The Diversity of Gut Microbiota at Weaning Is Altered in Prolactin Receptor-Null Mice
Highlights
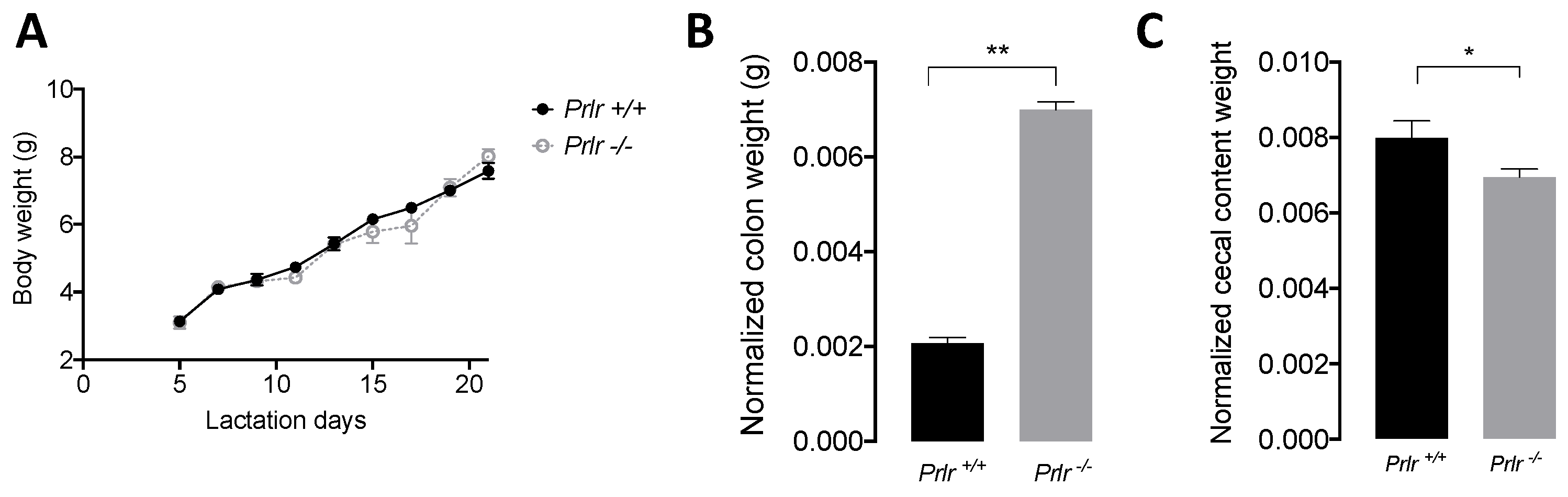
- Prolactin receptor-null lactating mice exhibited lower gut bacterial diversity (Simpson Evenness levels) compared to their wild-type counterparts.
