Metagenomic Sequencing Identified Specific Bacteriophage Signature Discriminating between Healthy and Diarrheal Neonatal Piglets
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experiment
2.2. DNA Extraction and Metagenomic Sequencing
2.3. Virus Identification
2.4. Viral Taxonomy Classification
2.5. Functional Annotation of Viral Contigs
2.6. Host Prediction
2.7. Statistics Analysis
3. Results
3.1. Viral Contig Statistics
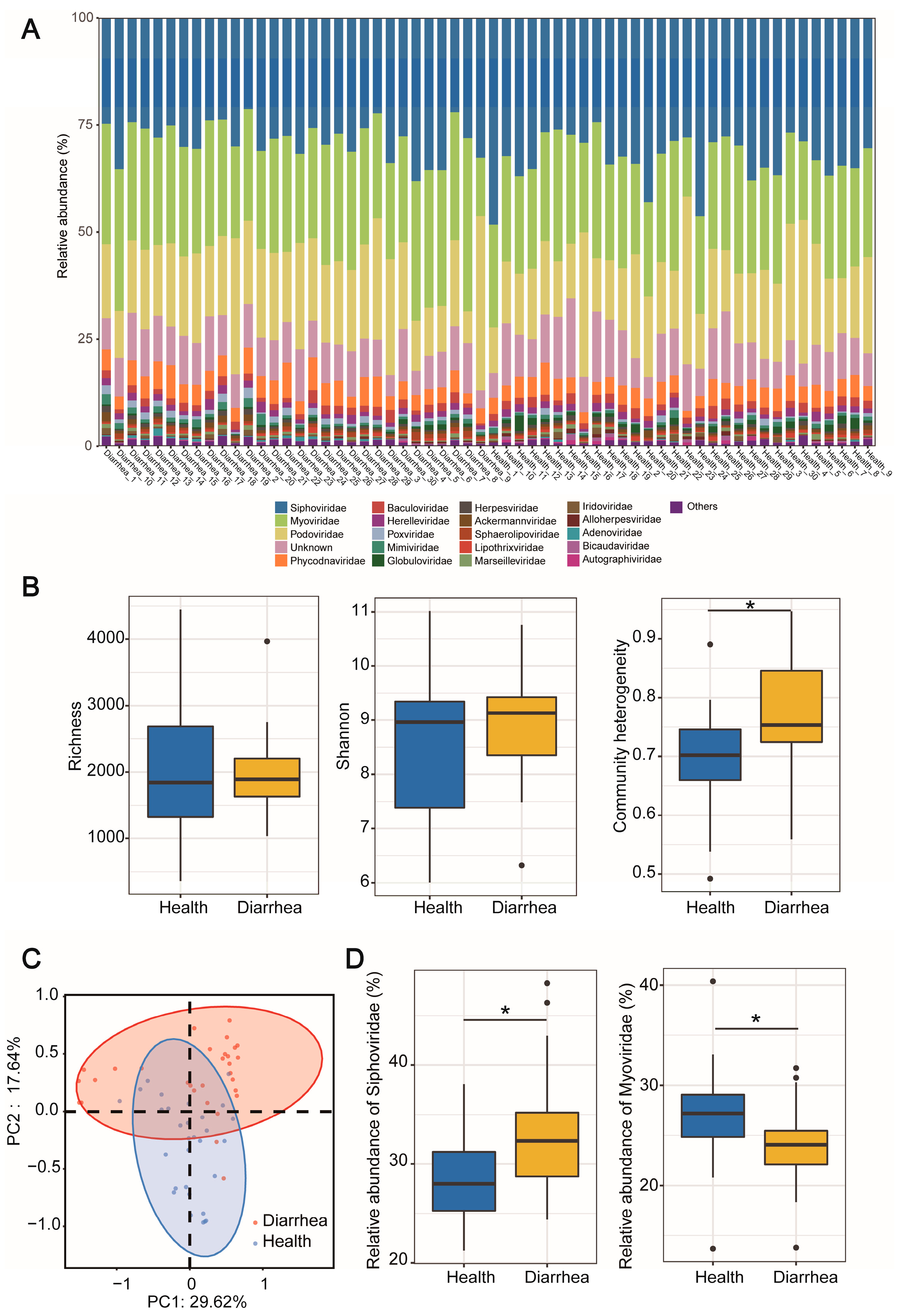
3.2. Taxonomy Composition of Piglet Gut Virome
3.3. Viral Diversity and Structure Differences between Gut Virome of Diarrheal and Healthy Piglets
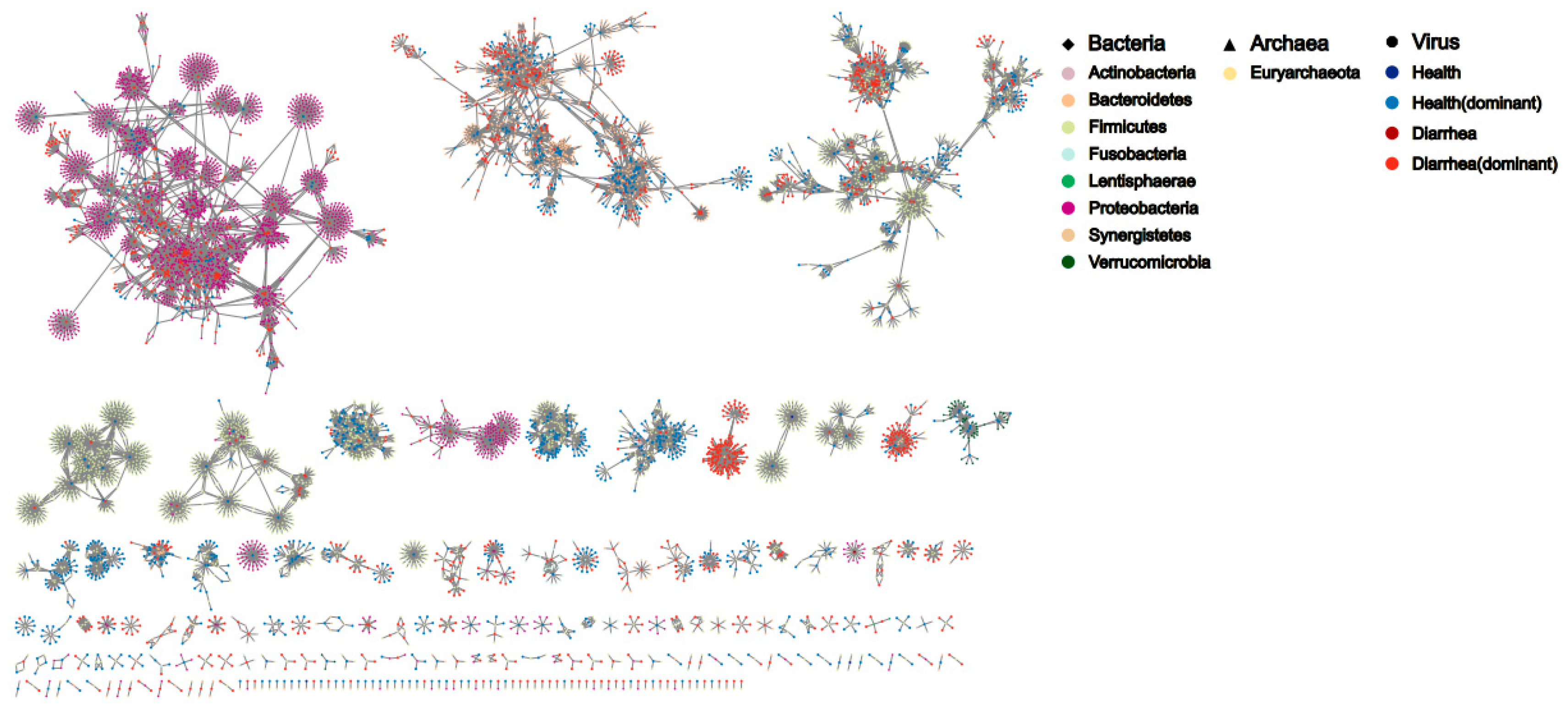
3.4. Virus–Host Interaction Differs between Diarrheal and Healthy Piglets
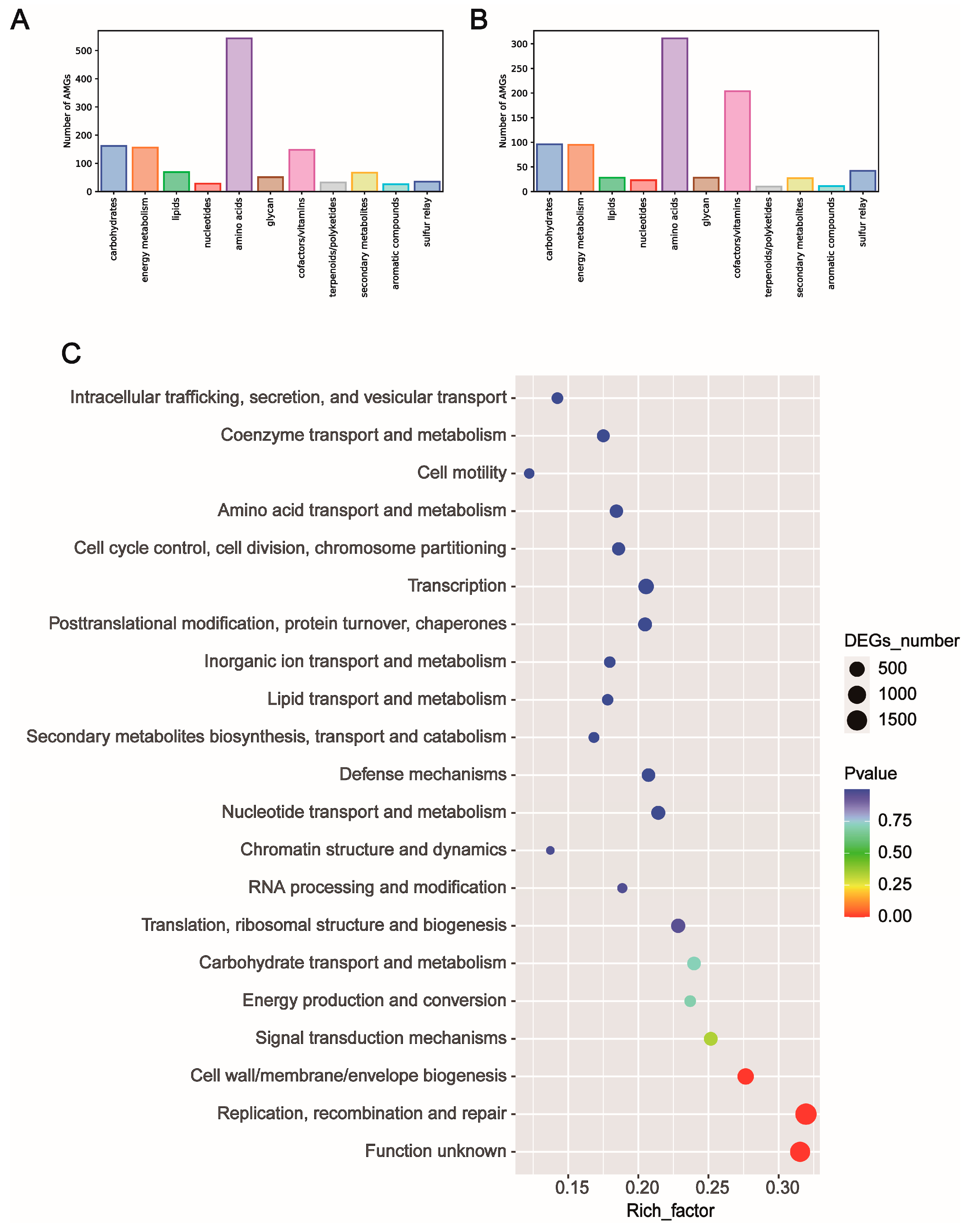
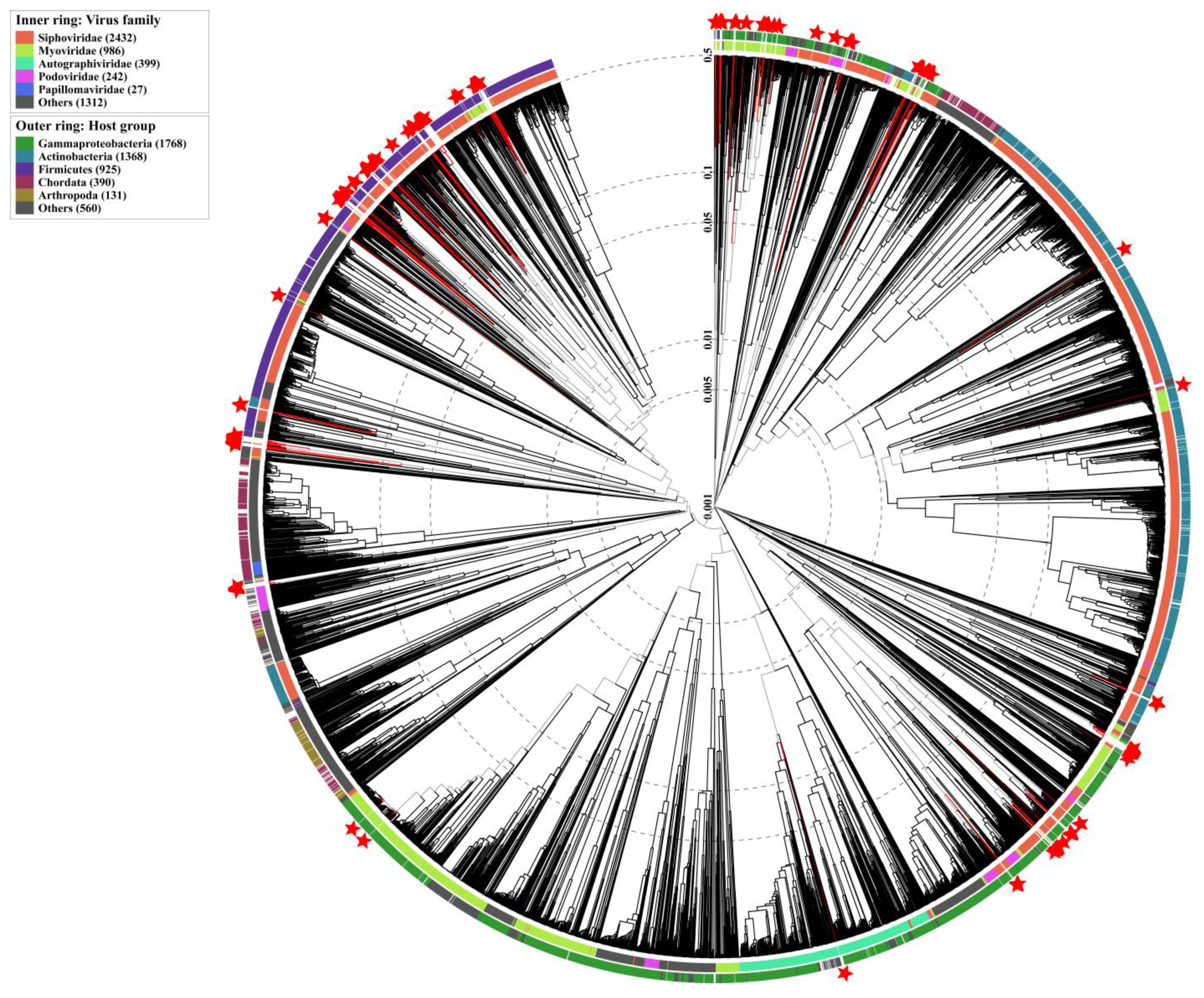
3.5. Functional Annotation and Phylogenetic Tree of Differential Viral Contigs
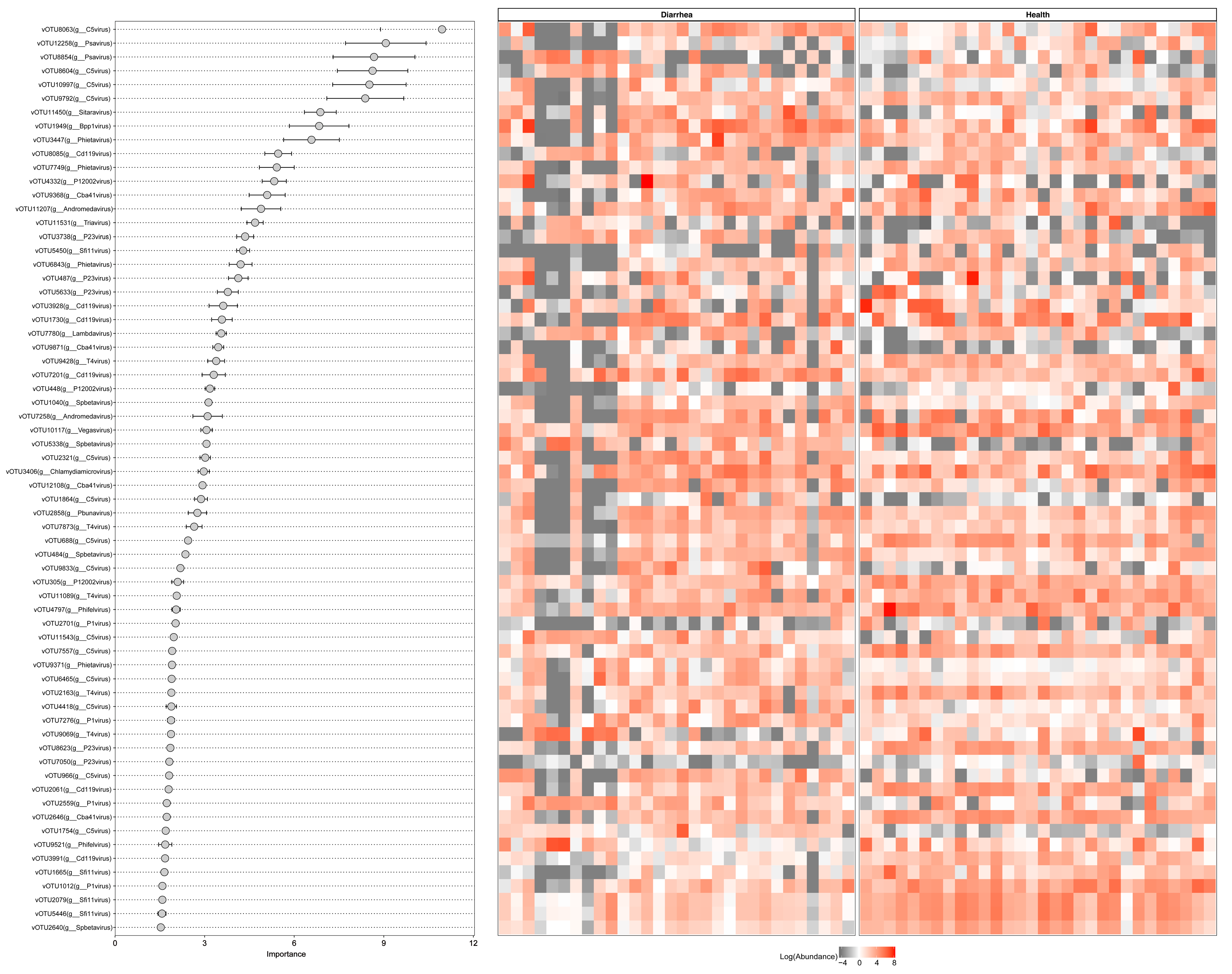
3.6. Viral Biomarkers Discriminating between Diarrheal Piglets and Healthy Piglets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Farthing, M.; Salam, M.A.; Lindberg, G.; Dite, P.; Khalif, I.; Salazar-Lindo, E.; Ramakrishna, B.S.; Goh, K.-L.; Thomson, A.; Khan, A.G.; et al. Acute Diarrhea in Adults and Children. J. Clin. Gastroenterol. 2013, 47, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Florez, I.D.; Niño-Serna, L.F.; Beltrán-Arroyave, C.P. Acute Infectious Diarrhea and Gastroenteritis in Children. Curr. Infect. Dis. Rep. 2020, 22, 4. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Nasrin, D.; Blackwelder, W.C.; Wu, Y.; Farag, T.; Panchalingham, S.; Sow, S.O.; Sur, D.; Zaidi, A.K.M.; Faruque, A.S.G.; et al. The Incidence, Aetiology, and Adverse Clinical Consequences of Less Severe Diarrhoeal Episodes among Infants and Children Residing in Low-Income and Middle-Income Countries: A 12-Month Case-Control Study as a Follow-on to the Global Enteric Multicenter St. Lancet Glob. Health 2019, 7, e568–e584. [Google Scholar] [CrossRef]
- Khalil, I.A.; Troeger, C.; Rao, P.C.; Blacker, B.F.; Brown, A.; Brewer, T.G.; Colombara, D.V.; De Hostos, E.L.; Engmann, C.; Guerrant, R.L.; et al. Morbidity, Mortality, and Long-Term Consequences Associated with Diarrhoea from Cryptosporidium Infection in Children Younger than 5 Years: A Meta-Analyses Study. Lancet Glob. Health 2018, 6, e758–e768. [Google Scholar] [CrossRef]
- Abba, K.; Sinfield, R.; Hart, C.A.; Garner, P. Pathogens Associated with Persistent Diarrhoea in Children in Low and Middle Income Countries: Systematic Review. BMC Infect. Dis. 2009, 9, 88. [Google Scholar] [CrossRef]
- Lanata, C.F.; Fischer-Walker, C.L.; Olascoaga, A.C.; Torres, C.X.; Aryee, M.J.; Black, R.E. Global Causes of Diarrheal Disease Mortality in Children <5 Years of Age: A Systematic Review. PLoS ONE 2013, 8, e72788. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Hill, C. Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host Microbe 2019, 25, 195–209. [Google Scholar] [CrossRef]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The Gut Virome: A New Microbiome Component in Health and Disease. eBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Brockhurst, M.A. Bacteria-Phage Coevolution as a Driver of Ecological and Evolutionary Processes in Microbial Communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef]
- Ping, D.; Wang, T.; Fraebel, D.T.; Maslov, S.; Sneppen, K.; Kuehn, S. Hitchhiking, Collapse, and Contingency in Phage Infections of Migrating Bacterial Populations. ISME J. 2020, 14, 2007–2018. [Google Scholar] [CrossRef]
- Gandon, S.; Buckling, A.; Decaestecker, E.; Day, T. Host-Parasite Coevolution and Patterns of Adaptation across Time and Space. J. Evol. Biol. 2008, 21, 1861–1866. [Google Scholar] [CrossRef]
- Watson, B.N.J.; Steens, J.A.; Staals, R.H.J.; Westra, E.R.; van Houte, S. Coevolution between Bacterial CRISPR-Cas Systems and Their Bacteriophages. Cell Host Microbe 2021, 29, 715–725. [Google Scholar] [CrossRef]
- Makau, D.N.; Lycett, S.; Michalska-Smith, M.; Paploski, I.A.D.; Cheeran, M.C.J.; Craft, M.E.; Kao, R.R.; Schroeder, D.C.; Doeschl-Wilson, A.; VanderWaal, K. Ecological and Evolutionary Dynamics of Multi-Strain RNA Viruses. Nat. Ecol. Evol. 2022, 6, 1414–1422. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, S.; Jiao, X.; Liu, X.F. Prevalence of Serogroups and Virulence Factors of Escherichia coli Strains Isolated from Pigs with Postweaning Diarrhoea in Eastern China. Vet. Microbiol. 