Antimicrobial Resistance Genes (ARGs), the Gut Microbiome, and Infant Nutrition
Abstract
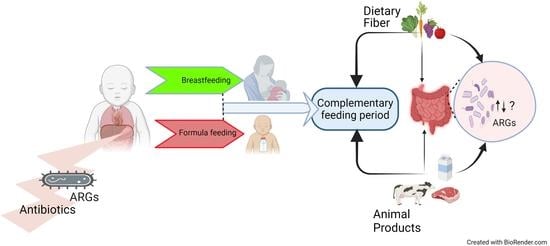
1. Introduction
2. Diet, Age, and Antimicrobial Resistance Gene Carriage
3. Diet and the Gut Microbiome of Infants (0 to 6 Months)
4. Infant Gut Microbiome, ARG Acquisition, and Breastmilk Feeding
5. Infant Diet during the Complimentary Feeding Period and the Gut Microbiome (Over 6 Months)
6. Complementary Feeding Period and ARG Carriage in Infants
7. Infant Gut Resistome as a Source of ARGs
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention; US Department of Health and Human Services. Antibiotic Resistance Threats in the United States 2013; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Centers for Disease Control and Prevention. CDC’s Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Nelson, R.E.; Hatfield, K.M.; Wolford, H.; Samore, M.H.; Scott, R.D.; Reddy, S.C.; Olubajo, B.; Paul, P.; Jernigan, J.A.; Baggs, J. National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin. Infect. Dis. 2021, 72, S17–S26. [Google Scholar] [CrossRef]
- Romandini, A.; Pani, A.; Schenardi, P.A.; Pattarino, G.A.C.; De Giacomo, C.; Scaglione, F. Antibiotic resistance in pediatric infections: Global emerging threats, predicting the near future. Antibiotics 2021, 10, 393. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.-A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- Chatterjee, A.; Modarai, M.; Naylor, N.R.; Boyd, S.E.; Atun, R.; Barlow, J.; Holmes, A.H.; Johnson, A.; Robotham, J.V. Quantifying drivers of antibiotic resistance in humans: A systematic review. Lancet Infect. Dis. 2018, 18, e368–e378. [Google Scholar] [CrossRef] [PubMed]
- Versporten, A.; Bielicki, J.; Drapier, N.; Sharland, M.; Goossens, H.; Group, A.P.; Calle, G.M.; Garrahan, J.P.; Clark, J.; Cooper, C. The Worldwide Antibiotic Resistance and Prescribing in European Children (ARPEC) point prevalence survey: Developing hospital-quality indicators of antibiotic prescribing for children. J. Antimicrob. Chemother. 2016, 71, 1106–1117. [Google Scholar] [CrossRef]
- Rabbitts, J.A.; Groenewald, C.B. Epidemiology of pediatric surgery in the United States. Pediatr. Anesthesia 2020, 30, 1083–1090. [Google Scholar] [CrossRef]
- Miller, G.F.; Coffield, E.; Leroy, Z.; Wallin, R. Prevalence and costs of five chronic conditions in children. J. Sch. Nurs. 2016, 32, 357–364. [Google Scholar] [CrossRef]
- Omling, E.; Jarnheimer, A.; Rose, J.; Björk, J.; Meara, J.; Hagander, L. Population-based incidence rate of inpatient and outpatient surgical procedures in a high-income country. J. Br. Surg. 2018, 105, 86–95. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Stanton, C.; Ross, R.P. Vertical transfer of antibiotics and antibiotic resistant strains across the mother/baby axis. Trends Microbiol. 2022, 30, 47–56. [Google Scholar] [CrossRef]
- Nogacka, A.; Salazar, N.; Suárez, M.; Milani, C.; Arboleya, S.; Solís, G.; Fernández, N.; Alaez, L.; Hernández-Barranco, A.M.; de Los Reyes-Gavilán, C.G. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome 2017, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tapiainen, T.; Brinkac, L.; Lorenzi, H.A.; Moncera, K.; Tejesvi, M.V.; Salo, J.; Nelson, K.E. Vertical transmission of gut microbiome and antimicrobial resistance genes in infants exposed to antibiotics at birth. J. Infect. Dis. 2021, 224, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.K.; Crofts, T.S.; Dantas, G. Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 2015, 27, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, C.; Morrow, A.L.; Lagomarcino, A.J.; Altaye, M.; Taft, D.H.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Schibler, K.R. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J. Pediatr. 2014, 165, 23–29. [Google Scholar] [CrossRef]