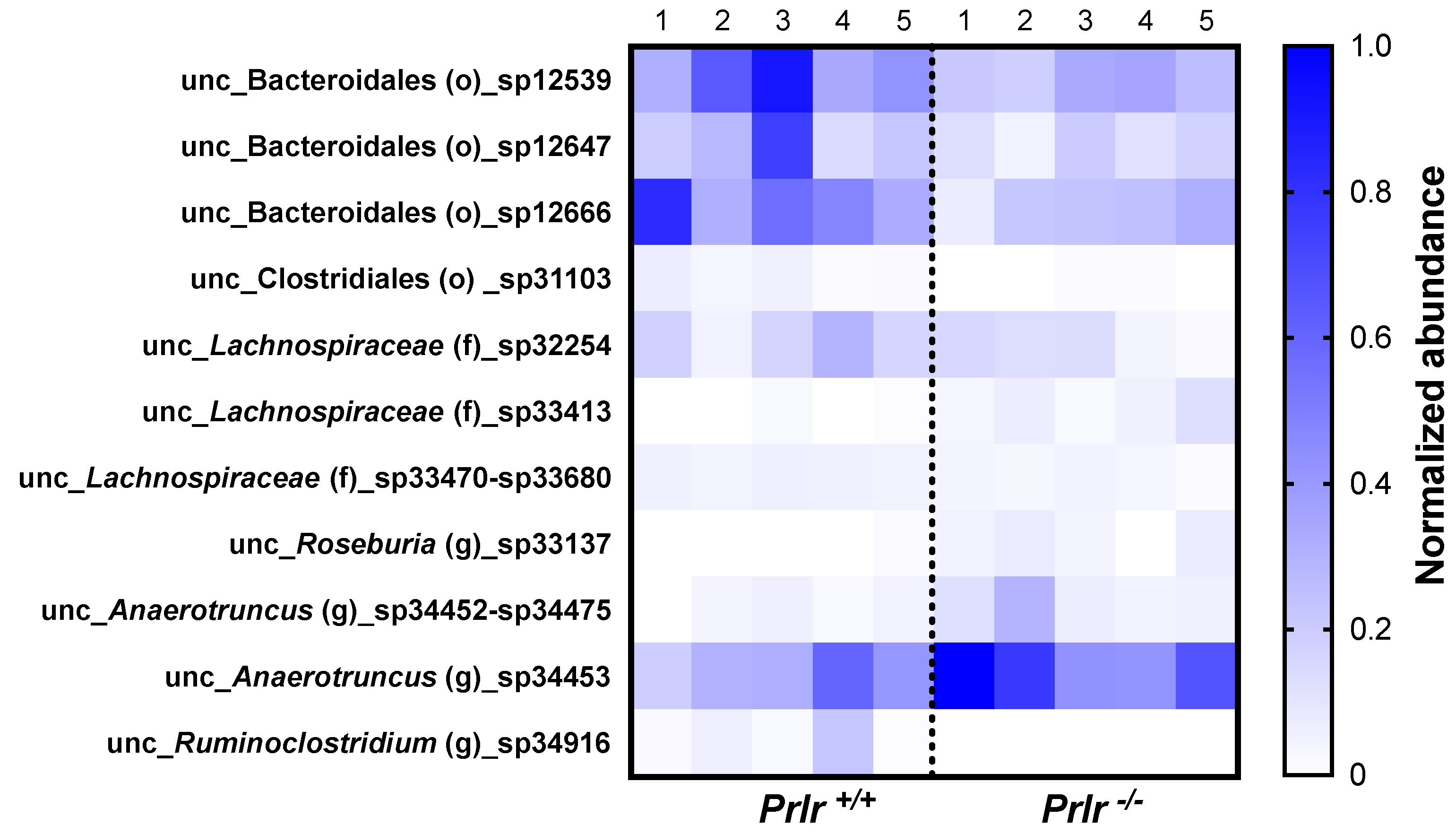
- Out of 469 bacterial species, 11 were differentially abundant based on the mice phenotype. Prlr −/− mice demonstrated the lowest abundances of bacteria from the Lachnospiraceae family and Roseburia genus.
