Eriodictyol Alleviated LPS/D-GalN-Induced Acute Liver Injury by Inhibiting Oxidative Stress and Cell Apoptosis via PI3K/AKT Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Network Pharmacology
2.2.1. Prediction of Eriodictyol Targets
2.2.2. Predict Targets of ALI
2.2.3. Eriodictyol-ALI Intersection Target
2.2.4. Construction of Protein Interaction (PPI) Network
2.2.5. GO, KEGG, and DO Analysis
2.3. Evaluating Expression Patterns of AKT1 and PI3K
2.4. Molecular Docking
2.5. Molecular Dynamics Simulation
2.6. Animals and Experimental Design
2.7. Organ Index
2.8. Biochemistry Analysis
2.9. Hematoxylin and Eosin (H&E) Staining
2.10. TdT-Mediated dUTP Nick End Labelling (TUNEL) Staining Analysis
2.11. Analysis of the Antioxidant System
2.12. Detection of Anti-Inflammatory Biomarkers
2.13. Immunohistochemistry
2.14. Quantitative Real-Time PCR (RT-PCR)
2.15. Statistical Analysis
3. Results
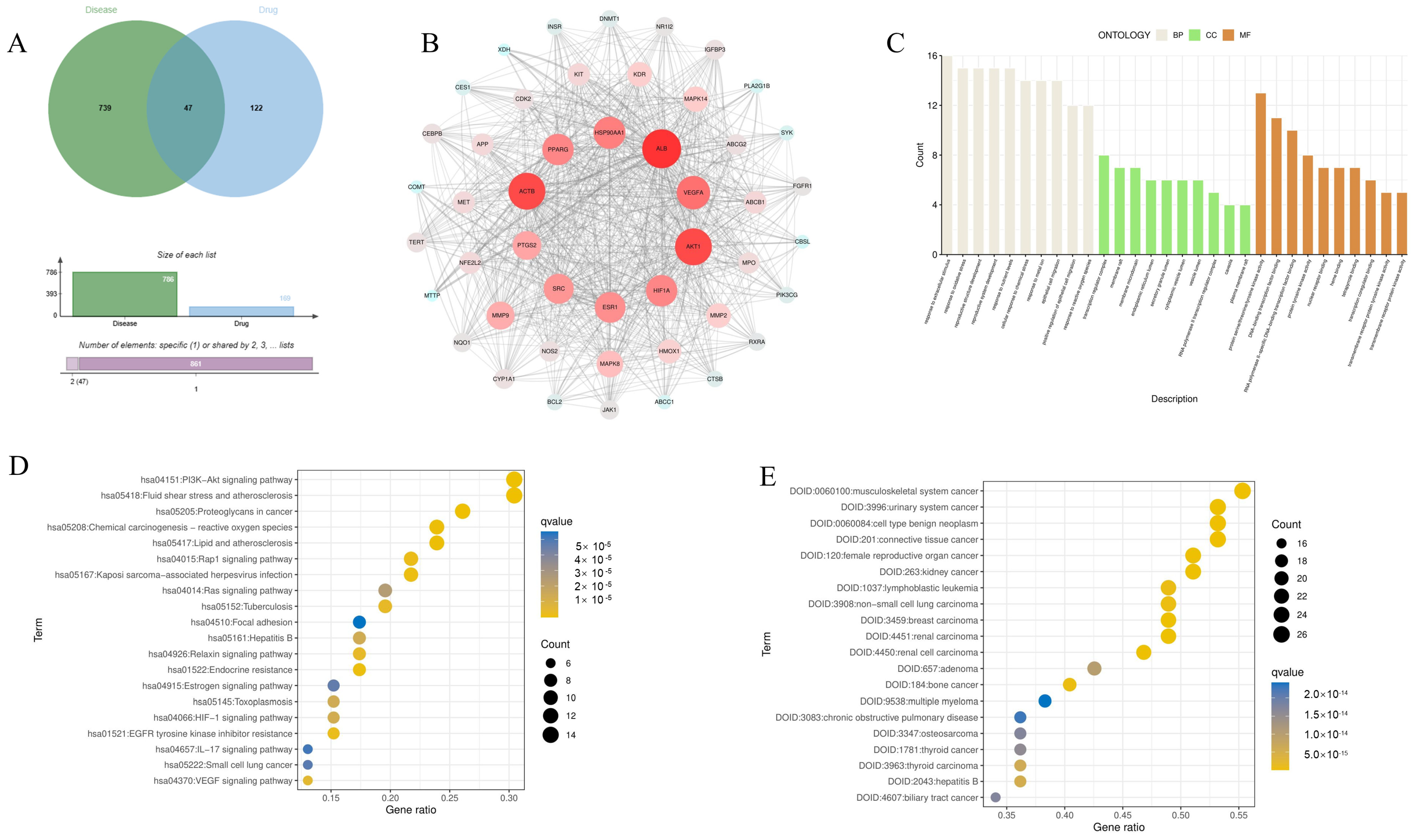
3.1. Identify the Potential Target of Eriodictyol in the Treatment of ALI
3.2. PPI Network Construction and Analysis
3.3. GO Function, KEGG Pathway Enrichment, and Disease Enrichment Analysis
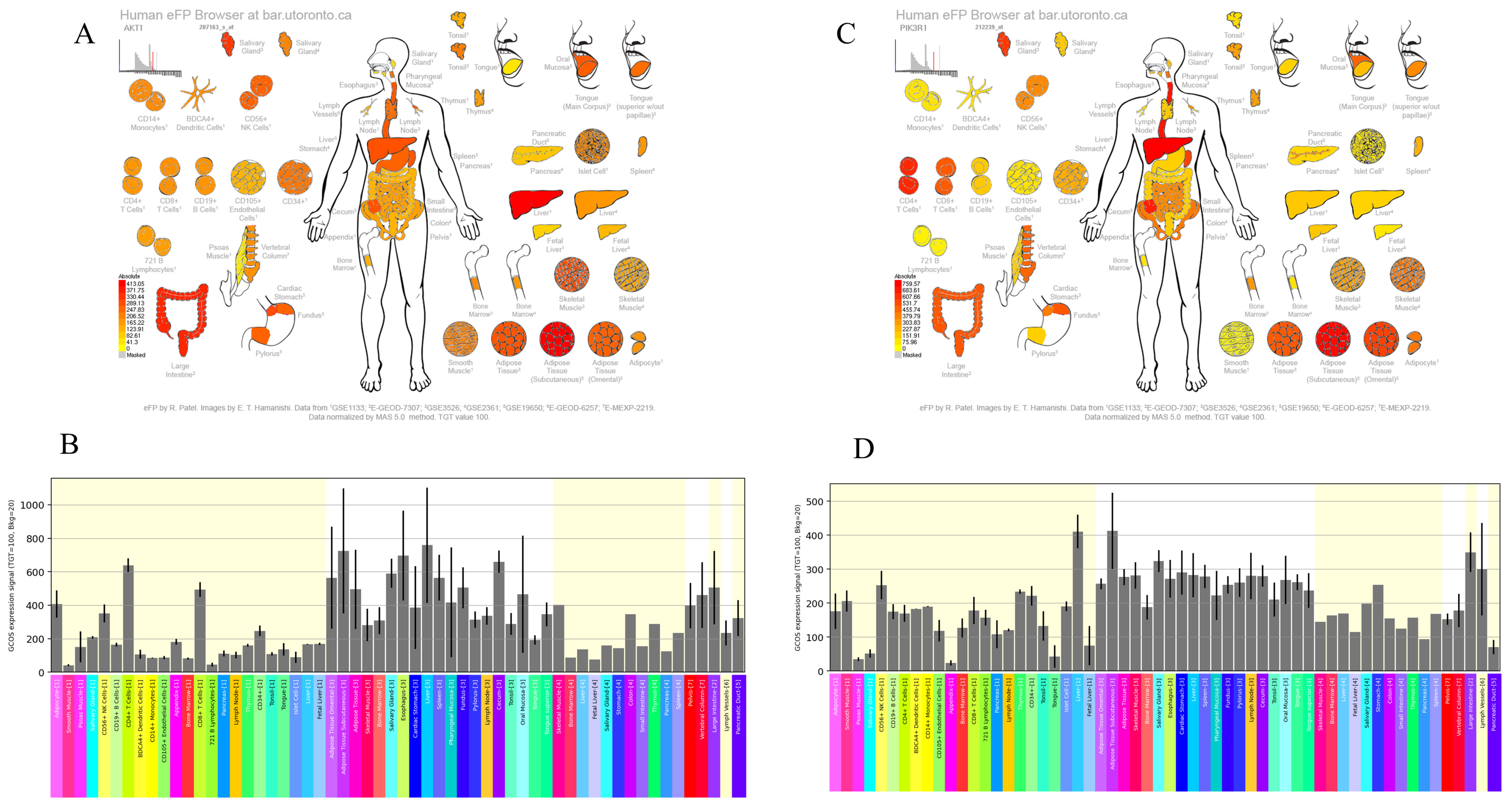
3.4. Expression Patterns of AKT1 and PI3K Genes in Liver Tissue
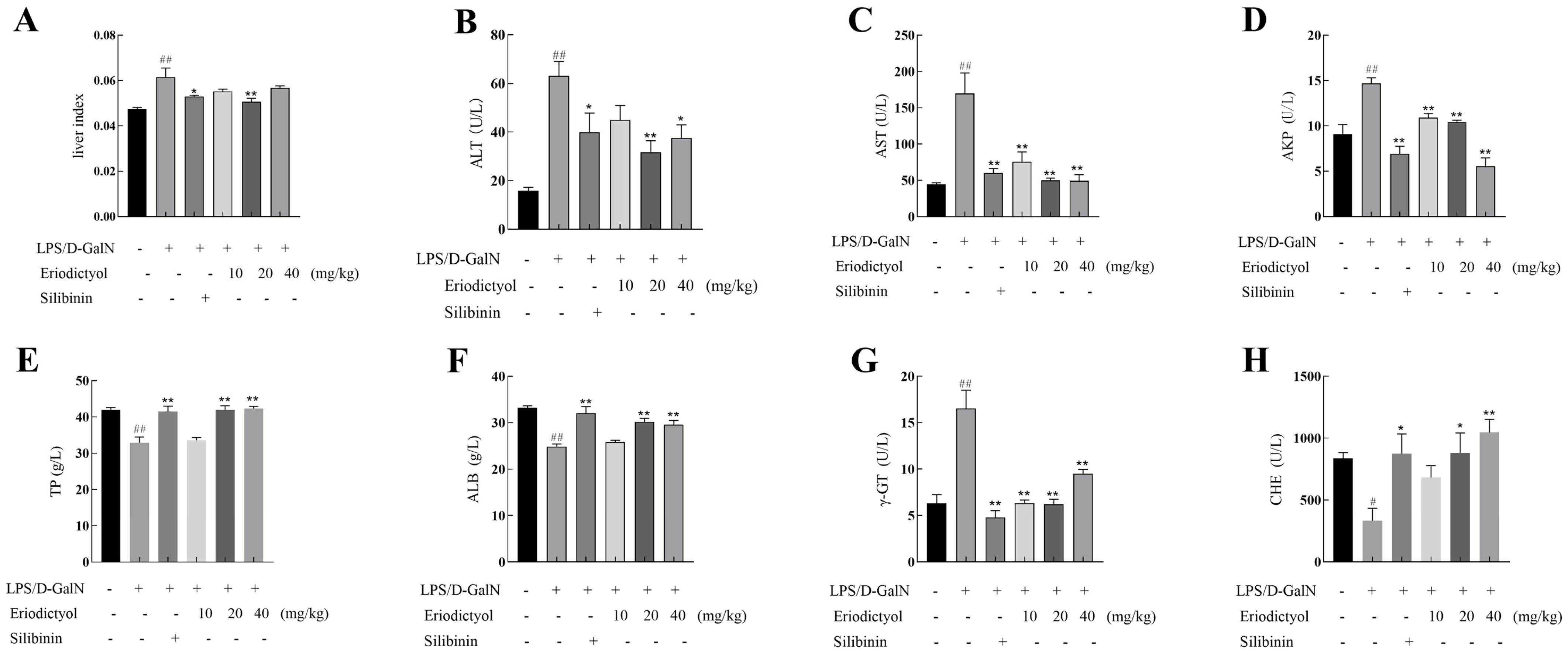
3.5. Eriodictyol Alleviated LPS/D-GalN-Induced Liver Injury in Mice
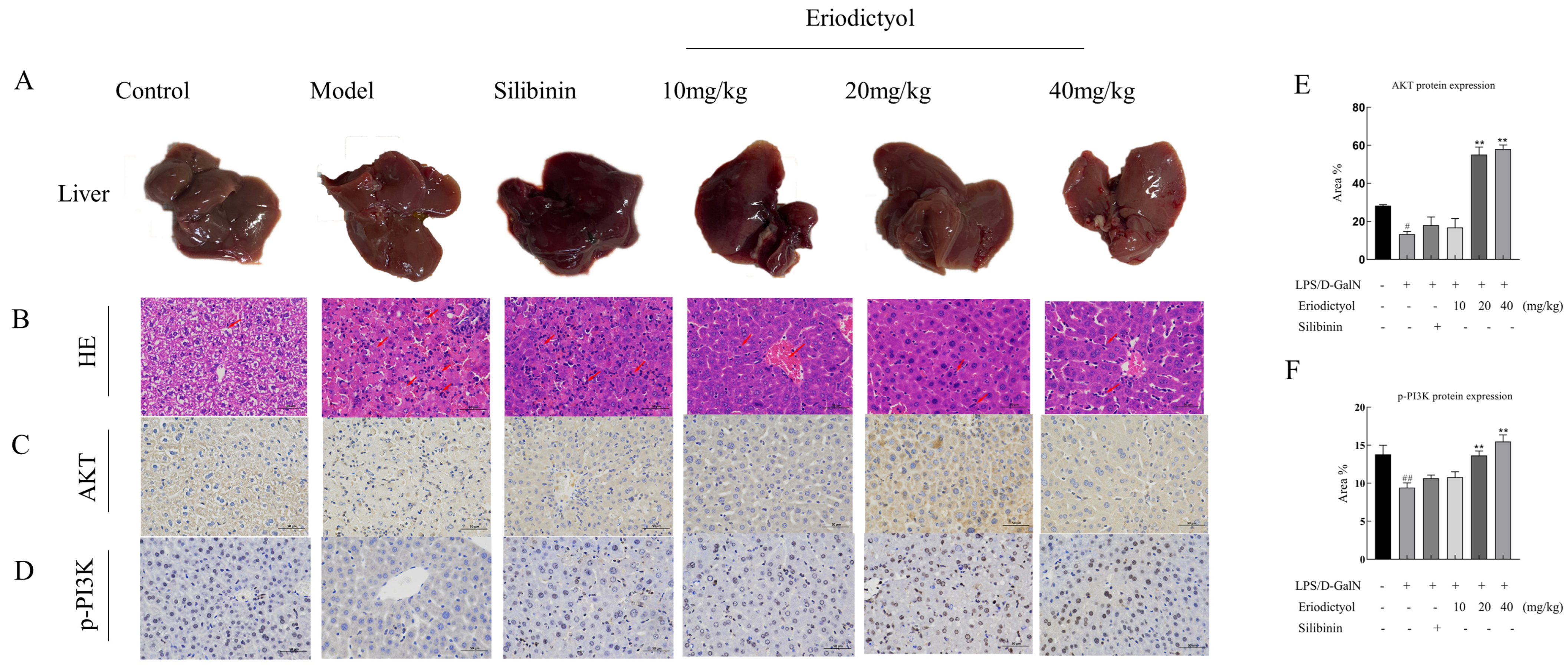
3.6. Eriodictyol Improves Liver Pathology and Apoptosis in Mice Induced by LPS/D-GalN
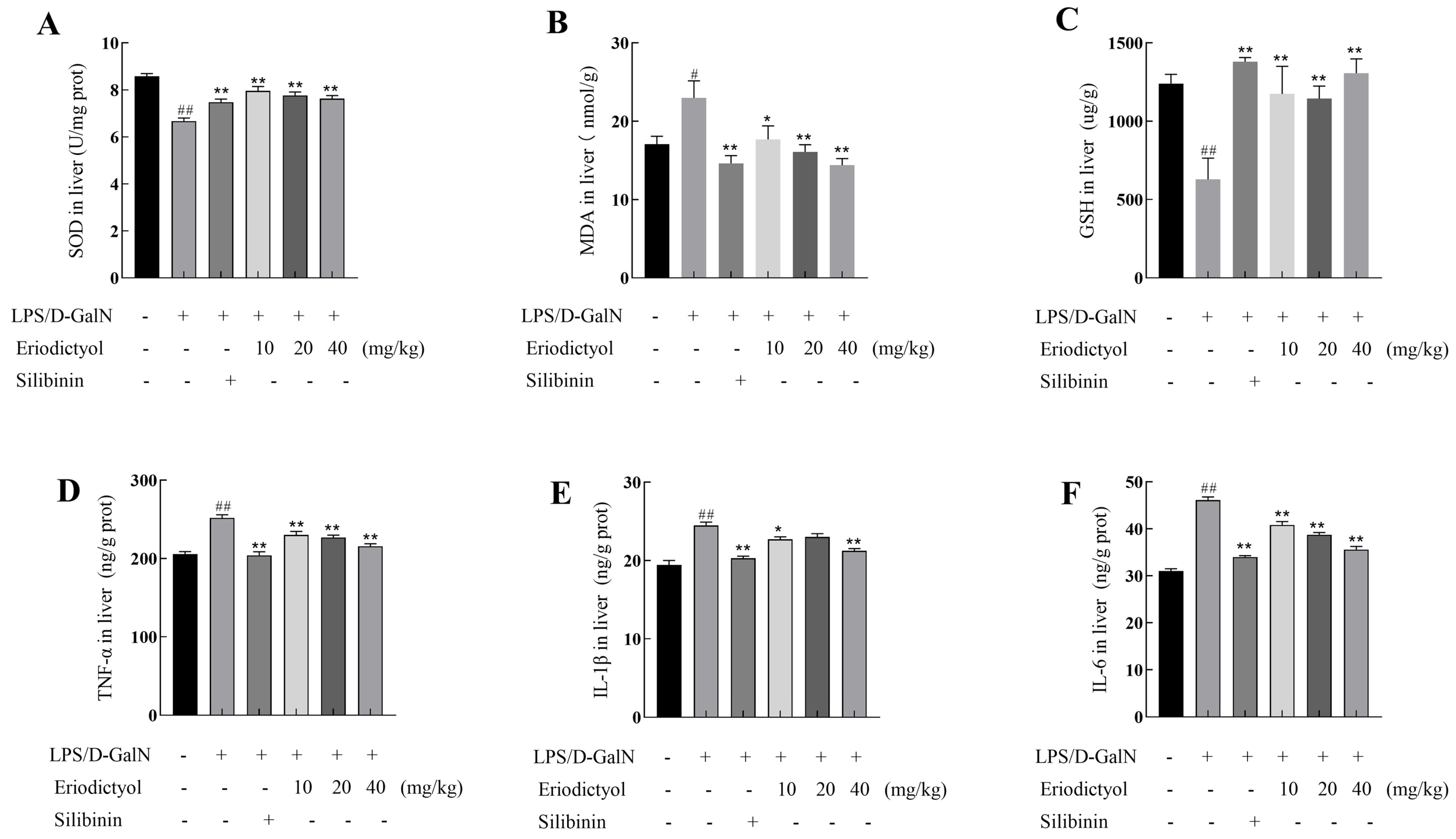
3.7. Eriodictyol Mitigated the Oxidative Stress and Inflammation Induced by LPS/D-GalN
