The Vegetable ‘Kale’ Protects against Dextran-Sulfate-Sodium-Induced Acute Inflammation through Moderating the Ratio of Proinflammatory and Anti-Inflammatory LPS-Producing Bacterial Taxa and Augmenting the Gut Barrier in C57BL6 Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Study
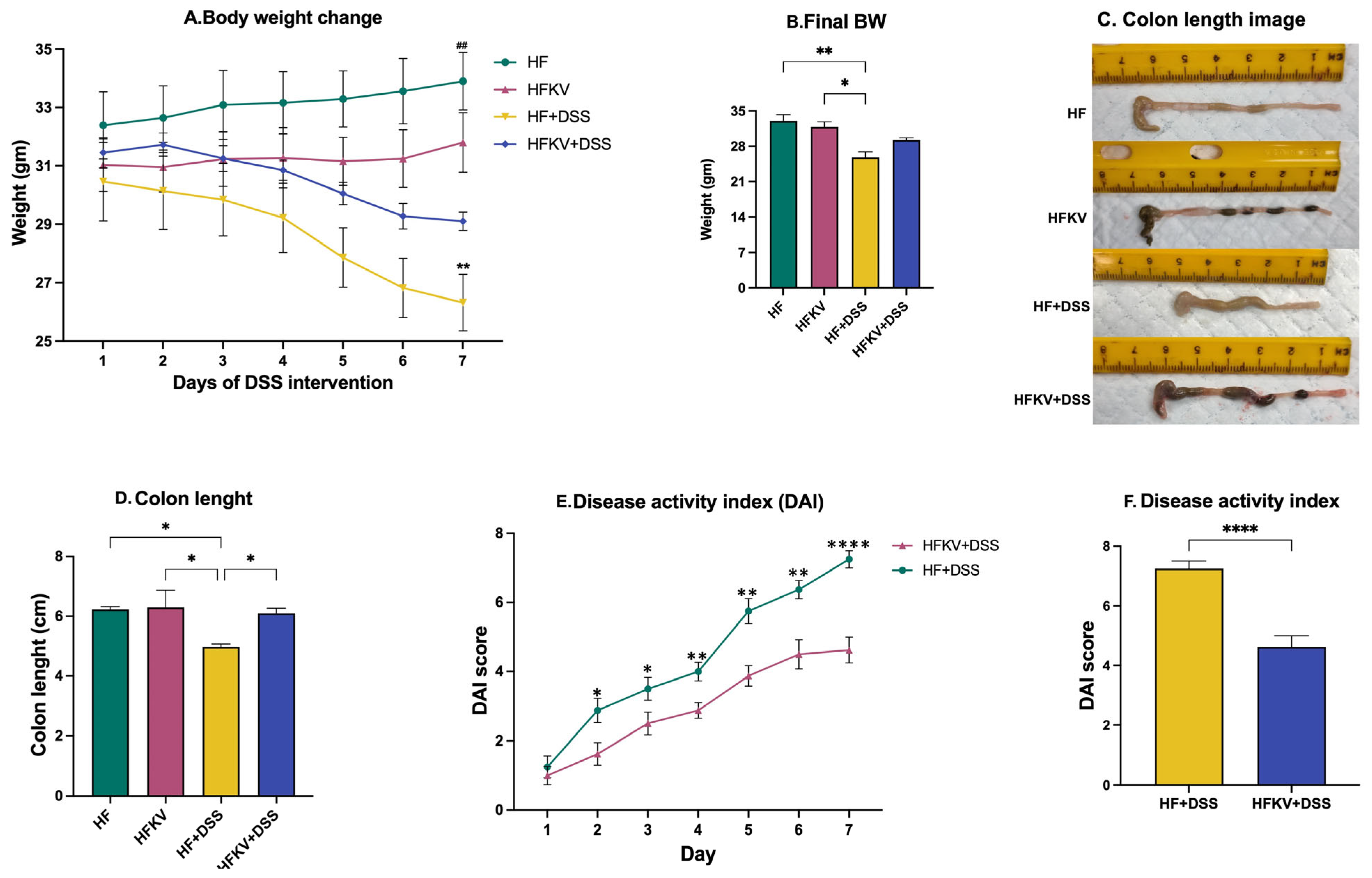
2.2. Disease Activity Index (DAI)
2.3. Euthanasia and Tissue Collection
2.4. Hematoxylin and Eosin (H & E) Staining
2.5. RNA Extraction and cDNA Preparation
2.6. Quantifying the Target Tight Junction Genes and Inflammation Markers by qPCR
2.7. Determination of Protein Expression by Western Blot
2.8. DNA Extraction and Amplification of the 16S Hypervariable Regions
2.9. Library Preparation, Template Preparation and Sequencing
3. 16S rRNA Gene Amplicon Sequencing Analysis
3.1. Linear Discriminant Analysis
3.2. PICRUSt2
3.3. Determination of LPS and LPB in Serum
3.4. Quantification of genus Turicibacter by qPCR
3.5. In Vitro Tests Using Kale Extract
4. Statistical Analysis
5. Results
5.1. Determination of DAI Score Parameters
5.2. Effects of Kale Supplementation on Histopathological Changes and Splenomegaly
5.3. Effects of Kale Supplementation on Gut Barrier Integrity
5.4. Effects of Kale Supplementation on Inflammatory Responses
5.5. Effects of Kale Supplementation on Gut Microbiota Composition and Diversity
5.5.1. Rarefaction
5.5.2. Alpha Diversity Measures
5.5.3. Beta Diversity
5.5.4. Comparative Analysis of the Gut Microbiota Taxa Composition
5.5.5. Linear Discriminant Analysis Effect Size (LEfSe)
5.6. Predicted Metabolic Functions
5.7. Effects of Kale Supplementation on LPS-Producing Bacteria and the LPS-Producing Pathway
5.8. Effects of Kale Supplementation on Genus Turicibacter and Tryptophan Metabolism Pathway
5.9. Effect of Kale Extract on LPS-induced Inflammation in RAW 264.7 Macrophages
6. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regionalregional, and National burden of inflammatory bowel disease in 195 countries and territories, 1990–2017. A Systematic analysis for the Global burden of disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Thomson, I.C.; Bryant, R.V.; Costello, S.P. Uncovering the cause of ulcerative colitis. Open Access J. Gastroenterol. Hepatol. 2019, 3, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Holman, J.; McKinstry, D.; Trindade, B.C.; Eaton, K.A.; Mendoza-Castrejon, J.; Ho, S.; Wells, E.; Yuan, H.; Wen, B.; et al. A steamed broccoli sprout diet preparation that reduces colitis via the gut microbiota. J. Nutr. Biochem. 2023, 112, 109215. [Google Scholar] [CrossRef]
- Migliozz, M.; Thavarajah, D.; Thavarajah, P.; Smith, P. Lentil, and kale: Complementary nutrient-rich whole food sources to combat micronutrient and calorie malnutrition. Nutrients 2015, 7, 9285–9298. [Google Scholar] [CrossRef] [PubMed]
- Di Noia, J. Defining powerhouse fruits and vegetables: A nutrient density approach. Prev. Chronic. Dis. 2014, 11, 130390. [Google Scholar] [CrossRef]