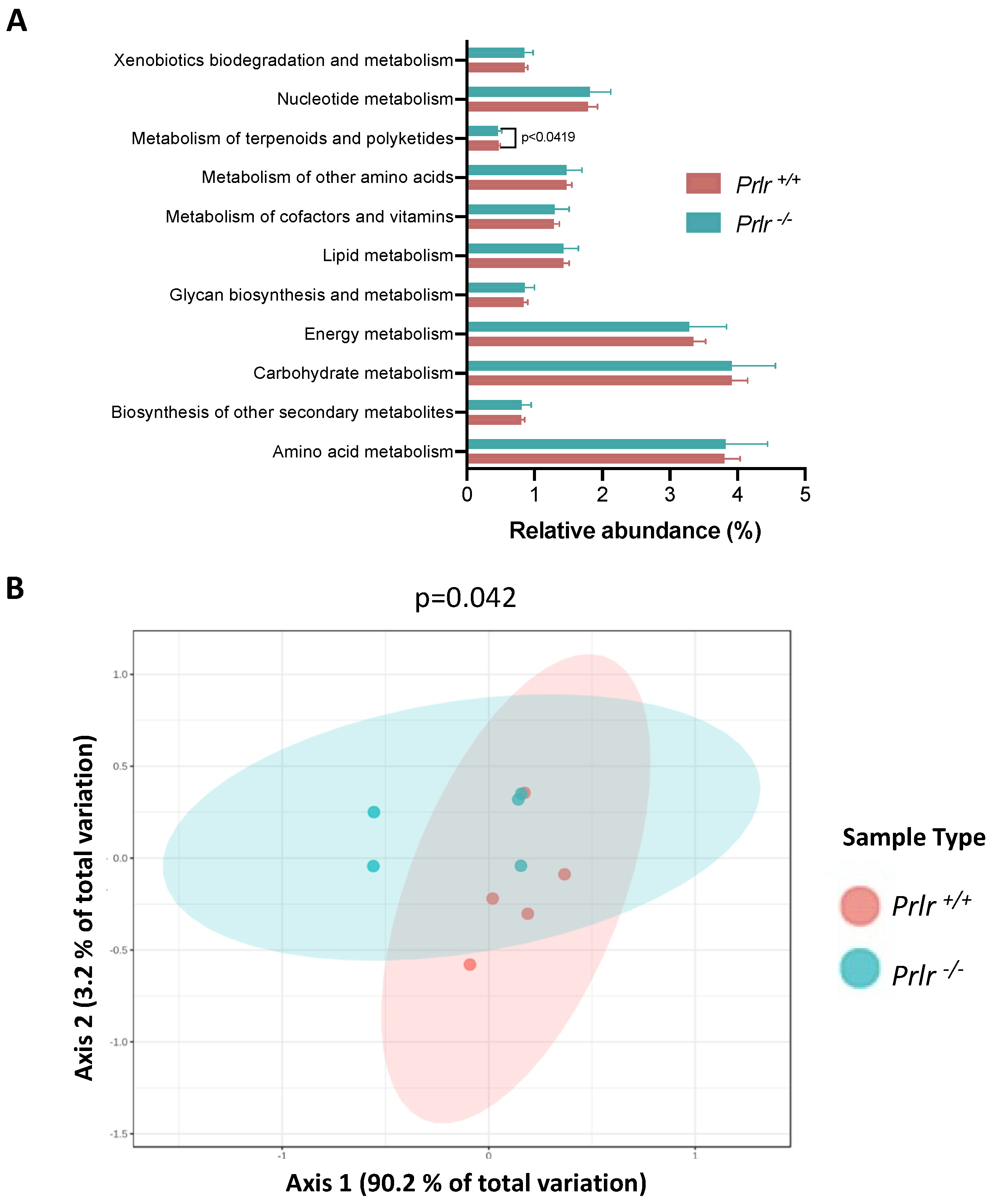
- Predictive analysis of metabolic performance indicated a reduction in the metabolism of terpenoids and polyketides in Prlr −/− mice.
- Prolactin is a crucial hormone during breastfeeding, and the absence of its signaling in vivo results in lower microbiota diversity and richness; this may contribute to an increased risk of pro-inflammatory conditions such as obesity and gastrointestinal diseases.