- Irfan, M.; Almotiri, A.; AlZeyadi, Z. Antimicrobial Resistance and Its Drivers—A Review. Antibiotics 2022, 11, 1362. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of antimicrobial resistance in bacteria. Am. J. Med. 2006, 119, S3–S10. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Pehrsson, E.C.; Forsberg, K.J.; Gibson, M.K.; Ahmadi, S.; Dantas, G. Novel resistance functions uncovered using functional metagenomic investigations of resistance reservoirs. Front. Microbiol. 2013, 4, 145. [Google Scholar] [CrossRef]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C. Editorial: Horizontal gene transfer mediated bacterial antibiotic resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef]
- Martínez, J.L.; Baquero, F. Emergence and spread of antibiotic resistance: Setting a parameter space. Upsala J. Med. Sci. 2014, 119, 68–77. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Li, J.; Lv, N.; Liu, F.; Wu, J.; Lin, I.Y.; Wu, N.; Weimer, B.C.; Gao, G.F. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl. Environ. Microbiol. 2016, 82, 6672–6681. [Google Scholar] [CrossRef]
- Galhano, B.S.P.; Ferrari, R.G.; Panzenhagen, P.; de Jesus, A.C.; Conte-Junior, C.A. Antimicrobial Resistance Gene Detection Methods for Bacteria in Animal-Based Foods: A Brief Review of Highlights and Advantages. Microorganisms 2021, 9, 923. [Google Scholar] [CrossRef]
- Landan, G.; Cohen, G.; Aharonowitz, Y.; Shuali, Y.; Graur, D.; Shiffman, D. Evolution of isopenicillin N synthase genes may have involved horizontal gene transfer. Mol. Biol. Evol. 1990, 7, 399–406. [Google Scholar] [CrossRef]
- Benveniste, R.; Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 1973, 70, 2276–2280. [Google Scholar] [CrossRef]
- Frye, J.G.; Jesse, T.; Long, F.; Rondeau, G.; Porwollik, S.; McClelland, M.; Jackson, C.R.; Englen, M.; Fedorka-Cray, P.J. DNA microarray detection of antimicrobial resistance genes in diverse bacteria. Int. J. Antimicrob. Agents 2006, 27, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Fluit, A.C.; Visser, M.R.; Schmitz, F.J. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 2001, 14, 836–871. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef]
- Lee, K.; Kim, D.-W.; Cha, C.-J. Overview of bioinformatic methods for analysis of antibiotic resistome from genome and metagenome data. J. Microbiol. 2021, 59, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Nowrotek, M.; Jałowiecki, Ł.; Harnisz, M.; Płaza, G.A. Culturomics and metagenomics: In understanding of environmental resistome. Front. Environ. Sci. Eng. 2019, 13, 40. [Google Scholar] [CrossRef]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial resistance: One Health approach. Veter-World 2022, 15, 743. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A. Antibiotic resistance: One health one world outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- World Health Organization. One Health Joint Plan of Action (2022–2026): Working Together for the Health of Humans, Animals, Plants and the Environment; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Destoumieux-Garzón, D.; Mavingui, P.; Boetsch, G.; Boissier, J.; Darriet, F.; Duboz, P.; Fritsch, C.; Giraudoux, P.; Le Roux, F.; Morand, S. The one health concept: 10 years old and a long road ahead. Front. Veter-Sci. 2018, 5, 14. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One health—Its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, K.; Acharya, K.P.; Shrestha, S. One health: The interface between veterinary and human health. Int. J. One Health 2018, 4, 8–14. [Google Scholar] [CrossRef]
