Aspartame and Its Metabolites Cause Oxidative Stress and Mitochondrial and Lipid Alterations in SH-SY5Y Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture and Treatment
2.3. Transmission Electron Microscopy
2.4. Detection of Cardiolipin
2.5. Detection of Oxidative Stress
2.6. Measurement of Lipid Species via Mass Spectrometry
2.6.1. Sample Preparation
2.6.2. Lipid Extraction
2.6.3. Mass Spectrometry
2.7. Analysis of Gene Expression
2.8. Detection of Cell Viability
2.9. Data and Statistical Analysis
3. Results
3.1. Transmission Electron Microcopy
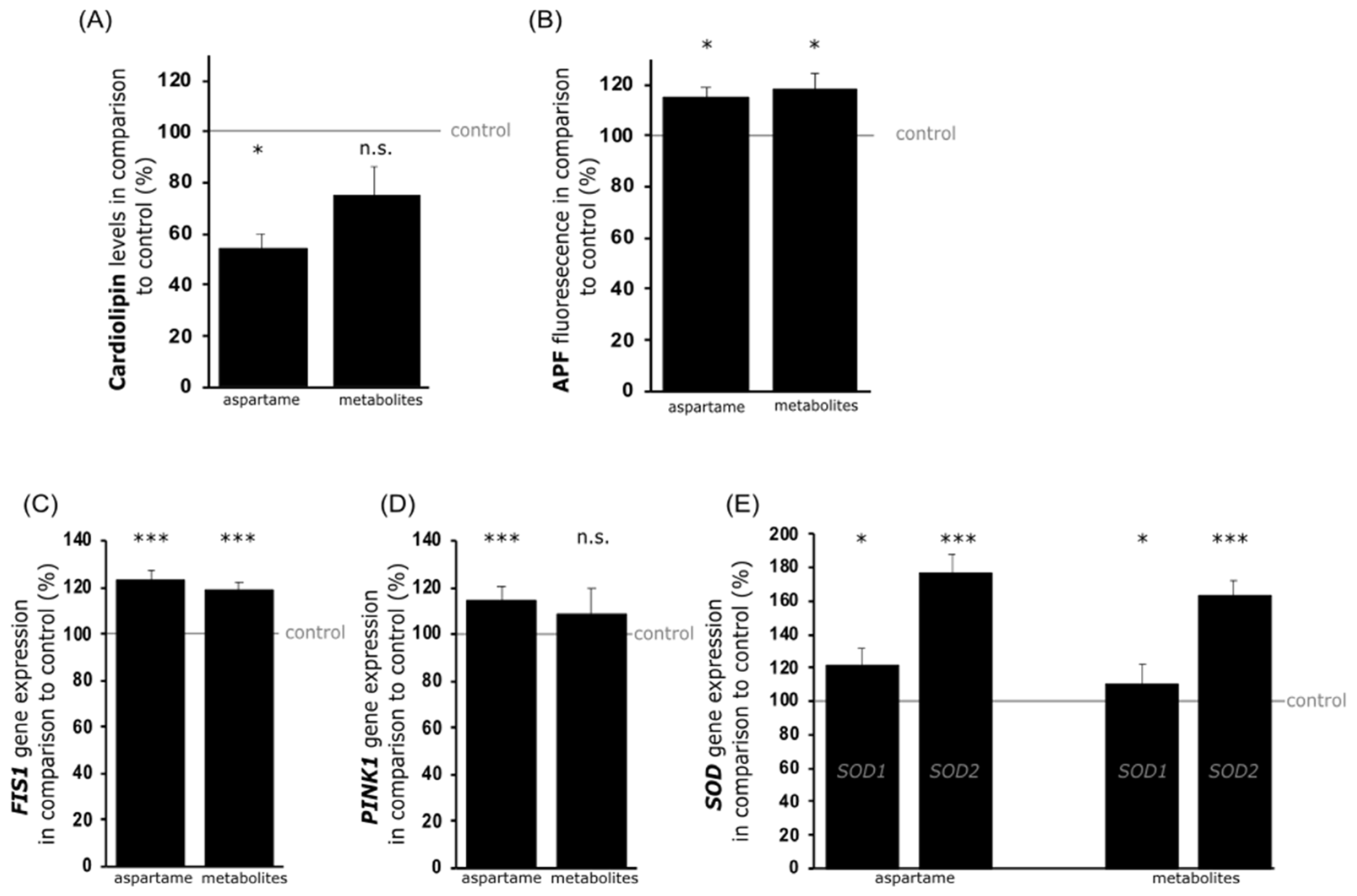
3.2. Analysis of Mitochondrial Damage
3.3. Lipid Analysis
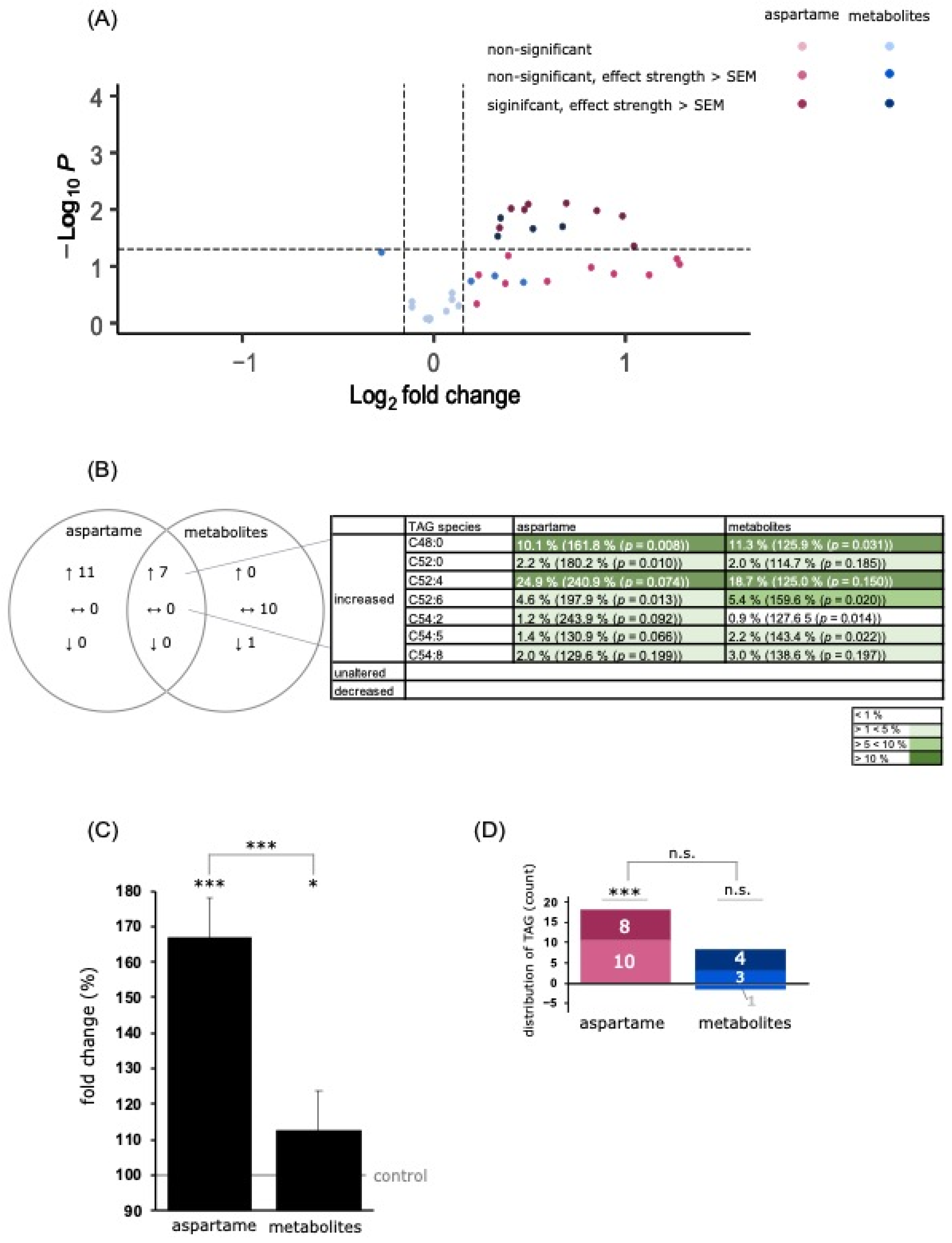
3.3.1. Analysis of Triacylglycerol Species
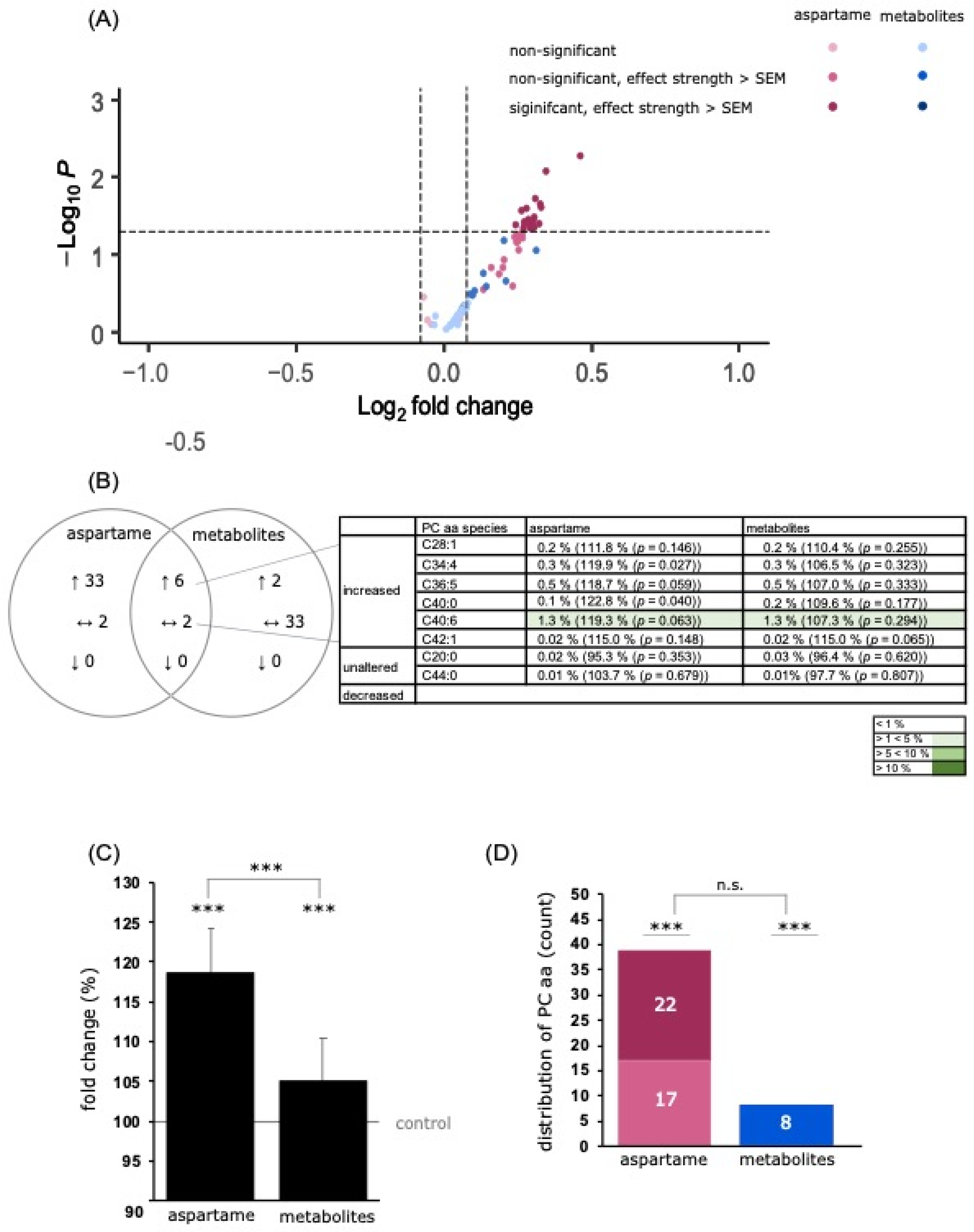
3.3.2. Analysis of Phospholipids
PC Species
PE Species
3.4. Analysis of Carnitine Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sylvetsky, A.C.; Rother, K.I. Trends in the consumption of low-calorie sweeteners. Physiol. Behav. 2016, 164, 446–450. [Google Scholar] [CrossRef]
- Reed, J.; Bain, S.; Kanamarlapudi, V. A review of current trends with type 2 diabetes epidemiology, aetiology, pathogenesis, treatments and future perspectives. Diabetes Metab. Syndr. Obes. 2021, 14, 3567–3602. [Google Scholar] [CrossRef] [PubMed]
- Kopp, W. How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab. Syndr. Obes. 2019, 12, 2221–2236. [Google Scholar] [CrossRef]
- Walbolt, J.; Koh, Y. Non-nutritive sweeteners and their associations with obesity and type 2 diabetes. J. Obes. Metab. Syndr. 2020, 29, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, B.A.; Burdock, G.A.; Doull, J.; Kroes, R.M.; Marsh, G.M.; Pariza, M.W.; Spencer, P.S.; Waddell, W.J.; Walker, R.; Williams, G.M. Aspartame: A safety evaluation based on current use levels, regulations, and toxicological and epidemiological studies. Crit. Rev. Toxicol. 2007, 37, 629–727. [Google Scholar] [CrossRef]
- Renwick, A.G.; Nordmann, H. First european conference on aspartame: Putting safety and benefits into perspective. Synopsis of presentations and conclusions. Food Chem. Toxicol. 2007, 45, 1308–1313. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Ghoshal, S.; Mukherjee, A. Genotoxicity testing of low-calorie sweeteners: Aspartame, acesulfame-k, and saccharin. Drug Chem. Toxicol. 2008, 31, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.K.; Lee, Y.Y. Neurophysiological symptoms and aspartame: What is the connection? Nutr. Neurosci. 2018, 21, 306–316. [Google Scholar] [CrossRef]