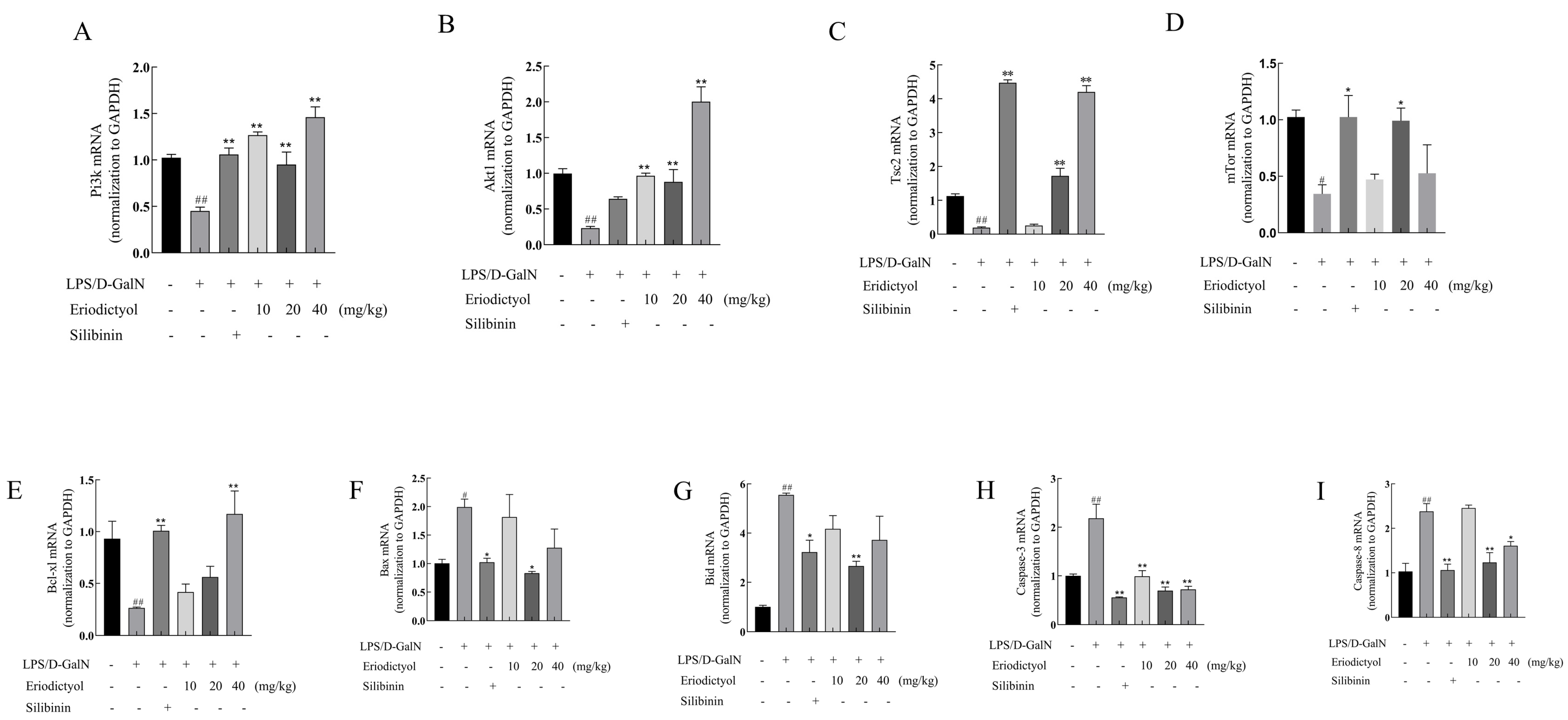
3.8. Eriodictyol Alleviates LPS/D-GalN-Induced ALI in Mice through Modulation of the PI3K/AKT Signaling Pathway and the Apoptosis Pathway
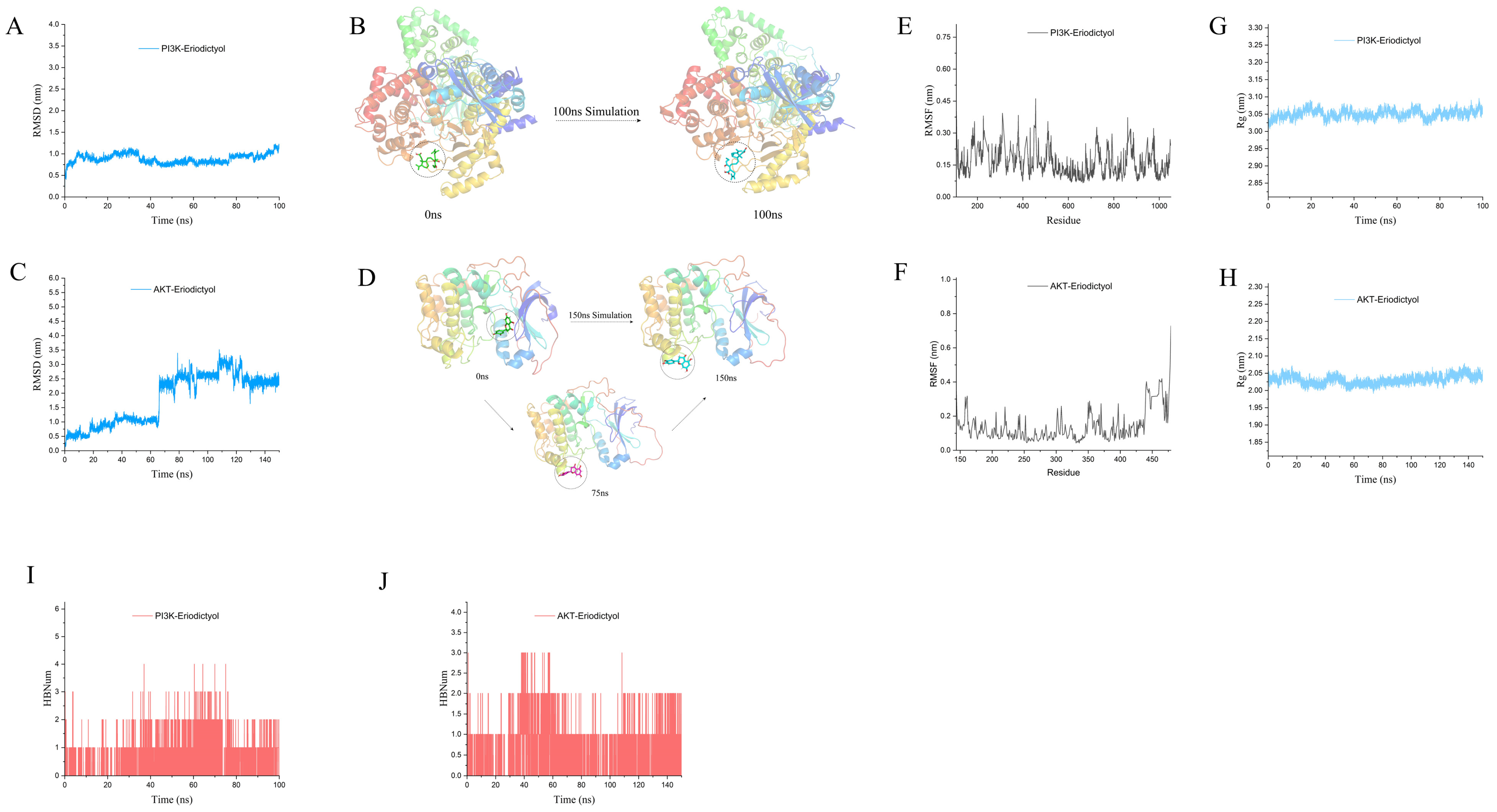
3.9. Molecular Docking Verification
3.10. MD Simulation to Explore the Interaction of Eriodictyol with PI3K and AKT
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Peng, J.; Li, J.; Huang, J.; Xu, P.; Huang, H.; Liu, Y.; Yu, L.; Yang, Y.; Zhou, B.; Jiang, H.; et al. p300/CBP inhibitor A-485 alleviates acute liver injury by regulating macrophage activation and polarization. Theranostics 2019, 9, 8344–8361. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Cao, M.; You, D.; Qin, G.; Liu, Z. Research Progress on the Animal Models of Drug-Induced Liver Injury: Current Status and Further Perspectives. BioMed Res. Int. 2019, 2019, 1283824. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Cho, W.C.; Upadhyay, G. Drug-Induced Liver Toxicity and Prevention by Herbal Antioxidants: An Overview. Front. Physiol. 2015, 6, 363. [Google Scholar] [CrossRef]
- Wu, Z.L.; Wang, J. Dioscin attenuates Bleomycin-Induced acute lung injury via inhibiting the inflammatory response in mice. Exp. Lung Res. 2019, 45, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, Y.; Du, S.; Li, S.; Zhang, Y.; Liu, F.; Chen, Y.; Weng, D.; Chen, J. Dioscin Exerts Protective Effects Against Crystalline Silica-induced Pulmonary Fibrosis in Mice. Theranostics 2017, 7, 4255–4275. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Liu, D.; Yuwen, M.; Koffas, M.A.; Zha, J. Biosynthesis of eriodictyol from tyrosine by Corynebacterium glutamicum. Microb. Cell Factories 2022, 21, 86. [Google Scholar] [CrossRef]
