewska, K.; Kretowski, A. Brown Adipose Tissue and Its Role in Insulin and Glucose Homeostasis. Int. J. Mol. Sci. 2021, 22, 1530. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef]
- Guglielmi, V.; Cardellini, M.; Cinti, F.; Corgosinho, F.; Cardolini, I.; D’Adamo, M.; Zingaretti, M.C.; Bellia, A.; Lauro, D.; Gentileschi, P.; et al. Omental adipose tissue fibrosis and insulin resistance in severe obesity. Nutr. Diabetes 2015, 5, e175. [Google Scholar] [CrossRef] [PubMed]
- Crewe, C.; An, Y.A.; Scherer, P.E. The ominous triad of adipose tissue dysfunction: Inflammation, fibrosis, and impaired angiogenesis. J. Clin. Investig. 2017, 127, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef]
- Flier, J.S. The adipocyte: Storage depot or node on the energy information superhighway? Cell 1995, 80, 15–18. [Google Scholar] [CrossRef]
- Barbarroja, N.; Lopez-Pedrera, C.; Garrido-Sanchez, L.; Mayas, M.D.; Oliva-Olivera, W.; Bernal-Lopez, M.R.; El Bekay, R.; Tinahones, F.J. Progression from high insulin resistance to type 2 diabetes does not entail additional visceral adipose tissue inflammation. PLoS ONE 2012, 7, e48155. [Google Scholar] [CrossRef]
- Giralt, M.; Villarroya, F. White, brown, beige/brite: Different adipose cells for different functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef]
- Shabalina, I.G.; Petrovic, N.; de Jong, J.M.; Kalinovich, A.V.; Cannon, B.; Nedergaard, J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013, 5, 1196–1203. [Google Scholar] [CrossRef]
- De Matteis, R.; Lucertini, F.; Guescini, M.; Polidori, E.; Zeppa, S.; Stocchi, V.; Cinti, S.; Cuppini, R. Exercise as a new physiological stimulus for brown adipose tissue activity. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 582–590. [Google Scholar] [CrossRef]
- Georgiev, A.; Granata, C.; Roden, M. The role of mitochondria in the pathophysiology and treatment of common metabolic diseases in humans. Am. J. Physiol. Cell Physiol. 2022, 322, C1248–C1259. [Google Scholar] [CrossRef] [PubMed]
- Goldenthal, M.J.; Marín-García, J. Mitochondrial signaling pathways: A receiver/integrator organelle. Mol. Cell Biochem. 2004, 262, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kakkar, P.; Singh, B.K. Mitochondria: A hub of redox activities and cellular distress control. Mol. Cell Biochem. 2007, 305, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.X.; Thompson, K.; Taylor, R.W.; Oláhová, M. Mitochondrial OXPHOS Biogenesis: Co-Regulation of Protein Synthesis, Import, and Assembly Pathways. Int. J. Mol. Sci. 2020, 21, 3820. [Google Scholar] [CrossRef]
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Ma, K.; Chen, G.; Li, W.; Kepp, O.; Zhu, Y.; Chen, Q. Mitophagy, Mitochondrial Homeostasis, and Cell Fate. Front. Cell Dev. Biol. 2020, 8, 467. [Google Scholar] [CrossRef]
- Kusminski, C.M.; Scherer, P.E. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 2012, 23, 435–443. [Google Scholar] [CrossRef]
- Heinonen, S.; Jokinen, R.; Rissanen, A.; Pietiläinen, K.H. White adipose tissue mitochondrial metabolism in health and in obesity. Obes. Rev. 2020, 21, e12958. [Google Scholar] [CrossRef]
- De Pauw, A.; Tejerina, S.; Raes, M.; Keijer, J.; Arnould, T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am. J. Pathol. 2009, 175, 927–939. [Google Scholar] [CrossRef]