- Stålsby Lundborg, C.; Diwan, V.; Pathak, A.; Purohit, M.R.; Shah, H.; Sharma, M.; Mahadik, V.K.; Tamhankar, A.J. Protocol: A ‘One health’two year follow-up, mixed methods study on antibiotic resistance, focusing children under 5 and their environment in rural India. BMC Public Health 2015, 15, 1321. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Neglected Tropical Diseases and One Health: Gearing up Against Antimicrobial Resistance to Secure the Safety of Future Generations: Meeting Report, 24 November 2020; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Zhang, A.-N.; Gaston, J.M.; Dai, C.L.; Zhao, S.; Poyet, M.; Groussin, M.; Yin, X.; Li, L.-G.; van Loosdrecht, M.; Topp, E. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021, 12, 4765. [Google Scholar] [CrossRef]
- Lederberg, J.; McCray, A.T. Ome SweetOmics--A genealogical treasury of words. Science 2001, 15, 8. [Google Scholar]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef]
- Anthony, W.E.; Burnham, C.-A.D.; Dantas, G.; Kwon, J.H. The gut microbiome as a reservoir for antimicrobial resistance. J. Infect. Dis. 2021, 223, S209–S213. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Wu, G.D.; Compher, C.; Chen, E.Z.; Smith, S.A.; Shah, R.D.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016, 65, 63–72. [Google Scholar] [CrossRef]
- Tanes, C.; Bittinger, K.; Gao, Y.; Friedman, E.S.; Nessel, L.; Paladhi, U.R.; Chau, L.; Panfen, E.; Fischbach, M.A.; Braun, J.; et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 2021, 29, 394–407.e395. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Xue, Z.; Villanueva, Y.T.; Durbin-Johnson, B.; Alkan, Z.; Taft, D.H.; Liu, J.; Korf, I.; Laugero, K.D.; Stephensen, C.B. Association of diet and antimicrobial resistance in healthy US adults. mBio 2022, 13, e00101-22. [Google Scholar] [CrossRef]
- Angulo, F.J.; Baker, N.L.; Olsen, S.J.; Anderson, A.; Barrett, T.J. Antimicrobial use in agriculture: Controlling the transfer of antimicrobial resistance to humans. Semin. Pediatr. Infect. Dis. 2004, 15, 78–85. [Google Scholar] [CrossRef]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. mBio 2016, 7, e02227-15. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef]
- Manges, A.R.; Smith, S.P.; Lau, B.J.; Nuval, C.J.; Eisenberg, J.N.; Dietrich, P.S.; Riley, L.W. Retail Meat Consumption and the Acquisition of Antimicrobial Resistant Escherichia coli Causing Urinary Tract Infections: A Case–Control Study. Foodborne Pathog. Dis. 2007, 4, 419–431. [Google Scholar] [CrossRef]
- da Silva, S.F.; Reis, I.B.; Monteiro, M.G.; Dias, V.C.; Machado, A.B.F.; da Silva, V.L.; Diniz, C.G. Influence of Human Eating Habits on Antimicrobial Resistance Phenomenon: Aspects of Clinical Resistome of Gut Microbiota in Omnivores, Ovolactovegetarians, and Strict Vegetarians. Antibiotics 2021, 10, 276. [Google Scholar] [CrossRef]
- Gosalbes, M.; Vallès, Y.; Jiménez-Hernández, N.; Balle, C.; Riva, P.; Miravet-Verde, S.; de Vries, L.E.; Llop, S.; Agersø, Y.; Sørensen, S.J. High frequencies of antibiotic resistance genes in infants’ meconium and early fecal samples. J. Dev. Orig. Health Dis. 2016, 7, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Taft, D.H.; Liu, J.; Maldonado-Gomez, M.X.; Akre, S.; Huda, M.N.; Ahmad, S.; Stephensen, C.B.; Mills, D.A. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. Msphere 2018, 3, e00441-18. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Report of the Expert Consultation of the Optimal Duration of Exclusive Breastfeeding; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