- The predicted reduction in colonic metabolism of terpenoids and polyketides in lactating mice lacking the prolactin receptor may suggest a diminished capacity to regulate colonic biosynthesis and oxidation of microbial metabolites. These results highlight the relevance of signals derived from maternal milk to the health of the lactating offspring.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Husbandry, Care, and Macroscopic Measurements
2.2. Fecal and Cecal DNA Extraction and Quality Control
2.3. Library Preparation, 16S rRNA Sequencing, and Diversity Index Analyses
2.4. Statistical Analysis
3. Results
3.1. Body Weight and Macroscopic Measurements
3.2. General Microbial Diversity Analyses of the Weaned Prlr-WT and Prlr-KO Mice
3.3. Taxonomical Bacterial Composition
3.4. PICRUSt Metabolic Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; Lieber, A.D.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, Birth Mode, and Diet Shape Microbiome Maturation during Early Life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokossou, G.A.G.; Kouakanou, L.; Schumacher, A.; Zenclussen, A.C. Human Breast Milk: From Food to Active Immune Response With Disease Protection in Infants and Mothers. Front. Immunol. 2022, 13, 849012. [Google Scholar] [CrossRef]
- Gopalakrishnan, L.; Doriya, K.; Kumar, D.S. Moringa Oleifera: A Review on Nutritive Importance and Its Medicinal Applications. Food Sci. Hum. Wellness 2016, 5, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Godhia, M.; Patel, N. Colostrum—Its Composition, Benefits As A Nutraceutical: A Review. Curr. Res. Nutr. Food Sci. J. 2013, 1, 37–47. [Google Scholar] [CrossRef]
- Macotela, Y.; Ruiz-Herrera, X.; Vázquez-Carrillo, D.I.; Ramírez-Hernandez, G.; Martínez de la Escalera, G.; Clapp, C. The Beneficial Metabolic Actions of Prolactin. Front. Endocrinol. 2022, 13, 1001703. [Google Scholar] [CrossRef]
- Macotela, Y.; Triebel, J.; Clapp, C. Time for a New Perspective on Prolactin in Metabolism. Trends Endocrinol. Metab. 2020, 31, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Luzardo-Ocampo, I.; Dena-Beltrán, J.L.; Ruiz-Herrera, X.; Ocampo-Ruiz, A.L.; Martínez de la Escalera, G.; Clapp, C.; Macotela, Y. Obesity-Derived Alterations in the Lactating Mammary Gland: Focus on Prolactin. Mol. Cell. Endocrinol. 2023, 559, 111810. [Google Scholar] [CrossRef]
- de los Ríos, E.A.; Ruiz-Herrera, X.; Tinoco-Pantoja, V.; López-Barrera, F.; Escalera, G.M.; Clapp, C.; Macotela, Y. Impaired Prolactin Actions Mediate Altered Offspring Metabolism Induced by Maternal High-fat Feeding during Lactation. FASEB J. 2018, 32, 3457–3470. [Google Scholar] [CrossRef] [Green Version]
- Rizzetto, L.; Fava, F.; Tuohy, K.M.; Selmi, C. Connecting the Immune System, Systemic Chronic Inflammation and the Gut Microbiome: The Role of Sex. J. Autoimmun. 2018, 92, 12–34. [Google Scholar] [CrossRef]
- Neuman, H.; Debelius, J.W.; Knight, R.; Koren, O. Microbial Endocrinology: The Interplay between the Microbiota and the Endocrine System. FEMS Microbiol. Rev. 2015, 39, 509–521. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-F.; Fu, S.-P.; Li, S.-N.; Hu, Z.-M.; Xue, W.-J.; Li, Z.-Q.; Huang, B.-X.; Lv, Q.-K.; Liu, J.-X.; Wang, W. Short-Chain Fatty Acids Inhibit Growth Hormone and Prolactin Gene Transcription via CAMP/PKA/CREB Signaling Pathway in Dairy Cow Anterior Pituitary Cells. Int. J. Mol. Sci. 2013, 14, 21474–21488. [Google Scholar] [CrossRef] [Green Version]