- Wilson-Fritch, L.; Burkart, A.; Bell, G.; Mendelson, K.; Leszyk, J.; Nicoloro, S.; Czech, M.; Corvera, S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell Biol. 2003, 23, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 2000, 16, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Wilson-Fritch, L.; Nicoloro, S.; Chouinard, M.; Lazar, M.A.; Chui, P.C.; Leszyk, J.; Straubhaar, J.; Czech, M.P.; Corvera, S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 2004, 114, 1281–1289. [Google Scholar] [CrossRef]
- Castro, J.P.; Grune, T.; Speckmann, B. The two faces of reactive oxygen species (ROS) in adipocyte function and dysfunction. Biol. Chem. 2016, 397, 709–724. [Google Scholar] [CrossRef]
- Koh, E.H.; Park, J.Y.; Park, H.S.; Jeon, M.J.; Ryu, J.W.; Kim, M.; Kim, S.Y.; Kim, M.S.; Kim, S.W.; Park, I.S.; et al. Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes 2007, 56, 2973–2981. [Google Scholar] [CrossRef]
- Nakajima, T.; Yokota, T.; Shingu, Y.; Yamada, A.; Iba, Y.; Ujihira, K.; Wakasa, S.; Ooka, T.; Takada, S.; Shirakawa, R.; et al. Impaired mitochondrial oxidative phosphorylation capacity in epicardial adipose tissue is associated with decreased concentration of adiponectin and severity of coronary atherosclerosis. Sci. Rep. 2019, 9, 3535. [Google Scholar] [CrossRef] [PubMed]
- Blanquer-Rosselló, M.M.; Santandreu, F.M.; Oliver, J.; Roca, P.; Valle, A. Leptin Modulates Mitochondrial Function, Dynamics and Biogenesis in MCF-7 Cells. J. Cell Biochem. 2015, 116, 2039–2048. [Google Scholar] [CrossRef]
- Deveaud, C.; Beauvoit, B.; Salin, B.; Schaeffer, J.; Rigoulet, M. Regional differences in oxidative capacity of rat white adipose tissue are linked to the mitochondrial content of mature adipocytes. Mol. Cell Biochem. 2004, 267, 157–166. [Google Scholar] [CrossRef]
- Lindinger, A.; Peterli, R.; Peters, T.; Kern, B.; von Flüe, M.; Calame, M.; Hoch, M.; Eberle, A.N.; Lindinger, P.W. Mitochondrial DNA content in human omental adipose tissue. Obes. Surg. 2010, 20, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S. Transdifferentiation properties of adipocytes in the adipose organ. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E977–E986. [Google Scholar] [CrossRef]
- Carpentier, A.C.; Blondin, D.P.; Virtanen, K.A.; Richard, D.; Haman, F.; Turcotte, É.E. Brown Adipose Tissue Energy Metabolism in Humans. Front. Endocrinol. 2018, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Forner, F.; Kumar, C.; Luber, C.A.; Fromme, T.; Klingenspor, M.; Mann, M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab. 2009, 10, 324–335. [Google Scholar] [CrossRef]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef] [PubMed]
- Nedergaard, J.; Cannon, B. The changed metabolic world with human brown adipose tissue: Therapeutic visions. Cell Metab. 2010, 11, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Chaves, V.E.; Frasson, D.; Kawashita, N.H. Several agents and pathways regulate lipolysis in adipocytes. Biochimie 2011, 93, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Knottnerus, S.J.G.; Bleeker, J.C.; Wüst, R.C.I.; Ferdinandusse, S.; IJlst, L.; Wijburg, F.A.; Wanders, R.J.A.; Visser, G.; Houtkooper, R.H. Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle. Rev. Endocr. Metab. Disord. 2018, 19, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Crichton, P.G.; Lee, Y.; Kunji, E.R. The molecular features of uncoupling protein 1 support a conventional mitochondrial carrier-like mechanism. Biochimie 2017, 134, 35–50. [Google Scholar] [CrossRef]