- Shahinozzaman, M.; Raychaudhuri, S.; Fan, S.; Obanda, D.N. Kale Attenuates Inflammation and Modulates Gut Microbial Composition and Function in C57BL/6J Mice with Diet Induced Obesity. Microorganisms 2021, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, S.; Fan, S.; Kraus, O.; Shahinozzaman, M.; Obanda, D.N. Kale supplementation during high fat feeding improves metabolic health in a mouse model of obesity and insulin resistance. PLoS ONE 2021, 16, e0256348. [Google Scholar] [CrossRef]
- Gronbach, K.; Flade, I.; Holst, O.; Lindner, B.; Ruscheweyh, H.J.; Wittmann, A.; Menz, S.; Schwiertz, A.; Adam, P.; Stecher, B.; et al. Endotoxicity of lipopolysaccharide as a determinant of T-cell-mediated colitis induction in mice. Gastroenterology 2014, 146, 765–775. [Google Scholar] [CrossRef]
- Steimle, A.; Autenrieth, I.B.; Frick, J.S. Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 2016, 306, 290–301. [Google Scholar] [CrossRef]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hamalainen, A.M.; et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 2014, 104, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemicaly induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.A. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Correction in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Xue, Z.; Coates, L.C.; Marco, M.L. Viable and Total Bacterial Populations Undergo Equipment- and Time-Dependent Shifts during Milk Processing. Appl. Environ. Microbiol. 2019, 85, e00270-19. [Google Scholar] [CrossRef]
- Lynch, J.B.; Gonzalez, E.L.; Choy, K.; Faull, K.F.; Jewell, T.; Arellano, A.; Liang, J.; Yu, K.B.; Paramo, J.; Hsiao, E.Y. Turicibacter modifies host bile acids and lipids in a strain-specific manner. bioRxiv 2022. [Google Scholar] [CrossRef]
- Oldham, A.L.; Duncan, K.E. Similar gene estimates from circular and linear standards in quantitative PCR analyses using the prokaryotic 16S rRNA Gene as a model. PLoS ONE 2012, 7, e51931. [Google Scholar] [CrossRef]
- Olsen, H.; Aaby, K.; Borge, G.I. Characterization, and quantification of flavonoids and hydroxycinnamic acids in curly kale (Brassica oleracea L. Convar. acephala Var. sabellica) by HPLC-DAD-ESI-MSn. J. Agric. Food. Chem. 2009, 57, 2816–2825. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Kostov, R.V. Glucosinolates and isothiocyanates in health and disease. Trends Mol. Med. 2012, 18, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kim, J.K.; Kim, H.; Kim, Y.J.; Park, Y.J.; Kim, S.J.; Kim, C.; Park, S.U. Transcriptome analysis and metabolic profiling of green and red kale (Brassica oleracea var. acephala) seedlings. Food Chem. 2018, 15, 241. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.K.; Pritt, B.S.; Alexander, M.P. Histopathologic review of granulomatous inflammation. J. Clin. Tuberc. Other Mycobact. Dis. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dos Anjos Cassado, A. F4/80 as a major macrophage marker: The case of the peritoneum and spleen. Results Probl. Cell Differ. 2017, 62, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Cochran, K.E.; Lamson, N.G.; Whitehead, K.A. Expanding the utility of the dextran sulfate sodium (DSS) mouse model to induce a clinically relevant loss of intestinal barrier function. PeerJ 2020, 8, e8681. [Google Scholar] [CrossRef]