2004, 103, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Fischetti, V.A. Detailed Genomic Analysis of the Wβ and γ Phages Infecting Bacillus Anthracis: Implications for Evolution of Environmental Fitness and Antibiotic Resistance. J. Bacteriol. 2006, 188, 3037–3051. [Google Scholar] [CrossRef]
- Zuo, T.; Lu, X.-J.; Zhang, Y.; Cheung, C.P.; Lam, S.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Zhao, R.; Chan, P.K.S.; et al. Gut Mucosal Virome Alterations in Ulcerative Colitis. Gut 2019, 68, 1169–1179. [Google Scholar] [CrossRef]
- Duerkop, B.A.; Kleiner, M.; Paez-Espino, D.; Zhu, W.; Bushnell, B.; Hassell, B.; Winter, S.E.; Kyrpides, N.C.; Hooper, L.V. Murine Colitis Reveals a Disease-Associated Bacteriophage Community. Nat. Microbiol. 2018, 3, 1023–1031. [Google Scholar] [CrossRef]
- Clooney, A.G.; Sutton, T.D.S.; Shkoporov, A.N.; Holohan, R.K.; Daly, K.M.; O’Regan, O.; Ryan, F.J.; Draper, L.A.; Plevy, S.E.; Ross, R.P.; et al. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe 2019, 26, 764–778.e5. [Google Scholar] [CrossRef]
- Tao, S.; Zou, H.; Li, J.; Wei, H. Landscapes of Enteric Virome Signatures in Early-Weaned Piglets. Microbiol. Spectr. 2022, 10, e01698-22. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Roux, S.; Chen, I.M.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Huntemann, M.; Reddy, T.B.K.; Pons, J.C.; Llabrés, M.; et al. IMG/VR v.2.0: An Integrated Data Management and Analysis System for Cultivated and Environmental Viral Genomes. Nucleic Acids Res. 2019, 47, D678–D686. [Google