- Lustig, R.H.; Collier, D.; Kassotis, C.; Roepke, T.A.; Kim, M.J.; Blanc, E.; Barouki, R.; Bansal, A.; Cave, M.C.; Chatterjee, S.; et al. Obesity I: Overview and molecular and biochemical mechanisms. Biochem. Pharmacol. 2022, 199, 115012. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, A.; Oh, K.J.; Lee, S.C.; Kim, W.K.; Bae, K.H. The Role of Adipose Tissue Mitochondria: Regulation of Mitochondrial Function for the Treatment of Metabolic Diseases. Int. J. Mol. Sci. 2019, 20, 4924. [Google Scholar] [CrossRef]
- Cedikova, M.; Kripnerová, M.; Dvorakova, J.; Pitule, P.; Grundmanova, M.; Babuska, V.; Mullerova, D.; Kuncova, J. Mitochondria in White, Brown, and Beige Adipocytes. Stem. Cells Int. 2016, 2016, 6067349. [Google Scholar] [CrossRef]
- Semple, R.K.; Crowley, V.C.; Sewter, C.P.; Laudes, M.; Christodoulides, C.; Considine, R.V.; Vidal-Puig, A.; O’Rahilly, S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obes. Relat. Metab. Disord. 2004, 28, 176–179. [Google Scholar] [CrossRef]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef]
- Srdić, B.; Stokić, E.; Korać, A.; Ukropina, M.; Veličković, K.; Breberina, M. Morphological characteristics of abdominal adipose tissue in normal-weight and obese women of different metabolic profiles. Exp. Clin. Endocrinol. Diabetes 2010, 118, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Pierce, K.A.; Jedrychowski, M.P.; Garrity, R.; Winther, S.; Vidoni, S.; Yoneshiro, T.; Spinelli, J.B.; Lu, G.Z.; Kazak, L.; et al. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature 2018, 560, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, R.S.; Delafiori, J.; Zuccoli, G.; Carregari, V.C.; Prado, T.P.; Morari, J.; Sidarta-Oliveira, D.; Solon, C.S.; Catharino, R.R.; Araujo, E.P.; et al. Exogenous succinate impacts mouse brown adipose tissue mitochondrial proteome and potentiates body mass reduction induced by liraglutide. Am. J. Physiol. Endocrinol. Metab. 2023, 324, E226–E240. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.; Rauch, B.; Szerafin, T.; Uray, K.; Tóth, A.; Bai, P. Nicotinamide-riboside shifts the differentiation of human primary white adipocytes to beige adipocytes impacting substrate preference and uncoupling respiration through SIRT1 activation and mitochondria-derived reactive species production. Front. Cell Dev. Biol. 2022, 10, 979330. [Google Scholar] [CrossRef] [PubMed]
- Da Eira, D.; Jani, S.; Ceddia, R.B. An obesogenic diet impairs uncoupled substrate oxidation and promotes whitening of the brown adipose tissue in rats. J. Physiol. 2023, 601, 69–82. [Google Scholar] [CrossRef]
- Flatt, J.P. Conversion of carbohydrate to fat in adipose tissue: An energy-yielding and, therefore, self-limiting process. J. Lipid Res. 1970, 11, 131–143. [Google Scholar] [CrossRef]
- Choi, C.S.; Ghoshal, P.; Srinivasan, M.; Kim, S.; Cline, G.; Patel, M.S. Liver-specific pyruvate dehydrogenase complex deficiency upregulates lipogenesis in adipose tissue and improves peripheral insulin sensitivity. Lipids 2010, 45, 987–995. [Google Scholar] [CrossRef]