- Llewellyn, S.R.; Britton, G.J.; Contijoch, E.J.; Vennaro, O.H.; Mortha, A. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology 2018, 154, 1037–1046.e2. [Google Scholar] [CrossRef]
- Forster, S.C.; Clare, S.; Beresford-Jones, B.S.; Harcourt, K.; Notley, G.; Stares, M.D.; Kumar, N.; Soderholm, A.T.; Adoum, A.; Wong, H.; et al. Identification of gut microbial species linked with disease variability in a widely used mouse model of colitis. Nat. Microbiol. 2022, 7, 590–599. [Google Scholar] [CrossRef]
- Vigsnæs, L.K.; Brynskov, J.; Steenholdt, C.; Wilcks, A.; Licht, T.R. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef. Microbes 2012, 3, 287–297. [Google Scholar] [CrossRef]
- Ciesielska, A.; Matyjek, M.; Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 2021, 78, 1233–1261. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Fischer, A.; Siegmund, B.; Kupz, A.; Niebergall, J.; Fuchs, D.; Jahn, H.-K.; Freudenberg, M.; Loddenkemper, C.; Batra, A.; et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS ONE 2007, 2, e662. [Google Scholar] [CrossRef] [PubMed]
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G.; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional characterization of inflammatory bowel disease–associated gut dysbiosis in gnotobiotic mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Kolho, K.L.; Korpela, K.; Jaakkola, T.; Pichai, M.V.A.; Zoetendal, E.G.; Salonen, A. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am. J. Gastroenterol. 2015, 110, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.M.; Margolis, K.G. Building community in the gut: A role for mucosal serotonin. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 6–8. [Google Scholar] [CrossRef]
- Scott, S.A.; Fu, J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387. [Google Scholar] [CrossRef]
- Coates, M.D.; Tekin, I.; Vrana, K.E.; Mawe, G.M. Review article: The many potential roles of intestinal serotonin (5-hydroxytryptamine, 5-HT) signalling in inflammatory bowel disease. Aliment Pharmacol. Ther. 2017, 46, 569–580. [Google Scholar] [CrossRef]








| Ingredients (g) | HF | HF with 4.5% Kale |
|---|---|---|
| Casein | 200 | 196 |
| L-cystine | 3 | 3 |
| Corn starch | 72.8 | 60.7 |
| Maltodextrin 10 | 100 | 100 |
| Sucrose | 172.8 | 172.8 |
| Cellulose, BW200 | 50 | 32.2 |
| Soybean oil | 25 | 24.24 |
| Lard | 177.5 | 177.5 |
| Mineral mix | 10 | 10 |
| Dicalcium phosphate | 13 | 13 |
| Calcium carbonate | 5.5 | 5.5 |
| Potassium citrate | 16.5 | 16.5 |
| Vitamin mix | 10 | 10 |
| Choline bitartrate | 2 | 2 |
| Kale dried powdered | 0 | 39 |
| Total (g) | 858.15 | 862.49 |
| Kcal from Macronutrients | ||
| Protein | 716 | 716 |