- Meek, J.Y.; Noble, L.; Section of Breastfeeding. Policy Statement: Breastfeeding and the Use of Human Milk. Pediatrics 2022, 150, e2022057988. [Google Scholar] [CrossRef]
- Andreas, N.J.; Kampmann, B.; Le-Doare, K.M. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Geddes, D.T. Immune cell–mediated protection of the mammary gland and the infant during breastfeeding. Adv. Nutr. 2015, 6, 267–275. [Google Scholar] [CrossRef]
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Laursen, M.F.; Bahl, M.I.; Licht, T.R. Settlers of our inner surface–factors shaping the gut microbiota from birth to toddlerhood. FEMS Microbiol. Rev. 2021, 45, fuab001. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.F. Gut microbiota development: Influence of diet from infancy to toddlerhood. Ann. Nutr. Metab. 2021, 77, 21–34. [Google Scholar] [CrossRef]
- Logan, W. The intestinal flora of infants and young children. J. Pathol. 1913, 18, 527–551. [Google Scholar] [CrossRef]
- Tannock, G.W.; Lawley, B.; Munro, K.; Gowri Pathmanathan, S.; Zhou, S.J.; Makrides, M.; Gibson, R.A.; Sullivan, T.; Prosser, C.G.; Lowry, D.; et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 2013, 79, 3040–3048. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. 2013, 60, 49–74. [Google Scholar]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef]
- Lewis, Z.T.; Totten, S.M.; Smilowitz, J.T.; Popovic, M.; Parker, E.; Lemay, D.G.; Van Tassell, M.L.; Miller, M.J.; Jin, Y.-S.; German, J.B. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015, 3, 13. [Google Scholar] [CrossRef]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef]
- Bode, L. The functional biology of human milk oligosaccharides. Early Hum. Dev. 2015, 91, 619–622. [Google Scholar] [CrossRef]
- Weichert, S.; Koromyslova, A.; Singh, B.K.; Hansman, S.; Jennewein, S.; Schroten, H.; Hansman, G.S. Structural basis for norovirus inhibition by human milk oligosaccharides. J. Virol. 2016, 90, 4843–4848. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Mills, D.A. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Taft, D.H.; Lewis, Z.T.; Nguyen, N.; Ho, S.; Masarweh, C.; Dunne-Castagna, V.; Tancredi, D.J.; Huda, M.N.; Stephensen, C.B.; Hinde, K. Bifidobacterium species colonization in infancy: A global cross-sectional comparison by population history of breastfeeding. Nutrients 2022, 14, 1423. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Hallowell, H.A.; Koutouvalis, K.; Suez, J. Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome. Gut Microbes 2022, 14, 2055944. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, K.M.; de Goffau, M.C.; Perez-Muñoz, M.E.; Arrieta, M.-C.; Bäckhed, F.; Bork, P.; Braun, T.; Bushman, F.D.; Dore, J.; de Vos, W.M.; et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature 2023, 613, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.M.; Ahmadi, S.; Patel, S.; Gibson, M.K.; Wang, B.; Ndao, I.M.; Deych, E.; Shannon, W.; Tarr, P.I.; Warner, B.B. Gut resistome development in healthy twin pairs in the first year of life. Microbiome 2015, 3, 27. [Google Scholar] [CrossRef]
- Bratzler, D.W.; Dellinger, E.P.; Olsen, K.M.; Perl, T.M.; Auwaerter, P.G.; Bolon, M.K.; Fish, D.N.; Napolitano, L.M.; Sawyer, R.G.; Slain, D.; et al. Clinical practice guidelines for antimicrobial prophylaxis in surgery. Am. J. Health-Syst. Pharm. 2013, 70, 195–283. [Google Scholar] [CrossRef]