- Yeh, L.A.; Lee, K.H.; Kim, K.H. Regulation of rat liver acetyl-CoA carboxylase. Regulation of phosphorylation and inactivation of acetyl-CoA carboxylase by the adenylate energy charge. J. Biol. Chem. 1980, 255, 2308–2314. [Google Scholar] [CrossRef]
- Pietiläinen, K.H.; Naukkarinen, J.; Rissanen, A.; Saharinen, J.; Ellonen, P.; Keränen, H.; Suomalainen, A.; Götz, A.; Suortti, T.; Yki-Järvinen, H.; et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: Pathways behind acquired obesity. PLoS Med. 2008, 5, e51. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Harada, Y.; Yoshikawa, T.; Dai, P. Mitochondrial Energy Metabolism in the Regulation of Thermogenic Brown Fats and Human Metabolic Diseases. Int. J. Mol. Sci. 2023, 24, 1352. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Dai, W.; Jang, M.J.; Medrano, L.; Li, Z.; Zhao, H.; Shao, M.; Tan, J.; Li, A.; Ning, T.; et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J. Clin. Investig. 2020, 130, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, L.; Zhang, L.; Yan, B.; Sun, T.; Guo, F.; Yin, X. Reduced Beige Adipogenic Potential in Subcutaneous Adipocytes Derived from Obese Chinese Individuals. Diabetes Metab. Syndr. Obes. 2020, 13, 2551–2562. [Google Scholar] [CrossRef]
- Saari, T.J.; Raiko, J.; U-Din, M.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.; Haaparanta-Solin, M.; Nuutila, P.; Virtanen, K.A. Basal and cold-induced fatty acid uptake of human brown adipose tissue is impaired in obesity. Sci. Rep. 2020, 10, 14373. [Google Scholar] [CrossRef]
- Derrien, M.; van Hylckama Vlieg, J.E. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Natividad, J.M.; Verdu, E.F. Modulation of intestinal barrier by intestinal microbiota: Pathological and therapeutic implications. Pharmacol. Res. 2013, 69, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Bastiaanssen, T.F.S.; Cowan, C.S.M.; Claesson, M.J.; Dinan, T.G.; Cryan, J.F. Making Sense of … the Microbiome in Psychiatry. Int. J. Neuropsychopharmacol. 2019, 22, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.H.; Woo, Y.S.; Lee, S.Y.; Bahk, W.M. The Brain-Gut-Microbiome Axis in Psychiatry. Int. J. Mol. Sci. 2020, 21, 7122. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F.; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Sepp, E.; Smidt, I.; Rööp, T.; Štšepetova, J.; Kõljalg, S.; Mikelsaar, M.; Soidla, I.; Ainsaar, M.; Kolk, H.; Vallas, M.; et al. Comparative Analysis of Gut Microbiota in Centenarians and Young People: Impact of Eating Habits and Childhood Living Environment. Front. Cell Infect Microbiol. 2022, 12, 851404. [Google Scholar] [CrossRef]
- Badal, V.D.; Vaccariello, E.D.; Murray, E.R.; Yu, K.E.; Knight, R.; Jeste, D.V.; Nguyen, T.T. The Gut Microbiome, Aging, and Longevity: A Systematic Review. Nutrients 2020, 12, 3759. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Cong, J.; Zhou, P.; Zhang, R. Intestinal Microbiota-Derived Short Chain Fatty Acids in Host Health and Disease. Nutrients 2022, 14, 1977. [Google Scholar] [CrossRef]
- McNeil, N.I.; Cummings, J.H.; James, W.P. Short chain fatty acid absorption by the human large intestine. Gut 1978, 19, 819–822. [Google Scholar] [CrossRef]
- Topping, D.L.; Clifton, P.M. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Stumpff, F. A look at the smelly side of physiology: Transport of short chain fatty acids. Pflug. Arch. 2018, 470, 571–598. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef]