- Rycerz, K.; Jaworska-Adamu, J.E. Effects of aspartame metabolites on astrocytes and neurons. Folia Neuropathol. 2013, 51, 10–17. [Google Scholar] [CrossRef]
- Iyaswamy, A.; Kammella, A.K.; Thavasimuthu, C.; Wankupar, W.; Dapkupar, W.; Shanmugam, S.; Rajan, R.; Rathinasamy, S. Oxidative stress evoked damages leading to attenuated memory and inhibition of nmdar-camkii-erk/creb signalling on consumption of aspartame in rat model. J. Food Drug Anal. 2018, 26, 903–916. [Google Scholar] [CrossRef]
- Abu-Taweel, G.M.; Zyadah, M.A.; Ajarem, J.S.; Ahmad, M. Cognitive and biochemical effects of monosodium glutamate and aspartame, administered individually and in combination in male albino mice. Neurotoxicol. Teratol. 2014, 42, 60–67. [Google Scholar] [CrossRef]
- Iyyaswamy, A.; Rathinasamy, S. Effect of chronic exposure to aspartame on oxidative stress in the brain of albino rats. J. Biosci. 2012, 37, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Ashok, I.; Sheeladevi, R.; Wankhar, D. Acute effect of aspartame-induced oxidative stress in wistar albino rat brain. J. Biomed. Res. 2015, 29, 390–396. [Google Scholar]
- Ayala, A.; Munoz, M.F.; Arguelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.K.; Pretorius, E. Revisiting the safety of aspartame. Nutr. Rev. 2017, 75, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Simintzi, I.; Schulpis, K.H.; Angelogianni, P.; Liapi, C.; Tsakiris, S. The effect of aspartame on acetylcholinesterase activity in hippocampal homogenates of suckling rats. Pharmacol. Res. 2007, 56, 155–159. [Google Scholar] [CrossRef]
- Aya-Ramos, L.; Contreras-Vargas, C.; Rico, J.L.; Duenas, Z. Early maternal separation induces preference for sucrose and aspartame associated with increased blood glucose and hyperactivity. Food Funct. 2017, 8, 2592–2600. [Google Scholar] [CrossRef]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules 2019, 24, 1583. [Google Scholar]
- Reitz, C.; Mayeux, R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 2014, 88, 640–651. [Google Scholar] [CrossRef]