- Kwon, E.Y.; Choi, M.S. Dietary Eriodictyol Alleviates Adiposity, Hepatic Steatosis, Insulin Resistance, and Inflammation in Diet-Induced Obese Mice. Int. J. Mol. Sci. 2019, 20, 1227. [Google Scholar] [CrossRef]
- He, P.; Yan, S.; Zheng, J.; Gao, Y.; Zhang, S.; Liu, Z.; Liu, X.; Xiao, C. Eriodictyol Attenuates LPS-Induced Neuroinflammation, Amyloidogenesis, and Cognitive Impairments via the Inhibition of NF-kappaB in Male C57BL/6J Mice and BV2 Microglial Cells. J. Agric. Food Chem. 2018, 66, 10205–10214. [Google Scholar] [CrossRef]
- Li, W.; Du, Q.; Li, X.; Zheng, X.; Lv, F.; Xi, X.; Huang, G.; Yang, J.; Liu, S. Eriodictyol Inhibits Proliferation, Metastasis and Induces Apoptosis of Glioma Cells via PI3K/Akt/NF-kappaB Signaling Pathway. Front. Pharmacol. 2020, 11, 114. [Google Scholar] [CrossRef]
- Wang, Z.; Lan, Y.; Chen, M.; Wen, C.; Hu, Y.; Liu, Z.; Ye, L. Eriodictyol, Not Its Glucuronide Metabolites, Attenuates Acetaminophen-Induced Hepatotoxicity. Mol. Pharm. 2017, 14, 2937–2951. [Google Scholar] [CrossRef]
- Xie, G.; Meng, X.; Wang, F.; Bao, Y.; Huo, J. Eriodictyol attenuates arsenic trioxide-induced liver injury by activation of Nrf2. Oncotarget 2017, 8, 68668–68674. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, W.J.; Zheng, X.R.; Liu, Q.L.; Du, Q.; Lai, Y.J.; Liu, S.Q. Eriodictyol ameliorates cognitive dysfunction in APP/PS1 mice by inhibiting ferroptosis via vitamin D receptor-mediated Nrf2 activation. Mol. Med. 2022, 28, 11. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zheng, X.; Wen, Q.; Zhang, Y.; Tang, H.; Zhao, L.; Shi, F.; Li, Y.; Yin, Z.; Zou, Y.; et al. Swertia cincta Burkill alleviates LPS/D-GalN-induced acute liver failure by modulating apoptosis and oxidative stress signaling pathways. Aging 2023, 15, 5887–5916. [Google Scholar] [CrossRef]
- Malle, S.; Eskandari, M.; Morrison, M.; Belzile, F. Genome-wide association identifies several QTLs controlling cysteine and methionine content in soybean seed including some promising candidate genes. Sci. Rep. 2020, 10, 21812. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Deng, R.; Dong, J.; Huang, L.; Li, J.; Zhang, B. Eriodictyol ameliorates lipopolysaccharide-induced acute lung injury by suppressing the inflammatory COX-2/NLRP3/NF-kappaB pathway in mice. J. Biochem. Mol. Toxicol. 2020, 34, e22434. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Greenblatt, M.B.; Brady, N.; Lotinun, S.; Zhai, B.; de Rivera, H.; Singh, A.; Sun, J.; Gygi, S.P.; Baron, R.; et al. The microtubule-associated protein DCAMKL1 regulates osteoblast function via repression of Runx2. J. Exp. Med. 2013, 210, 1793–1806. [Google Scholar] [CrossRef]
- Zhao, J.; Lin, F.; Liang, G.; Han, Y.; Xu, N.; Pan, J.; Luo, M.; Yang, W.; Zeng, L. Exploration of the Molecular Mechanism of Polygonati Rhizoma in the Treatment of Osteoporosis Based on Network Pharmacology and Molecular Docking. Front. Endocrinol. 2021, 12, 815891. [Google Scholar] [CrossRef]