- Psichas, A.; Sleeth, M.L.; Murphy, K.G.; Brooks, L.; Bewick, G.A.; Hanyaloglu, A.C.; Ghatei, M.A.; Bloom, S.R.; Frost, G. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int. J. Obes. 2015, 39, 424–429. [Google Scholar] [CrossRef]
- Theodorakis, M.J.; Carlson, O.; Michopoulos, S.; Doyle, M.E.; Juhaszova, M.; Petraki, K.; Egan, J.M. Human duodenal enteroendocrine cells: Source of both incretin peptides, GLP-1 and GIP. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E550–E559. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Rey, F.E.; Faith, J.J.; Bain, J.; Muehlbauer, M.J.; Stevens, R.D.; Newgard, C.B.; Gordon, J.I. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 2010, 285, 22082–22090. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Lordan, C.; Thapa, D.; Ross, R.P.; Cotter, P.D. Potential for enriching next-generation health-promoting gut bacteria through prebiotics and other dietary components. Gut Microbes 2020, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Shortt, C.; Hasselwander, O.; Meynier, A.; Nauta, A.; Fernández, E.N.; Putz, P.; Rowland, I.; Swann, J.; Türk, J.; Vermeiren, J.; et al. Systematic review of the effects of the intestinal microbiota on selected nutrients and non-nutrients. Eur. J. Nutr. 2018, 57, 25–49. [Google Scholar] [CrossRef]
- Felig, P.; Marliss, E.; Cahill, G.F., Jr. Plasma amino acid levels and insulin secretion in obesity. N. Engl. J. Med. 1969, 281, 811–816. [Google Scholar] [CrossRef]
- Siddik, M.A.B.; Shin, A.C. Recent Progress on Branched-Chain Amino Acids in Obesity, Diabetes, and Beyond. Endocrinol. Metab. 2019, 34, 234–246. [Google Scholar] [CrossRef]
- Dai, Z.L.; Wu, G.; Zhu, W.Y. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768–1786. [Google Scholar] [CrossRef]
- Gojda, J.; Cahova, M. Gut Microbiota as the Link between Elevated BCAA Serum Levels and Insulin Resistance. Biomolecules 2021, 11, 1414. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef]
- Zacarías, M.F.; Collado, M.C.; Gómez-Gallego, C.; Flinck, H.; Aittoniemi, J.; Isolauri, E.; Salminen, S. Pregestational overweight and obesity are associated with differences in gut microbiota composition and systemic inflammation in the third trimester. PLoS ONE 2018, 13, e0200305. [Google Scholar] [CrossRef]
- Patil, D.P.; Dhotre, D.P.; Chavan, S.G.; Sultan, A.; Jain, D.S.; Lanjekar, V.B.; Gangawani, J.; Shah, P.S.; Todkar, J.S.; Shah, S.; et al. Molecular analysis of gut microbiota in obesity among Indian individuals. J. Biosci. 2012, 37, 647–657. [Google Scholar] [CrossRef]
- Davis, S.C.; Yadav, J.S.; Barrow, S.D.; Robertson, B.K. Gut microbiome diversity influenced more by the Westernized dietary regime than the body mass index as assessed using effect size statistic. Microbiologyopen 2017, 6, e00476. [Google Scholar] [CrossRef]
- Duncan, S.H.; Lobley, G.E.; Holtrop, G.; Ince, J.; Johnstone, A.M.; Louis, P.; Flint, H.J. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. 2008, 32, 1720–1724. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Van Hul, M.; Cani, P.D. The gut microbiota in obesity and weight management: Microbes as friends or foe? Nat. Rev. Endocrinol. 2023, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Yun, K.E.; Kim, J.; Park, E.; Chang, Y.; Ryu, S.; Kim, H.L.; Kim, H.N. Gut microbiota and metabolic health among overweight and obese individuals. Sci. Rep. 2020, 10, 19417. [Google Scholar] [CrossRef]