- Al Nabhani, Z.; Dulauroy, S.; Marques, R.; Cousu, C.; Al Bounny, S.; Déjardin, F.; Sparwasser, T.; Bérard, M.; Cerf-Bensussan, N.; Eberl, G. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity 2019, 50, 1276–1288.e5. [Google Scholar] [CrossRef]
- Noce, A.; Marrone, G.; Di Daniele, F.; Ottaviani, E.; Wilson Jones, G.; Bernini, R.; Romani, A.; Rovella, V. Impact of Gut Microbiota Composition on Onset and Progression of Chronic Non-Communicable Diseases. Nutrients 2019, 11, 1073. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.H.; Cho, Y.-S. Fecal Microbiota Transplantation: Current Applications, Effectiveness, and Future Perspectives. Clin. Endosc. 2016, 49, 257–265. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Yin, L.; Wan, Y.-D.; Pan, X.-T.; Zhou, C.-Y.; Lin, N.; Ma, C.-T.; Yao, J.; Su, Z.; Wan, C.; Yu, Y.-W.; et al. Association Between Gut Bacterial Diversity and Mortality in Septic Shock Patients: A Cohort Study. Med. Sci. Monit. 2019, 25, 7376–7382. [Google Scholar] [CrossRef]
- Moore, J.C. Diversity, Taxonomic versus Functional. In Encyclopedia of Biodiversity; Levin, S.A., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2013; pp. 648–656. ISBN 978-0-12-384720-1. [Google Scholar]
- Ormandy, C.J.; Camus, A.; Barra, J.; Damotte, D.; Lucas, B.; Buteau, H.; Edery, M.; Brousse, N.; Babinet, C.; Binart, N.; et al. Null Mutation of the Prolactin Receptor Gene Produces Multiple Reproductive Defects in the Mouse. Genes Dev. 1997, 11, 167–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horseman, N.D. Defective Mammopoiesis, but Normal Hematopoiesis, in Mice with a Targeted Disruption of the Prolactin Gene. EMBO J. 1997, 16, 6926–6935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, M.P.; Jensen, E.R.; Montecino-Rodriguez, E.; Leathers, H.; Horseman, N.; Dorshkind, K. Humoral and Cell-Mediated Immunity in Mice with Genetic Deficiencies of Prolactin, Growth Hormone, Insulin-like Growth Factor-I, and Thyroid Hormone. Clin. Immunol. 2000, 96, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, B.; Ormandy, C.J.; Di Santo, J.P.; Kelly, P.A. Immune System Development and Function in Prolactin Receptor-Deficient Mice. J. Immunol. 1999, 163, 576–582. [Google Scholar] [CrossRef]
- Ruiz-Herrera, X.; de los Ríos, E.A.; Díaz, J.M.; Lerma-Alvarado, R.M.; de la Escalera, L.M.; López-Barrera, F.; Lemini, M.; Arnold, E.; de la Escalera, G.M.; Clapp, C.; et al. Prolactin Promotes Adipose Tissue Fitness and Insulin Sensitivity in Obese Males. Endocrinology 2017, 158, 56–68. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Carranza, B.; Bravo-Manríquez, M.; Baez, A.; Ledesma-Colunga, M.G.; Ruiz-Herrera, X.; Reyes-Ortega, P.; de los Ríos, E.A.; Macotela, Y.; Martínez de la Escalera, G.; Clapp, C. Prolactin Regulates Liver Growth during Postnatal Development in Mice. Am. J. Physiol. Integr. Comp. Physiol. 2018, 314, R902–R908. [Google Scholar] [CrossRef] [Green Version]
- Ben-Jonathan, N.; LaPensee, C.R.; LaPensee, E.W. What Can We Learn from Rodents about Prolactin in Humans? Endocr. Rev. 2008, 29, 1–41. [Google Scholar] [CrossRef] [Green Version]