- Wu, Z.; Han, M.; Chen, T.; Yan, W.; Ning, Q. Acute liver failure: Mechanisms of immune-mediated liver injury. Liver Int. 2010, 30, 782–794. [Google Scholar] [CrossRef]
- Lv, H.; Yang, H.; Wang, Z.; Feng, H.; Deng, X.; Cheng, G.; Ci, X. Nrf2 signaling and autophagy are complementary in protecting lipopolysaccharide/d-galactosamine-induced acute liver injury by licochalcone A. Cell Death Dis. 2019, 10, 313. [Google Scholar] [CrossRef]
- Zimmermann, H.W.; Trautwein, C.; Tacke, F. Functional role of monocytes and macrophages for the inflammatory response in acute liver injury. Front. Physiol. 2012, 3, 56. [Google Scholar] [CrossRef]
- Li, H.; Sureda, A.; Devkota, H.P.; Pittalà, V.; Barreca, D.; Silva, A.S.; Tewari, D.; Xu, S.; Nabavi, S.M. Curcumin, the golden spice in treating cardiovascular diseases. Biotechnol. Adv. 2020, 38, 107343. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, L.; Wang, Y.; Zhao, T.; Khan, A.; Wang, Y.; Cao, J.; Cheng, G. Protective effect of Que Zui tea hot-water and aqueous ethanol extract against acetaminophen-induced liver injury in mice via inhibition of oxidative stress, inflammation, and apoptosis. Food Funct. 2021, 12, 2468–2480. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Lei, Z.; Yao, L.; Jiang, P.; Gu, T.; Ren, F.; Liu, Y.; Gou, C.; Li, X.; Wen, T. Circulating histones are major mediators of systemic inflammation and cellular injury in patients with acute liver failure. Cell Death Dis. 2016, 7, e2391. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Ai, Q.; Lin, L.; Dai, J.; Jia, M.; Zhou, D.; Che, Q.; Wan, J.; Jiang, R.; Zhang, L. Protective effects of garcinol in mice with lipopolysaccharide/D-galactosamine-induced apoptotic liver injury. Int. Immunopharmacol. 2014, 19, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yin, L.; Quan, H.; Chu, Y.; Lu, J. Hepatoprotective Effects of Kaempferol-3-O-alpha-l-Arabinopyranosyl-7-O-alpha-l-Rhamnopyranoside on d-Galactosamine and Lipopolysaccharide Caused Hepatic Failure in Mice. Molecules 2017, 22, 1755. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Lv, P.; Yu, J.; Xu, X.; Lu, T.; Xu, F. Eriodictyol inhibits high glucose-induced oxidative stress and inflammation in retinal ganglial cells. J. Cell. Biochem. 2019, 120, 5644–5651. [Google Scholar] [CrossRef]
- Long, M.; Yang, S.; Li, P.; Song, X.; Pan, J.; He, J.; Zhang, Y.; Wu, R. Combined Use of C. butyricum Sx-01 and L. salivarius C-1-3 Improves Intestinal Health and Reduces the Amount of Lipids in Serum via Modulation of Gut Microbiota in Mice. Nutrients 2018, 10, 810. [Google Scholar] [CrossRef]
- Fayard, E.; Xue, G.; Parcellier, A.; Bozulic, L.; Hemmings, B.A. Protein kinase B (PKB/Akt), a key mediator of the PI3K signaling pathway. Curr. Top. Microbiol. Immunol. 2010, 346, 31–56. [Google Scholar]
- Stark, A.K.; Sriskantharajah, S.; Hessel, E.M.; Okkenhaug, K. PI3K inhibitors in inflammation, autoimmunity and cancer. Curr. Opin. Pharmacol. 2015, 23, 82–91. [Google Scholar] [CrossRef]