- Tremaroli, V.; Karlsson, F.; Werling, M.; Ståhlman, M.; Kovatcheva-Datchary, P.; Olbers, T.; Fändriks, L.; le Roux, C.W.; Nielsen, J.; Bäckhed, F. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab. 2015, 22, 228–238. [Google Scholar] [CrossRef]
- Palleschi, S.; Guglielmi, V.; Nisticò, L.; Ferreri, C.; Tabolacci, C.; Facchiano, F.; Iorio, E.; Giuliani, A.; Brescianini, S.; Medda, E.; et al. A multi-marker integrative analysis reveals benefits and risks of bariatric surgery. Sci. Rep. 2022, 12, 18877. [Google Scholar] [CrossRef]
- Martínez-Sánchez, M.A.; Balaguer-Román, A.; Fernández-Ruiz, V.E.; Almansa-Saura, S.; García-Zafra, V.; Ferrer-Gómez, M.; Frutos, M.D.; Queipo-Ortuño, M.I.; Ruiz-Alcaraz, A.J.; Núñez-Sánchez, M.Á.; et al. Plasma short-chain fatty acid changes after bariatric surgery in patients with severe obesity. Surg. Obes. Relat. Dis. 2023. [Google Scholar] [CrossRef]
- Ballard, J.W.O.; Towarnicki, S.G. Mitochondria, the gut microbiome and ROS. Cell Signal 2020, 75, 109737. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Duan, L. The role of microbiota-mitochondria crosstalk in pathogenesis and therapy of intestinal diseases. Pharmacol. Res. 2022, 186, 106530. [Google Scholar] [CrossRef]
- Houghton, M.J.; Kerimi, A.; Mouly, V.; Tumova, S.; Williamson, G. Gut microbiome catabolites as novel modulators of muscle cell glucose metabolism. Faseb J. 2019, 33, 1887–1898. [Google Scholar] [CrossRef] [PubMed]
- Vezza, T.; Abad-Jiménez, Z.; Marti-Cabrera, M.; Rocha, M.; Víctor, V.M. Microbiota-Mitochondria Inter-Talk: A Potential Therapeutic Strategy in Obesity and Type 2 Diabetes. Antioxidants 2020, 9, 848. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wang, H.; Xie, L.; Hu, F. Cross-Talk between Gut Microbiota and Adipose Tissues in Obesity and Related Metabolic Diseases. Front. Endocrinol. 2022, 13, 908868. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-Chain Fatty Acids Protect against High-Fat Diet-Induced Obesity via a PPARγ-Dependent Switch from Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef]
- Hu, J.; Kyrou, I.; Tan, B.K.; Dimitriadis, G.K.; Ramanjaneya, M.; Tripathi, G.; Patel, V.; James, S.; Kawan, M.; Chen, J.; et al. Short-Chain Fatty Acid Acetate Stimulates Adipogenesis and Mitochondrial Biogenesis via GPR43 in Brown Adipocytes. Endocrinology 2016, 157, 1881–1894. [Google Scholar] [CrossRef]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. A review on gut microbiota: A central factor in the pathophysiology of obesity. Lipids Health Dis. 2021, 20, 65. [Google Scholar] [CrossRef]
- Li, X.; Huang, J.; Yun, J.; Zhang, G.; Zhang, Y.; Zhao, M.; Zabed, H.M.; Ravikumar, Y.; Qi, X. d-Arabitol Ameliorates Obesity and Metabolic Disorders via the Gut Microbiota-SCFAs-WAT Browning Axis. J. Agric. Food Chem. 2023, 71, 522–534. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Jiang, Q.; Yin, Y. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol. Metab. 2021, 32, 159–169. [Google Scholar] [CrossRef]
- Tirosh, A.; Calay, E.S.; Tuncman, G.; Claiborn, K.C.; Inouye, K.E.; Eguchi, K.; Alcala, M.; Rathaus, M.; Hollander, K.S.; Ron, I.; et al. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci. Transl. Med. 2019, 11, eaav0120. [Google Scholar] [CrossRef]