- Atri, A. The alzheimer’s disease clinical spectrum: Diagnosis and management. Med. Clin. N. Am. 2019, 103, 263–293. [Google Scholar] [CrossRef]
- Sery, O.; Povova, J.; Misek, I.; Pesak, L.; Janout, V. Molecular mechanisms of neuropathological changes in alzheimer’s disease: A review. Folia Neuropathol. 2013, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tonnies, E.; Trushina, E. Oxidative stress, synaptic dysfunction, and alzheimer’s disease. J. Alzheimer’s Dis. 2017, 57, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, R.H.; Burns, J.M.; Khan, S.M. The alzheimer’s disease mitochondrial cascade hypothesis: Progress and perspectives. Biochim. Biophys. Acta 2014, 1842, 1219–1231. [Google Scholar] [CrossRef] [PubMed]
- Basson, A.R.; Rodriguez-Palacios, A.; Cominelli, F. Artificial sweeteners: History and new concepts on inflammation. Front. Nutr. 2021, 8, 746247. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, M.; Enkhtaivan, G.; Kim, D.H. Cytotoxic effects of aspartame on human cervical carcinoma cells. Toxicol. Res. 2016, 5, 45–52. [Google Scholar] [CrossRef]
- Rencuzogullari, E.; Tuylu, B.A.; Topaktas, M.; Ila, H.B.; Kayraldiz, A.; Arslan, M.; Diler, S.B. Genotoxicity of aspartame. Drug Chem. Toxicol. 2004, 27, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, M.; Enkhtaivan, G.; Mistry, B.; Chandrasekaran, M.; Noorzai, R.; Kim, D.H. Investigation of role of aspartame on apoptosis process in hela cells -->. Saudi J. Biol. Sci. 2016, 23, 503–506. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Lauer, A.A.; Nguyen, V.T.T.; Janitschke, D.; Dos Santos Guilherme, M.; Bachmann, C.M.; Grimm, H.S.; Hartmann, T.; Endres, K.; Grimm, M.O.W. The influence of acitretin on brain lipidomics in adolescent mice-implications for pediatric and adolescent dermatological therapy. Int. J. Mol. Sci. 2022, 23, 15535. [Google Scholar] [CrossRef]
- Pfeiffer, K.; Gohil, V.; Stuart, R.A.; Hunte, C.; Brandt, U.; Greenberg, M.L.; Schagger, H. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003, 278, 52873–52880. [Google Scholar] [CrossRef]
- Shi, Y. Emerging roles of cardiolipin remodeling in mitochondrial dysfunction associated with diabetes, obesity, and cardiovascular diseases. J. Biomed. Res. 2010, 24, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.H.; Reddy, T.P.; Manczak, M.; Calkins, M.J.; Shirendeb, U.; Mao, P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res. Rev. 2011, 67, 103–118. [Google Scholar] [CrossRef]