- van den Elsen, L.W.J.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the Gut Microbiota by Breastfeeding: The Gateway to Allergy Prevention? Front. Pediatr. 2019, 7, 47. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Sakamoto, K.; Seo, S.-U.; Pickard, J.M.; Gillilland, M.G.; Pudlo, N.A.; Hoostal, M.; Li, X.; Wang, T.D.; Feehley, T.; et al. Neonatal Acquisition of Clostridia Species Protects against Colonization by Bacterial Pathogens. Science 2017, 356, 315–319. [Google Scholar] [CrossRef] [Green Version]
- McKenney, P.T.; Pamer, E.G. From Hype to Hope: The Gut Microbiota in Enteric Infectious Disease. Cell 2015, 163, 1326–1332. [Google Scholar] [CrossRef] [Green Version]
- Christian, P.; Smith, E.R.; Lee, S.E.; Vargas, A.J.; Bremer, A.A.; Raiten, D.J. The Need to Study Human Milk as a Biological System. Am. J. Clin. Nutr. 2021, 113, 1063–1072. [Google Scholar] [CrossRef]
- Erliana, U.D.; Fly, A.D. The Function and Alteration of Immunological Properties in Human Milk of Obese Mothers. Nutrients 2019, 11, 1284. [Google Scholar] [CrossRef] [Green Version]
- Yuen, B.H. Prolactin in Human Milk: The Influence of Nursing and the Duration of Postpartum Lactation. Am. J. Obstet. Gynecol. 1988, 158, 583–586. [Google Scholar] [CrossRef]
- Jensen, E.A.; Young, J.A.; Mathes, S.C.; List, E.O.; Carroll, R.K.; Kuhn, J.; Onusko, M.; Kopchick, J.J.; Murphy, E.R.; Berryman, D.E. Crosstalk between the Growth Hormone/Insulin-like Growth Factor-1 Axis and the Gut Microbiome: A New Frontier for Microbial Endocrinology. Growth Horm. IGF Res. 2020, 53–54, 101333. [Google Scholar] [CrossRef]
- Tran, H.Q.; Bretin, A.; Adeshirlarijaney, A.; Yeoh, B.S.; Vijay-Kumar, M.; Zou, J.; Denning, T.L.; Chassaing, B.; Gewirtz, A.T. “Western Diet”-Induced Adipose Inflammation Requires a Complex Gut Microbiota. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 313–333. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Schubert, A.M.; Zackular, J.P.; Iverson, K.D.; Young, V.B.; Petrosino, J.F. Stabilization of the Murine Gut Microbiome Following Weaning. Gut Microbes 2012, 3, 383–393. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L. Assessing and Improving Methods Used in Operational Taxonomic Unit-Based Approaches for 16S RRNA Gene Sequence Analysis. Appl. Environ. Microbiol. 2011, 77, 3219–3226. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Lou, J.; Yang, L.; Wang, H.; Wu, L.; Xu, J. Assessing Soil Bacterial Community and Dynamics by Integrated High-Throughput Absolute Abundance Quantification. PeerJ 2018, 6, e4514. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Triebel, J.; Robles, J.P.; Zamora, M.; Martínez de la Escalera, G.; Bertsch, T.; Clapp, C. Regulator of Angiogenesis and Vascular Function: A 2019 Update of the Vasoinhibin Nomenclature. Front. Endocrinol. 2019, 10, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devost, D.; Boutin, J.-M. Autoregulation of the Rat Prolactin Gene in Lactotrophs. Mol. Cell. Endocrinol. 1999, 158, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Grases-Pintó, B.; Abril-Gil, M.; Castell, M.; Rodríguez-Lagunas, M.J.; Burleigh, S.; Fåk Hållenius, F.; Prykhodko, O.; Pérez-Cano, F.J.; Franch, À. Influence of Leptin and Adiponectin Supplementation on Intraepithelial Lymphocyte and Microbiota Composition in Suckling Rats. Front. Immunol. 2019, 10, 2369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marungruang, N.; Arévalo Sureda, E.; Lefrançoise, A.; Weström, B.; Nyman, M.; Prykhodko, O.; Fåk Hållenius, F. Impact of Dietary Induced Precocious Gut Maturation on Cecal Microbiota and Its Relation to the Blood-Brain Barrier during the Postnatal Period in Rats. Neurogastroenterol. Motil. 2018, 30, e13285. [Google Scholar] [CrossRef] [Green Version]