- Jia, Z.; Chen, X.; Chen, J.; Zhang, L.; Oprescu, S.N.; Luo, N.; Xiong, Y.; Yue, F.; Kuang, S. ACSS3 in brown fat drives propionate catabolism and its deficiency leads to autophagy and systemic metabolic dysfunction. Clin. Transl. Med. 2022, 12, e665. [Google Scholar] [CrossRef]
- Hernández, M.A.G.; Canfora, E.E.; Jocken, J.W.E.; Blaak, E.E. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients 2019, 11, 1943. [Google Scholar] [CrossRef]
- Li, Z.; Gurung, M.; Rodrigues, R.R.; Padiadpu, J.; Newman, N.K.; Manes, N.P.; Pederson, J.W.; Greer, R.L.; Vasquez-Perez, S.; You, H.; et al. Microbiota and adipocyte mitochondrial damage in type 2 diabetes are linked by Mmp12+ macrophages. J. Exp. Med. 2022, 219, e20220017. [Google Scholar] [CrossRef]
- Moreno-Navarrete, J.M.; Fernandez-Real, J.M. The gut microbiota modulates both browning of white adipose tissue and the activity of brown adipose tissue. Rev. Endocr. Metab. Disord. 2019, 20, 387–397. [Google Scholar] [CrossRef]
- Lundgren, P.; Thaiss, C.A. The microbiome-adipose tissue axis in systemic metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G717–G724. [Google Scholar] [CrossRef] [PubMed]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L.; et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11, eaav1892. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Navarrete, J.M.; Serino, M.; Blasco-Baque, V.; Azalbert, V.; Barton, R.H.; Cardellini, M.; Latorre, J.; Ortega, F.; Sabater-Masdeu, M.; Burcelin, R.; et al. Gut Microbiota Interacts with Markers of Adipose Tissue Browning, Insulin Action and Plasma Acetate in Morbid Obesity. Mol. Nutr. Food. Res. 2018, 62, 1700721. [Google Scholar] [CrossRef] [PubMed]
| Mitochondrial Characteristics | WAT | BAT |
|---|---|---|
| Content | Lower | Higher |
| Dimension * | Smaller | Bigger |
| Shape * | Elongated | Spherical |
| Inner membrane cristae * | Less packed | More packed |
| UCP-1 expression | Lower | Higher |
| Main function | Energy storage and lipid homeostasis | Non-shivering thermogenesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colangeli, L.; Escobar Marcillo, D.I.; Simonelli, V.; Iorio, E.; Rinaldi, T.; Sbraccia, P.; Fortini, P.; Guglielmi, V. The Crosstalk between Gut Microbiota and White Adipose Tissue Mitochondria in Obesity. Nutrients 2023, 15, 1723. https://doi.org/10.3390/nu15071723
Colangeli L, Escobar Marcillo DI, Simonelli V, Iorio E, Rinaldi T, Sbraccia P, Fortini P, Guglielmi V. The Crosstalk between Gut Microbiota and White Adipose Tissue Mitochondria in Obesity. Nutrients. 2023; 15(7):1723. https://doi.org/10.3390/nu15071723
Chicago/Turabian StyleColangeli, Luca, David Israel Escobar Marcillo, Valeria Simonelli, Egidio Iorio, Tommaso Rinaldi, Paolo Sbraccia, Paola Fortini, and Valeria Guglielmi. 2023. "The Crosstalk between Gut Microbiota and White Adipose Tissue Mitochondria in Obesity" Nutrients 15, no. 7: 1723. https://doi.org/10.3390/nu15071723
APA StyleColangeli, L., Escobar Marcillo, D. I., Simonelli, V., Iorio, E., Rinaldi, T., Sbraccia, P., Fortini, P., & Guglielmi, V. (2023). The Crosstalk between Gut Microbiota and White Adipose Tissue Mitochondria in Obesity. Nutrients, 15(7), 1723. https://doi.org/10.3390/nu15071723