- Yoo, S.M.; Jung, Y.K. A molecular approach to mitophagy and mitochondrial dynamics. Mol. Cells 2018, 41, 18–26. [Google Scholar] [PubMed]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the cuzn-sod (sod1), mn-sod (sod2), and ec-sod (sod3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Farese, R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012, 81, 687–714. [Google Scholar] [CrossRef] [PubMed]
- Islimye, E.; Girard, V.; Gould, A.P. Functions of stress-induced lipid droplets in the nervous system. Front. Cell Dev. Biol. 2022, 10, 863907. [Google Scholar] [CrossRef]
- Naudi, A.; Cabre, R.; Jove, M.; Ayala, V.; Gonzalo, H.; Portero-Otin, M.; Ferrer, I.; Pamplona, R. Lipidomics of human brain aging and alzheimer’s disease pathology. Int. Rev. Neurobiol. 2015, 122, 133–189. [Google Scholar]
- Jakubec, M.; Barias, E.; Kryuchkov, F.; Hjornevik, L.V.; Halskau, O. Fast and quantitative phospholipidomic analysis of sh-sy5y neuroblastoma cell cultures using liquid chromatography-tandem mass spectrometry and 31P nuclear magnetic resonance. ACS Omega 2019, 4, 21596–21603. [Google Scholar] [CrossRef]
- Choi, J.; Yin, T.; Shinozaki, K.; Lampe, J.W.; Stevens, J.F.; Becker, L.B.; Kim, J. Comprehensive analysis of phospholipids in the brain, heart, kidney, and liver: Brain phospholipids are least enriched with polyunsaturated fatty acids. Mol. Cell. Biochem. 2018, 442, 187–201. [Google Scholar] [CrossRef]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-Schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A.; et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef]
- Lee, S.J.; Zhang, J.; Choi, A.M.; Kim, H.P. Mitochondrial dysfunction induces formation of lipid droplets as a generalized response to stress. Oxid. Med. Cell. Longev. 2013, 2013, 327167. [Google Scholar] [CrossRef]
- Zafar, U.; Khaliq, S.; Ahmad, H.U.; Manzoor, S.; Lone, K.P. Metabolic syndrome: An update on diagnostic criteria, pathogenesis, and genetic links. Hormones 2018, 17, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, V.E.; Herrera, P.F.; Laura, R. Effect of nutrition on neurodegenerative diseases. A systematic review. Nutr. Neurosci. 2021, 24, 810–834. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, J.A.; Muldoon, E.; Ranney, R.E. Metabolism of aspartame in monkeys. J. Nutr. 1973, 103, 1454–1459. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, J.A.; Ranney, R.E. The metabolism of aspartate in infant and adult mice. J. Environ. Pathol. Toxicol. 1979, 2, 987–998. [Google Scholar] [PubMed]