- Schrag, S.J.; Verani, J.R. Intrapartum antibiotic prophylaxis for the prevention of perinatal group B streptococcal disease: Experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine 2013, 31, D20–D26. [Google Scholar] [CrossRef]
- Puopolo, K.M.; Lynfield, R.; Cummings, J.J.; Hand, I.; Adams-Chapman, I.; Poindexter, B.; Stewart, D.L.; Aucott, S.W.; Goldsmith, J.P.; Mowitz, M. Management of infants at risk for group B streptococcal disease. Pediatrics 2019, 144, e20191881. [Google Scholar] [CrossRef]
- Donovan, B.M.; Abreo, A.; Ding, T.; Gebretsadik, T.; Turi, K.N.; Yu, C.; Ding, J.; Dupont, W.D.; Stone, C.A.; Hartert, T.V.; et al. Dose, Timing, and Type of Infant Antibiotic Use and the Risk of Childhood Asthma. Clin. Infect. Dis. 2019, 70, 1658–1665. [Google Scholar] [CrossRef]
- Stam, J.; van Stuijvenberg, M.; Grüber, C.; Mosca, F.; Arslanoglu, S.; Chirico, G.; Braegger, C.P.; Riedler, J.; Boehm, G.; Sauer, P.J.; et al. Antibiotic use in infants in the first year of life in five European countries. Acta Paediatr. 2012, 101, 929–934. [Google Scholar] [CrossRef]
- Thänert, R.; Sawhney, S.S.; Schwartz, D.J.; Dantas, G. The resistance within: Antibiotic disruption of the gut microbiome and resistome dynamics in infancy. Cell Host Microbe 2022, 30, 675–683. [Google Scholar] [CrossRef]
- Cox, L.M.; Blaser, M.J. Antibiotics in early life and obesity. Nat. Rev. Endocrinol. 2015, 11, 182–190. [Google Scholar] [CrossRef]
- Gasparrini, A.J.; Crofts, T.S.; Gibson, M.K.; Tarr, P.I.; Warner, B.B.; Dantas, G. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes 2016, 7, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Loo, E.X.L.; Zain, A.; Yap, G.C.; Purbojati, R.W.; Drautz-Moses, D.I.; Koh, Y.Q.; Chong, Y.S.; Tan, K.H.; Gluckman, P.D.; Yap, F.; et al. Longitudinal assessment of antibiotic resistance gene profiles in gut microbiomes of infants at risk of eczema. BMC Infect. Dis. 2020, 20, 312. [Google Scholar] [CrossRef] [PubMed]
- Reichler, M.R.; Allphin, A.A.; Breiman, R.F.; Schreiber, J.R.; Arnold, J.E.; McDougal, L.K.; Facklam, R.R.; Boxerbaum, B.; May, D.; Walton, R.O.; et al. The Spread of Multiply Resistant Streptococcus pneumoniae at a Day Care Center in Ohio. J. Infect. Dis. 1992, 166, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Loeber, M.E.; Jacobson, A. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 2004, 98, 23–27. [Google Scholar] [CrossRef]
- Wolfs, T.F.; Duim, B.; Geelen, S.P.; Rigter, A.; Thomson-Carter, F.; Fleer, A.; Wagenaar, J.A. Neonatal sepsis by Campylobacter jejuni: Genetically proven transmission from a household puppy. Clin. Infect. Dis. 2001, 32, E97–E99. [Google Scholar] [CrossRef]
- Pärnänen, K.; Karkman, A.; Hultman, J.; Lyra, C.; Bengtsson-Palme, J.; Larsson, D.G.J.; Rautava, S.; Isolauri, E.; Salminen, S.; Kumar, H.; et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 2018, 9, 3891. [Google Scholar] [CrossRef]
- Wajda, Ł.; Ostrowski, A.; Błasiak, E.; Godowska, P. Enterococcus faecium Isolates Present in Human Breast Milk Might Be Carriers of Multi-Antibiotic Resistance Genes. Bacteria 2022, 1, 66–87. [Google Scholar] [CrossRef]
- Kozak, K.; Charbonneau, D.; Sanozky-Dawes, R.; Klaenhammer, T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 2015, 6, 341–351. [Google Scholar] [CrossRef]
- Pärnänen, K.M.M.; Hultman, J.; Markkanen, M.; Satokari, R.; Rautava, S.; Lamendella, R.; Wright, J.; McLimans, C.J.; Kelleher, S.L.; Virta, M.P. Early-life formula feeding is associated with infant gut microbiota alterations and an increased antibiotic resistance load. Am. J. Clin. Nutr. 2021, 115, 407–421. [Google Scholar] [CrossRef]