| Carbohydrate | 1422.4 | 1422.4 |
| Fat | 1822.5 | 1822.5 |
| Total Kcal | 3960.9 | 3961 |
| DIET Group (Weeks 1–3) | Definition | Drinking Water in Week 3 |
|---|---|---|
| HF (control diet) | HF diet with 45% fat | Tap water |
| HFKV | HF diet with 45% fat supplemented with 4.5% kale | Tap water |
| HF-DSS | HF diet with 45% fat | 3.0% DSS in tap water |
| HFKV-DSS | HF diet with 45% fat supplemented with 4.5% kale | 3.0% DSS in tap water |
| Gene | Forward | Reverse |
|---|---|---|
| TLR4 | AGT GCC CCG CTT TCA CCT CT | TCC GGC TCT TGT GGA AGC CT |
| iNOS | CAC CTT GGA GTT CAC CCA GT | ACC ACT CGT ACT TGG GAT GC |
| TNF-a | TAC TGA ACT TCG GGG TGA TTG GTC C | CAG CCT TGT CCC TTG AAG AGA ACC |
| NFKb | GAG TTT GCG GAA GGA TGT CT | TGT CTG CCT CTC TCG TCT T |
| IL-1ß | CCA GCT TCA AAT CTC ACA GCA G | CCA GCT TCA AAT CTC ACA GCA G |
| IL-6 | TCC AGT TGC CTT CTT GGG AC | GTA CTC CAG AAG ACC AGA GG |
| TJP1 | GCC ACT ACA GTA TGA CCA TCC | AAT GAA TAA TAT CAG CAC CAT GCC |
| MUC 2 | TCA AAG TGC TCT CCA AAC TCT C | CCT CTC AGA ATT CCA CAC TCT T |
| Claudin-1 | GTT TGC AGA GAC CCC ATC AC | AGA AGC CAG GAT GAA ACC CA |
| Occludin | CTC CCA TCC GAG TTT CAG GT | GCT GTC GCC TAA GGA AAG AG |
| F4/80 | GGA AGG AAA TGG AGA GAA AG | GAA GAT CTA CCC TGG TGA AT |
| TBP | CCA GAA CTG AAA ATC AAC GCA G | TGT ATC TAC CGT GAA TCT TGG C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raychaudhuri, S.; Shahinozzaman, M.; Subedi, U.; Fan, S.; Ogedengbe, O.; Obanda, D.N. The Vegetable ‘Kale’ Protects against Dextran-Sulfate-Sodium-Induced Acute Inflammation through Moderating the Ratio of Proinflammatory and Anti-Inflammatory LPS-Producing Bacterial Taxa and Augmenting the Gut Barrier in C57BL6 Mice. Nutrients 2023, 15, 3222. https://doi.org/10.3390/nu15143222
Raychaudhuri S, Shahinozzaman M, Subedi U, Fan S, Ogedengbe O, Obanda DN. The Vegetable ‘Kale’ Protects against Dextran-Sulfate-Sodium-Induced Acute Inflammation through Moderating the Ratio of Proinflammatory and Anti-Inflammatory LPS-Producing Bacterial Taxa and Augmenting the Gut Barrier in C57BL6 Mice. Nutrients. 2023; 15(14):3222. https://doi.org/10.3390/nu15143222
Chicago/Turabian StyleRaychaudhuri, Samnhita, Md Shahinozzaman, Ujjwol Subedi, Si Fan, Opeyemi Ogedengbe, and Diana N. Obanda. 2023. "The Vegetable ‘Kale’ Protects against Dextran-Sulfate-Sodium-Induced Acute Inflammation through Moderating the Ratio of Proinflammatory and Anti-Inflammatory LPS-Producing Bacterial Taxa and Augmenting the Gut Barrier in C57BL6 Mice" Nutrients 15, no. 14: 3222. https://doi.org/10.3390/nu15143222
APA StyleRaychaudhuri, S., Shahinozzaman, M., Subedi, U., Fan, S., Ogedengbe, O., & Obanda, D. N. (2023). The Vegetable ‘Kale’ Protects against Dextran-Sulfate-Sodium-Induced Acute Inflammation through Moderating the Ratio of Proinflammatory and Anti-Inflammatory LPS-Producing Bacterial Taxa and Augmenting the Gut Barrier in C57BL6 Mice. Nutrients, 15(14), 3222. https://doi.org/10.3390/nu15143222