- Stegink, L.D.; Brummel, M.C.; McMartin, K.; Martin-Amat, G.; Filer, L.J., Jr.; Baker, G.L.; Tephly, T.R. Blood methanol concentrations in normal adult subjects administered abuse doses of aspartame. J. Toxicol. Environ. Health 1981, 7, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Stegink, L.D.; Filer, L.J., Jr.; Baker, G.L. Plasma and erythrocyte concentrations of free amino acids in adult humans administered abuse doses of aspartame. J. Toxicol. Environ. Health 1981, 7, 291–305. [Google Scholar] [CrossRef]
- Stegink, L.D.; Koch, R.; Blaskovics, M.E.; Filer, L.J., Jr.; Baker, G.L.; McDonnell, J.E. Plasma phenylalanine levels in phenylketonuric heterozygous and normal adults administered aspartame at 34 mg/kg body weight. Toxicology 1981, 20, 81–90. [Google Scholar] [CrossRef]
- Collaboration, N.C.D.R.F. Height and body-mass index trajectories of school-aged children and adolescents from 1985 to 2019 in 200 countries and territories: A pooled analysis of 2181 population-based studies with 65 million participants. Lancet 2020, 396, 1511–1524. [Google Scholar]
- Finamor, I.A.; Ourique, G.M.; Pes, T.S.; Saccol, E.M.; Bressan, C.A.; Scheid, T.; Baldisserotto, B.; Llesuy, S.F.; Partata, W.A.; Pavanato, M.A. The protective effect of n-acetylcysteine on oxidative stress in the brain caused by the long-term intake of aspartame by rats. Neurochem. Res. 2014, 39, 1681–1690. [Google Scholar] [CrossRef]
- Finamor, I.; Pavanato, M.A.; Pes, T.; Ourique, G.; Saccol, E.; Schiefelbein, S.; Llesuy, S.; Partata, W. N-acetylcysteine protects the rat kidney against aspartame-induced oxidative stress. Free Radic. Biol. Med. 2014, 75 (Suppl. S1), S30. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, B. Oxidative stress and the pathogenesis of alzheimer’s disease. Oxid. Med. Cell. Longev. 2013, 2013, 316523. [Google Scholar] [CrossRef]
- Beal, M.F. Oxidative damage as an early marker of alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 2005, 26, 585–586. [Google Scholar] [CrossRef]