- Casaburi, G.; Duar, R.M.; Vance, D.P.; Mitchell, R.; Contreras, L.; Frese, S.A.; Smilowitz, J.T.; Underwood, M.A. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob. Resist. Infect. Control 2019, 8, 131. [Google Scholar] [CrossRef]
- Siimes, M.A.; Salmenperä, L.; Perheentupa, J. Exclusive breast-feeding for 9 months: Risk of iron deficiency. J. Pediatr. 1984, 104, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Dewey, K.G. The challenge of meeting nutrient needs of infants and young children during the period of complementary feeding: An evolutionary perspective. J. Nutr. 2013, 143, 2050–2054. [Google Scholar] [PubMed]
- Arimond, M.; Ruel, M.T. Dietary diversity is associated with child nutritional status: Evidence from 11 demographic and health surveys. J. Nutr. 2004, 134, 2579–2585. [Google Scholar] [CrossRef]
- Woo, J.G.; Herbers, P.M.; McMahon, R.J.; Davidson, B.S.; Ruiz-Palacios, G.M.; Peng, Y.-M.; Morrow, A.L. Longitudinal development of infant complementary diet diversity in 3 international cohorts. J. Pediatr. 2015, 167, 969–974.e961. [Google Scholar] [CrossRef]
- Cummins, A.; Thompson, F. Effect of breast milk and weaning on epithelial growth of the small intestine in humans. Gut 2002, 51, 748–754. [Google Scholar] [CrossRef]
- Wen, J.; Xu, Q.; Zhao, W.; Hu, C.; Zou, X.; Dong, X. Effects of early weaning on intestinal morphology, digestive enzyme activity, antioxidant status, and cytokine status in domestic pigeon squabs (Columba livia). Poult. Sci. 2022, 101, 101613. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Laursen, M.F.; Andersen, L.B.; Michaelsen, K.F.; Mølgaard, C.; Trolle, E.; Bahl, M.I.; Licht, T.R. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. Msphere 2016, 1, e00069-15. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Corwin, E.J.; Brennan, P.A.; Jordan, S.; Murphy, J.R.; Dunlop, A. The infant microbiome: Implications for infant health and neurocognitive development. Nurs. Res. 2016, 65, 76. [Google Scholar] [CrossRef] [PubMed]
- Homann, C.-M.; Rossel, C.A.; Dizzell, S.; Bervoets, L.; Simioni, J.; Li, J.; Gunn, E.; Surette, M.G.; de Souza, R.J.; Mommers, M. Infants’ first solid foods: Impact on gut microbiota development in two intercontinental cohorts. Nutrients 2021, 13, 2639. [Google Scholar] [CrossRef]
- Marrs, T.; Jo, J.-H.; Perkin, M.R.; Rivett, D.W.; Witney, A.A.; Bruce, K.D.; Logan, K.; Craven, J.; Radulovic, S.; Versteeg, S.A. Gut microbiota development during infancy: Impact of introducing allergenic foods. J. Allergy Clin. Immunol. 2021, 147, 613–621.e619. [Google Scholar] [CrossRef]
- Smilowitz, J.; Amicucci, M.; Nandita, E.; Galermo, A.; Tu, D.; Taft, D.; Meier, A.; Kurudimov, K.; Oakes, L.; Mills, D.; et al. The Introduction of Plant-Derived Glycans in Exclusively 6-Month Old Breastfed Infants Alters Fecal Glycan Profiles and Microbial Metabolism (IMiND Study). Curr. Dev. Nutr. 2020, 4, 1082. [Google Scholar] [CrossRef]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The effects of vegetarian and vegan diets on gut microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104, S1–S63. [Google Scholar] [CrossRef] [PubMed]
- Parkar, S.G.; Frost, J.K.T.; Rosendale, D.; Stoklosinski, H.M.; Jobsis, C.M.H.; Hedderley, D.I.; Gopal, P. The sugar composition of the fibre in selected plant foods modulates weaning infants’ gut microbiome composition and fermentation metabolites in vitro. Sci. Rep. 2021, 11, 9292. [Google Scholar] [CrossRef]