Scholar] [CrossRef]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: A Novel k-Mer Based Tool for Identifying Viral Sequences from Assembled Metagenomic Data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A Multi-Classifier, Expert-Guided Approach to Detect Diverse DNA and RNA Viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A Fast and Versatile Genome Alignment System. PLoS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Pons, J.C.; Paez-Espino, D.; Riera, G.; Ivanova, N.; Kyrpides, N.C.; Llabrés, M. VPF-Class: Taxonomic Assignment and Host Prediction of Uncultivated Viruses Based on Viral Protein Families. Bioinformatics 2021, 37, 1805–1813. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. DbCAN2: A Meta Server for Automated Carbohydrate-Active Enzyme Annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR Recognition Tool (CRT): A Tool for Automatic Detection of Clustered Regularly Interspaced Palindromic Repeats. BMC Bioinform. 2007, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Garmaeva, S.; Gulyaeva, A.; Sinha, T.; Shkoporov, A.N.; Clooney, A.G.; Stockdale, S.R.; Spreckels, J.E.; Sutton, T.D.S.; Draper, L.A.; Dutilh, B.E.; et al. Stability of the Human Gut Virome and Effect of Gluten-Free Diet. Cell Rep. 2021, 35, 109132. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 2019, 26, 527–541.e5. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 2020, 28, 724–740.e8. [Google Scholar] [CrossRef] [PubMed]