- Lemas, D.J.; Young, B.E.; Baker, P.R.; Tomczik, A.C.; Soderborg, T.K.; Hernandez, T.L.; de la Houssaye, B.A.; Robertson, C.E.; Rudolph, M.C.; Ir, D.; et al. Alterations in Human Milk Leptin and Insulin Are Associated with Early Changes in the Infant Intestinal Microbiome. Am. J. Clin. Nutr. 2016, 103, 1291–1300. [Google Scholar] [CrossRef] [Green Version]
- Adlercreutz, H.; Pulkkinen, M.O.; Hämäläinen, E.K.; Korpela, J.T. Studies on the Role of Intestinal Bacteria in Metabolism of Synthetic and Natural Steroid Hormones. J. Steroid Biochem. 1984, 20, 217–229. [Google Scholar] [CrossRef]
- Léonhardt, M.; Lesage, J.; Croix, D.; Dutriez-Casteloot, I.; Beauvillain, J.C.; Dupouy, J.P. Effects of Perinatal Maternal Food Restriction on Pituitary-Gonadal Axis and Plasma Leptin Level in Rat Pup at Birth and Weaning and on Timing of Puberty. Biol. Reprod. 2003, 68, 390–400. [Google Scholar] [CrossRef] [Green Version]
- Wostmann, B.S. Morphology and Physiology, Endocrinology and Biochemistry. In Germfree and Gnotobiotic Animal Models; Wostmann, B.S., Ed.; CRC Press: London, UK, 1996; pp. 39–66. ISBN 9780138753320. [Google Scholar]
- Pelzer, E.S.; Allan, J.A.; Theodoropoulos, C.; Ross, T.; Beagley, K.W.; Knox, C.L. Hormone-Dependent Bacterial Growth, Persistence and Biofilm Formation—A Pilot Study Investigating Human Follicular Fluid Collected during IVF Cycles. PLoS ONE 2012, 7, e49965. [Google Scholar] [CrossRef] [Green Version]
- Ohwaki, M.; Suganuma, N.; Seo, H.; Nawa, A.; Kikkawa, F.; Narita, O.; Matsui, N.; Tomoda, Y. Source of Prolactin in Human Follicular Fluid. Endocrinol. Jpn. 1992, 39, 601–607. [Google Scholar] [CrossRef] [Green Version]
- Menon, R.; Watson, S.E.; Thomas, L.N.; Allred, C.D.; Dabney, A.; Azcarate-Peril, M.A.; Sturino, J.M. Diet Complexity and Estrogen Receptor β Status Affect the Composition of the Murine Intestinal Microbiota. Appl. Environ. Microbiol. 2013, 79, 5763–5773. [Google Scholar] [CrossRef] [Green Version]
- Ridlon, J.M.; Ikegawa, S.; Alves, J.M.P.; Zhou, B.; Kobayashi, A.; Iida, T.; Mitamura, K.; Tanabe, G.; Serrano, M.; De Guzman, A.; et al. Clostridium Scindens: A Human Gut Microbe with a High Potential to Convert Glucocorticoids into Androgens. J. Lipid Res. 2013, 54, 2437–2449. [Google Scholar] [CrossRef] [Green Version]
- Clarke, M.B.; Hughes, D.T.; Zhu, C.; Boedeker, E.C.; Sperandio, V. The QseC Sensor Kinase: A Bacterial Adrenergic Receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Susic, D.F.; Wang, L.; Roberts, L.M.; Bai, M.; Gia, A.; McGovern, E.; Jiang, X.-T.; Davis, G.K.; El-Omar, E.; Henry, A. The P4 Study: Postpartum Maternal and Infant Faecal Microbiome 6 Months After Hypertensive Versus Normotensive Pregnancy. Front. Cell. Infect. Microbiol. 2022, 12, 646165. [Google Scholar] [CrossRef]
- Bailén, M.; Bressa, C.; Martínez-López, S.; González-Soltero, R.; Montalvo Lominchar, M.G.; San Juan, C.; Larrosa, M. Microbiota Features Associated With a High-Fat/Low-Fiber Diet in Healthy Adults. Front. Nutr. 2020, 7, 583608. [Google Scholar] [CrossRef]