- Tang, L.; Wang, F.; Xiao, L.; Shen, M.; Xia, S.; Zhang, Z.; Zhang, F.; Zheng, S.; Tan, S. Yi-Qi-Jian-Pi formula modulates the PI3K/AKT signaling pathway to attenuate acute-on-chronic liver failure by suppressing hypoxic injury and apoptosis in vivo and in vitro. J. Ethnopharmacol. 2021, 280, 114411. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Lv, H.; Li, J.; Che, Y.; Xu, B.; Tao, Z.; Jiang, W. Roles of Nrf2/HO-1 and HIF-1alpha/VEGF in lung tissue injury and repair following cerebral ischemia/reperfusion injury. J. Cell. Physiol. 2019, 234, 7695–7707. [Google Scholar] [CrossRef] [PubMed]
- Codispoti, B.; Makeeva, I.; Sied, J.; Benincasa, C.; Scacco, S.; Tatullo, M. Should we reconsider the apoptosis as a strategic player in tissue regeneration? Int. J. Biol. Sci. 2019, 15, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Zhou, J.; Qin, X.; Shi, X.; Zeng, Q.; Liu, J.; Yan, S.; Zhang, L. Reduction of apoptosis by proanthocyanidin-induced autophagy in the human gastric cancer cell line MGC-803. Oncol. Rep. 2016, 35, 649–658. [Google Scholar] [CrossRef]


| Gene | Primer Sequence | |
|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |
| Pi3k | CCGTGATGGAAAATATGGCTT | AGCTAAAGACTCATTCCGGTA |
| Akt1 | CGGTTCTTTGCCAACATCGT | CCTCATCGAAATACCTGGTGT |
| mTor | ATCCTGCACATTGACTTTGGG | ATGTGGTTCTGTAGTTGCCAT |
| Tsc2 | CTGCCTCTGTTCATTATCACC | TTACGCATCAACTTCCAGCAA |
| Bax | ATGCGTCCACCAAGAAGC | CAGTTGAAGTTGCCATCAGC |
| Bcl-xl | TCGACTTTCTCTCCTACAAGC | GCCTCAGTCCTATTCTCTTCG |
| Caspase3 | CTCTGGGATCTATCTGGACA | GATGACATTCCAGTGCTC |
| Caspase8 | CTTCGAGCAACAGAACCACAC | TTCTTCACCGTAGCCATTCCC |
| Bid | GTTCATGAATGGCAGCCTGT | TGGAAGACATCACGGAGCAA |
| GAPDH | CATCCGTAAAGACCTCTATGCCAAC | ATGGAGCCACCGATCCACA |