- Macdonald, R.; Barnes, K.; Hastings, C.; Mortiboys, H. Mitochondrial abnormalities in parkinson’s disease and alzheimer’s disease: Can mitochondria be targeted therapeutically? Biochem. Soc. Trans. 2018, 46, 891–909. [Google Scholar] [CrossRef]
- Calkins, M.J.; Manczak, M.; Mao, P.; Shirendeb, U.; Reddy, P.H. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of alzheimer’s disease. Hum. Mol. Genet. 2011, 20, 4515–4529. [Google Scholar] [CrossRef]
- Kikuchi, A.; Takeda, A.; Onodera, H.; Kimpara, T.; Hisanaga, K.; Sato, N.; Nunomura, A.; Castellani, R.J.; Perry, G.; Smith, M.A.; et al. Systemic increase of oxidative nucleic acid damage in parkinson’s disease and multiple system atrophy. Neurobiol. Dis. 2002, 9, 244–248. [Google Scholar] [CrossRef]
- Bosco, D.A.; Fowler, D.M.; Zhang, Q.; Nieva, J.; Powers, E.T.; Wentworth, P., Jr.; Lerner, R.A.; Kelly, J.W. Elevated levels of oxidized cholesterol metabolites in lewy body disease brains accelerate alpha-synuclein fibrilization. Nat. Chem. Biol. 2006, 2, 249–253. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase pink1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The roles of pink1, parkin, and mitochondrial fidelity in parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef]
- Cooper, O.; Seo, H.; Andrabi, S.; Guardia-Laguarta, C.; Graziotto, J.; Sundberg, M.; McLean, J.R.; Carrillo-Reid, L.; Xie, Z.; Osborn, T.; et al. Pharmacological rescue of mitochondrial deficits in ipsc-derived neural cells from patients with familial parkinson’s disease. Sci. Transl. Med. 2012, 4, 141ra190. [Google Scholar] [CrossRef]
- Lushchak, V.I. Interplay between bioenergetics and oxidative stress at normal brain aging. Aging as a result of increasing disbalance in the system oxidative stress-energy provision. Pflug. Arch. 2021, 473, 713–722. [Google Scholar] [CrossRef]
- Schiavone, S.; Colaianna, M.; Curtis, L. Impact of early life stress on the pathogenesis of mental disorders: Relation to brain oxidative stress. Curr. Pharm. Des. 2015, 21, 1404–1412. [Google Scholar] [CrossRef]
- Gubern, A.; Barcelo-Torns, M.; Casas, J.; Barneda, D.; Masgrau, R.; Picatoste, F.; Balsinde, J.; Balboa, M.A.; Claro, E. Lipid droplet biogenesis induced by stress involves triacylglycerol synthesis that depends on group via phospholipase a2. J. Biol. Chem. 2009, 284, 5697–5708. [Google Scholar] [CrossRef]