- Ferrenberg, S.; O’Neill, S.P.; Knelman, J.E.; Todd, B.; Duggan, S.; Bradley, D.; Robinson, T.; Schmidt, S.K.; Townsend, A.R.; Williams, M.W.; et al. Changes in Assembly Processes in Soil Bacterial Communities Following a Wildfire Disturbance. ISME J. 2013, 7, 1102–1111. [Google Scholar] [CrossRef]
- Lee, S.-H.; Sorensen, J.W.; Grady, K.L.; Tobin, T.C.; Shade, A. Divergent Extremes but Convergent Recovery of Bacterial and Archaeal Soil Communities to an Ongoing Subterranean Coal Mine Fire. ISME J. 2017, 11, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Bae, J.-W. Lysogeny Is Prevalent and Widely Distributed in the Murine Gut Microbiota. ISME J. 2018, 12, 1127–1141. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Turkington, C.J.; Hill, C. Mutualistic Interplay between Bacteriophages and Bacteria in the Human Gut. Nat. Rev. Microbiol. 2022, 20, 737–749. [Google Scholar] [CrossRef]
- Schokker, D.; Zhang, J.; Zhang, L.; Vastenhouw, S.A.; Heilig, H.G.H.J.; Smidt, H.; Rebel, J.M.J.; Smits, M.A. Early-Life Environmental Variation Affects Intestinal Microbiota and Immune Development in New-Born Piglets. PLoS ONE 2014, 9, e100040. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, J.; Ren, E.; Su, Y.; Zhu, W. Co-Occurrence of Early Gut Colonization in Neonatal Piglets with Microbiota in the Maternal and Surrounding Delivery Environments. Anaerobe 2018, 49, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zheng, Z.; Yu, L.; Wu, S.; Sun, L.; Wu, S.; Xu, Q.; Cai, S.; Qin, N.; Bao, W. Examination of the Temporal and Spatial Dynamics of the Gut Microbiome in Newborn Piglets Reveals Distinct Microbial Communities in Six Intestinal Segments. Sci. Rep. 2019, 9, 3453. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, X.; Zhang, G.; Hou, C.; Li, N.; Yu, H.; Shang, L.; Zhang, X.; Trevisi, P.; Yang, F.; et al. Maternal Milk and Fecal Microbes Guide the Spatiotemporal Development of Mucosa-Associated Microbiota and Barrier Function in the Porcine Neonatal Gut. BMC Biol. 2019, 17, 106. [Google Scholar] [CrossRef]
- Jin, M.; Guo, X.; Zhang, R.; Qu, W.; Gao, B.; Zeng, R. Diversities and Potential Biogeochemical Impacts of Mangrove Soil Viruses. Microbiome 2019, 7, 58. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Silveira, C.B.; Gregoracci, G.B.; Thompson, C.C.; Edwards, R.A.; Brussaard, C.P.D.; Dutilh, B.E.; Thompson, F.L. Marine Viruses Discovered via Metagenomics Shed Light on Viral Strategies throughout the Oceans. Nat. Commun. 2017, 8, 15955. [Google Scholar] [CrossRef]
- Cook, R.; Hooton, S.; Trivedi, U.; King, L.; Dodd, C.E.R.; Hobman, J.L.; Stekel, D.J.; Jones, M.A.; Millard, A.D. Hybrid Assembly of an Agricultural Slurry Virome Reveals a Diverse and Stable Community with the Potential to Alter the Metabolism and Virulence of Veterinary Pathogens. Microbiome 2021, 9, 65. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; Hallam, S.J.; Sullivan, M.B. Metabolic Reprogramming by Viruses in the Sunlit and Dark Ocean. Genome Biol. 2013, 14, R123. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; U’Ren, J.M. Viral Metabolic Reprogramming in Marine Ecosystems. Curr. Opin. Microbiol. 2016, 31, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Emmerling, M.; Dauner, M.; Ponti, A.; Fiaux, J.; Hochuli, M.; Szyperski, T.; Wüthrich, K.; Bailey, J.E.; Sauer, U. Metabolic Flux Responses to Pyruvate Kinase Knockout in Escherichia coli. J. Bacteriol. 2002, 184, 152–164. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Li, J.; Ma, L.; Liu, X.; Wei, H.; Xiao, Y.; Tao, S. Metagenomic Sequencing Identified Specific Bacteriophage Signature Discriminating between Healthy and Diarrheal Neonatal Piglets. Nutrients 2023, 15, 1616. https://doi.org/10.3390/nu15071616
Wang Z, Li J, Ma L, Liu X, Wei H, Xiao Y, Tao S. Metagenomic Sequencing Identified Specific Bacteriophage Signature Discriminating between Healthy and Diarrheal Neonatal Piglets. Nutrients. 2023; 15(7):1616. https://doi.org/10.3390/nu15071616
Chicago/Turabian StyleWang, Zhenyu, Jingjing Li, Lingyan Ma, Xiangdong Liu, Hong Wei, Yingping Xiao, and Shiyu Tao. 2023. "Metagenomic Sequencing Identified Specific Bacteriophage Signature Discriminating between Healthy and Diarrheal Neonatal Piglets" Nutrients 15, no. 7: 1616. https://doi.org/10.3390/nu15071616
APA StyleWang, Z., Li, J., Ma, L., Liu, X., Wei, H., Xiao, Y., & Tao, S. (2023). Metagenomic Sequencing Identified Specific Bacteriophage Signature Discriminating between Healthy and Diarrheal Neonatal Piglets. Nutrients, 15(7), 1616. https://doi.org/10.3390/nu15071616