- Fang, S.; Chen, X.; Zhou, L.; Wang, C.; Chen, Q.; Lin, R.; Xiao, T.; Gan, Q. Faecal Microbiota and Functional Capacity Associated with Weaning Weight in Meat Rabbits. Microb. Biotechnol. 2019, 12, 1441–1452. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [Green Version]
- Luzardo-Ocampo, I.; Loarca-Piña, G.; Gonzalez de Mejia, E. Gallic and Butyric Acids Modulated NLRP3 Inflammasome Markers in a Co-Culture Model of Intestinal Inflammation. Food Chem. Toxicol. 2020, 146, 111835. [Google Scholar] [CrossRef]
- Younge, N.E.; Newgard, C.B.; Cotten, C.M.; Goldberg, R.N.; Muehlbauer, M.J.; Bain, J.R.; Stevens, R.D.; O’Connell, T.M.; Rawls, J.F.; Seed, P.C.; et al. Disrupted Maturation of the Microbiota and Metabolome among Extremely Preterm Infants with Postnatal Growth Failure. Sci. Rep. 2019, 9, 8167. [Google Scholar] [CrossRef] [Green Version]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017, 171, 647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular Mechanisms of Cell Death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Ağagündüz, D.; Cemali, Ö.; Çelik, E. Interaction Between Natural Products and Gut Microbiota. Curr. Pharmacol. Rep. 2022, 9, 7–31. [Google Scholar] [CrossRef]
- Heinken, A.; Ravcheev, D.A.; Baldini, F.; Heirendt, L.; Fleming, R.M.T.; Thiele, I. Systematic Assessment of Secondary Bile Acid Metabolism in Gut Microbes Reveals Distinct Metabolic Capabilities in Inflammatory Bowel Disease. Microbiome 2019, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- Oldfield, E.; Lin, F.-Y. Terpene Biosynthesis: Modularity Rules. Angew. Chem. Int. Ed. 2012, 51, 1124–1137. [Google Scholar] [CrossRef] [Green Version]
- Fobofou, S.A.; Savidge, T. Microbial Metabolites: Cause or Consequence in Gastrointestinal Disease? Am. J. Physiol. Liver Physiol. 2022, 322, G535–G552. [Google Scholar] [CrossRef]
- Tang, X.; Kudo, Y.; Baker, J.L.; LaBonte, S.; Jordan, P.A.; McKinnie, S.M.K.; Guo, J.; Huan, T.; Moore, B.S.; Edlund, A. Cariogenic Streptococcus Mutans Produces Tetramic Acid Strain-Specific Antibiotics That Impair Commensal Colonization. ACS Infect. Dis. 2020, 6, 563–571. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luzardo-Ocampo, I.; Ocampo-Ruiz, A.L.; Dena-Beltrán, J.L.; Martínez de la Escalera, G.; Clapp, C.; Macotela, Y. The Diversity of Gut Microbiota at Weaning Is Altered in Prolactin Receptor-Null Mice. Nutrients 2023, 15, 3447. https://doi.org/10.3390/nu15153447
Luzardo-Ocampo I, Ocampo-Ruiz AL, Dena-Beltrán JL, Martínez de la Escalera G, Clapp C, Macotela Y. The Diversity of Gut Microbiota at Weaning Is Altered in Prolactin Receptor-Null Mice. Nutrients. 2023; 15(15):3447. https://doi.org/10.3390/nu15153447
Chicago/Turabian StyleLuzardo-Ocampo, Ivan, Ana Luisa Ocampo-Ruiz, José Luis Dena-Beltrán, Gonzalo Martínez de la Escalera, Carmen Clapp, and Yazmín Macotela. 2023. "The Diversity of Gut Microbiota at Weaning Is Altered in Prolactin Receptor-Null Mice" Nutrients 15, no. 15: 3447. https://doi.org/10.3390/nu15153447
APA StyleLuzardo-Ocampo, I., Ocampo-Ruiz, A. L., Dena-Beltrán, J. L., Martínez de la Escalera, G., Clapp, C., & Macotela, Y. (2023). The Diversity of Gut Microbiota at Weaning Is Altered in Prolactin Receptor-Null Mice. Nutrients, 15(15), 3447. https://doi.org/10.3390/nu15153447