- Cabodevilla, A.G.; Sanchez-Caballero, L.; Nintou, E.; Boiadjieva, V.G.; Picatoste, F.; Gubern, A.; Claro, E. Cell survival during complete nutrient deprivation depends on lipid droplet-fueled beta-oxidation of fatty acids. J. Biol. Chem. 2013, 288, 27777–27788. [Google Scholar] [CrossRef]
- Bailey, A.P.; Koster, G.; Guillermier, C.; Hirst, E.M.; MacRae, J.I.; Lechene, C.P.; Postle, A.D.; Gould, A.P. Antioxidant role for lipid droplets in a stem cell niche of drosophila. Cell 2015, 163, 340–353. [Google Scholar] [CrossRef]
- Ackerman, D.; Tumanov, S.; Qiu, B.; Michalopoulou, E.; Spata, M.; Azzam, A.; Xie, H.; Simon, M.C.; Kamphorst, J.J. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 2018, 24, 2596–2605.e5. [Google Scholar] [CrossRef]
- Cohen, S.; Valm, A.M.; Lippincott-Schwartz, J. Interacting organelles. Curr. Opin. Cell Biol. 2018, 53, 84–91. [Google Scholar] [CrossRef]
- Schuldiner, M.; Bohnert, M. A different kind of love—lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1188–1196. [Google Scholar] [CrossRef]
- Nguyen, T.B.; Louie, S.M.; Daniele, J.R.; Tran, Q.; Dillin, A.; Zoncu, R.; Nomura, D.K.; Olzmann, J.A. Dgat1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 2017, 42, 9–21 e25. [Google Scholar] [CrossRef]
- Shil, A.; Olusanya, O.; Ghufoor, Z.; Forson, B.; Marks, J.; Chichger, H. Artificial sweeteners disrupt tight junctions and barrier function in the intestinal epithelium through activation of the sweet taste receptor, t1r3. Nutrients 2020, 12, 1862. [Google Scholar] [CrossRef]
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef]
- Fasano, A. Leaky gut and autoimmune diseases. Clin. Rev. Allergy Immunol. 2012, 42, 71–78. [Google Scholar] [CrossRef]
- Filer, L.J., Jr.; Stegink, L.D. Aspartame metabolism in normal adults, phenylketonuric heterozygotes, and diabetic subjects. Diabetes Care 1989, 12, 67–74. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-brain barrier: From physiology to disease and back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.D.; Sagare, A.P.; Zlokovic, B.V. Blood-brain barrier breakdown in alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018, 14, 133–150. [Google Scholar] [CrossRef]





| Name | Provider |
|---|---|
| Phosphatidylcholine (06:0 PC (DHPC)) | Avanti Polar Lipids (850305P) |