- Bergström, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Mølgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of intestinal microbiota during early life: A longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014, 80, 2889–2900. [Google Scholar] [CrossRef]
- Ogbo, F.A.; Nguyen, H.; Naz, S.; Agho, K.E.; Page, A. The association between infant and young child feeding practices and diarrhoea in Tanzanian children. Trop. Med. Health 2018, 46, 2. [Google Scholar] [CrossRef]
- Michaelsen, K.F.; Friis, H. Complementary feeding: A global perspective. Nutrition 1998, 14, 763–766. [Google Scholar] [CrossRef]
- Xu, X.; Feng, Q.; Zhang, T.; Cheng, Q.; Gao, Y.; Zhang, W.; Wu, Q.; Xu, K.; Li, Y.; Nguyen, N. Infant age negatively correlates with the overall load of gut resistome reflecting modifications of carbohydrate metabolism during early life. Res. Sq. 2023, 1–33. [Google Scholar] [CrossRef]
- Hetzer, B.; Orth-Höller, D.; Würzner, R.; Kreidl, P.; Lackner, M.; Müller, T.; Knabl, L.; Geisler-Moroder, D.R.; Mellmann, A.; Sesli, Ö.; et al. Enhanced acquisition of antibiotic-resistant intestinal E. coli during the first year of life assessed in a prospective cohort study. Antimicrob. Resist. Infect. Control 2019, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Theophilus, R.J.; Miller, M.; Oldewage-Theron, W.H.; Dawson, J. The winning weaning food (WWF): The development of a complementary food for food-insecure infants and young children in Malawi. Nutrients 2019, 11, 2292. [Google Scholar] [PubMed]
- Tang, M. The impact of complementary feeding foods of animal origin on growth and the risk of overweight in infants. Anim. Front. 2019, 9, 5–11. [Google Scholar] [CrossRef]
- Hu, Y.; Rubin, J.; Mussio, K.; Riley, L.W. Risk factors for faecal carriage of multidrug-resistant Escherichia coli in a college community: A penalised regression model. J. Glob. Antimicrob. Resist. 2021, 26, 166–173. [Google Scholar] [CrossRef]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar] [CrossRef]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Chari, R.S.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. Can. Med. Assoc. J. 2013, 185, 385–394. [Google Scholar] [CrossRef]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Wu, D.; Jin, L.; Xie, J.; Liu, H.; Zhao, J.; Ye, D.; Li, X.-d. Inhalable antibiotic resistomes emitted from hospitals: Metagenomic insights into bacterial hosts, clinical relevance, and environmental risks. Microbiome 2022, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Holmes, S.J.; Morrow, A.L.; Pickering, L.K. Child-care practices: Effects of social change on the epidemiology of infectious diseases and antibiotic resistance. Epidemiol. Rev. 1996, 18, 10–28. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theophilus, R.J.; Taft, D.H. Antimicrobial Resistance Genes (ARGs), the Gut Microbiome, and Infant Nutrition. Nutrients 2023, 15, 3177. https://doi.org/10.3390/nu15143177
Theophilus RJ, Taft DH. Antimicrobial Resistance Genes (ARGs), the Gut Microbiome, and Infant Nutrition. Nutrients. 2023; 15(14):3177. https://doi.org/10.3390/nu15143177
Chicago/Turabian StyleTheophilus, Rufus J., and Diana Hazard Taft. 2023. "Antimicrobial Resistance Genes (ARGs), the Gut Microbiome, and Infant Nutrition" Nutrients 15, no. 14: 3177. https://doi.org/10.3390/nu15143177
APA StyleTheophilus, R. J., & Taft, D. H. (2023). Antimicrobial Resistance Genes (ARGs), the Gut Microbiome, and Infant Nutrition. Nutrients, 15(14), 3177. https://doi.org/10.3390/nu15143177