| Number | Name | Degree |
|---|---|---|
| 1 | ALB | 84 |
| 2 | ACTB | 78 |
| 3 | AKT1 | 78 |
| 4 | VEGFA | 68 |
| 5 | HSP90AA1 | 64 |
| 6 | HIF1A | 62 |
| 7 | PPARG | 62 |
| 8 | ESR1 | 60 |
| 9 | SRC | 58 |
| 10 | PTGS2 | 54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Wu, X.; Wen, Q.; Tang, H.; Zhao, L.; Shi, F.; Li, Y.; Yin, Z.; Zou, Y.; Song, X.; et al. Eriodictyol Alleviated LPS/D-GalN-Induced Acute Liver Injury by Inhibiting Oxidative Stress and Cell Apoptosis via PI3K/AKT Signaling Pathway. Nutrients 2023, 15, 4349. https://doi.org/10.3390/nu15204349
Zheng X, Wu X, Wen Q, Tang H, Zhao L, Shi F, Li Y, Yin Z, Zou Y, Song X, et al. Eriodictyol Alleviated LPS/D-GalN-Induced Acute Liver Injury by Inhibiting Oxidative Stress and Cell Apoptosis via PI3K/AKT Signaling Pathway. Nutrients. 2023; 15(20):4349. https://doi.org/10.3390/nu15204349
Chicago/Turabian StyleZheng, Xiaomei, Xinyan Wu, Qiqi Wen, Huaqiao Tang, Ling Zhao, Fei Shi, Yinglun Li, Zhongqiong Yin, Yuanfeng Zou, Xu Song, and et al. 2023. "Eriodictyol Alleviated LPS/D-GalN-Induced Acute Liver Injury by Inhibiting Oxidative Stress and Cell Apoptosis via PI3K/AKT Signaling Pathway" Nutrients 15, no. 20: 4349. https://doi.org/10.3390/nu15204349
APA StyleZheng, X., Wu, X., Wen, Q., Tang, H., Zhao, L., Shi, F., Li, Y., Yin, Z., Zou, Y., Song, X., Li, L., Zhao, X., & Ye, G. (2023). Eriodictyol Alleviated LPS/D-GalN-Induced Acute Liver Injury by Inhibiting Oxidative Stress and Cell Apoptosis via PI3K/AKT Signaling Pathway. Nutrients, 15(20), 4349. https://doi.org/10.3390/nu15204349