| Carnitines (octanoyl- and palmitoyl-L-carnitine d3) | Supelco Analytical (53230, 55107) |
| Triacylglycerides (15:0-18:1(d7)-15:0) | Avanti Polar Lipids (330709) |
| Parameter | |
|---|---|
| Curtain gas (CUR) | 20 psi |
| Temperature (TEM) | 200 °C |
| Ion source gas 1 (GS1) | 40 psi |
| Ion source gas 2 (GS2) | 50 psi |
| Interface heater (ihe) | On |
| Collisionally activated dissociation gas (CAD) | Medium |
| Ion spray voltage (IS) | 5500 V |
| Entrance potential (EP) | 10 V |
| Collision cell exit potential (cxp) | 15 V |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| HPRT | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
| FIS1 | TACGTCCGCGGGTTGCT | CCAGTTCCTTGGCCTGGTT |
| PINK1 | GGACGCTGTTCCTCGTTA | ATCTGCGATCACCAGCCA |
| SOD1 | CAGCAGGCTGTACCAGTGC | ACATTGCCCAAGTCTCCAAC |
| SOD2 | TACGTGAACAACCTGAACGT | CAAGCCATGTATCTTTCAGTTA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griebsch, L.V.; Theiss, E.L.; Janitschke, D.; Erhardt, V.K.J.; Erhardt, T.; Haas, E.C.; Kuppler, K.N.; Radermacher, J.; Walzer, O.; Lauer, A.A.; et al. Aspartame and Its Metabolites Cause Oxidative Stress and Mitochondrial and Lipid Alterations in SH-SY5Y Cells. Nutrients 2023, 15, 1467. https://doi.org/10.3390/nu15061467
Griebsch LV, Theiss EL, Janitschke D, Erhardt VKJ, Erhardt T, Haas EC, Kuppler KN, Radermacher J, Walzer O, Lauer AA, et al. Aspartame and Its Metabolites Cause Oxidative Stress and Mitochondrial and Lipid Alterations in SH-SY5Y Cells. Nutrients. 2023; 15(6):1467. https://doi.org/10.3390/nu15061467
Chicago/Turabian StyleGriebsch, Lea Victoria, Elena Leoni Theiss, Daniel Janitschke, Vincent Konrad Johannes Erhardt, Tobias Erhardt, Elodie Christiane Haas, Konstantin Nicolas Kuppler, Juliane Radermacher, Oliver Walzer, Anna Andrea Lauer, and et al. 2023. "Aspartame and Its Metabolites Cause Oxidative Stress and Mitochondrial and Lipid Alterations in SH-SY5Y Cells" Nutrients 15, no. 6: 1467. https://doi.org/10.3390/nu15061467
APA StyleGriebsch, L. V., Theiss, E. L., Janitschke, D., Erhardt, V. K. J., Erhardt, T., Haas, E. C., Kuppler, K. N., Radermacher, J., Walzer, O., Lauer, A. A., Matschke, V., Hartmann, T., Grimm, M. O. W., & Grimm, H. S. (2023). Aspartame and Its Metabolites Cause Oxidative Stress and Mitochondrial and Lipid Alterations in SH-SY5Y Cells. Nutrients, 15(6), 1467. https://doi.org/10.3390/nu15061467








