Gestational Weight Gain Relates to DNA Methylation in Umbilical Cord, Which, In Turn, Associates with Offspring Obesity-Related Parameters
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Samples
2.2. Clinical Assessments
2.3. DNA Methylome Analysis
2.4. Pyrosequencing Analysis
2.5. Gene Expression Analysis
2.6. Statistics
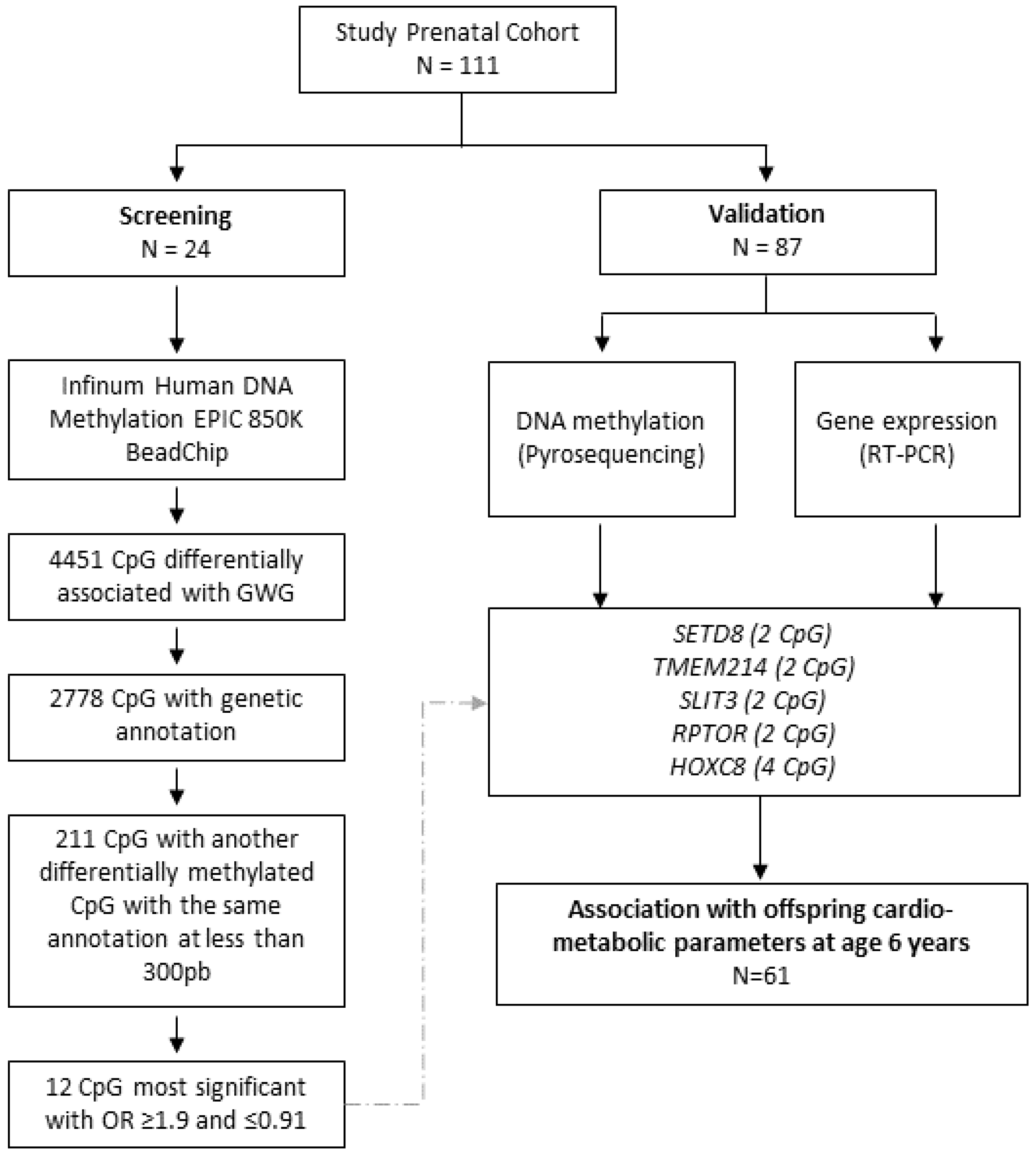
3. Results
3.1. DNA Methylome Analysis
3.2. Selected CpGs and Association with GWG
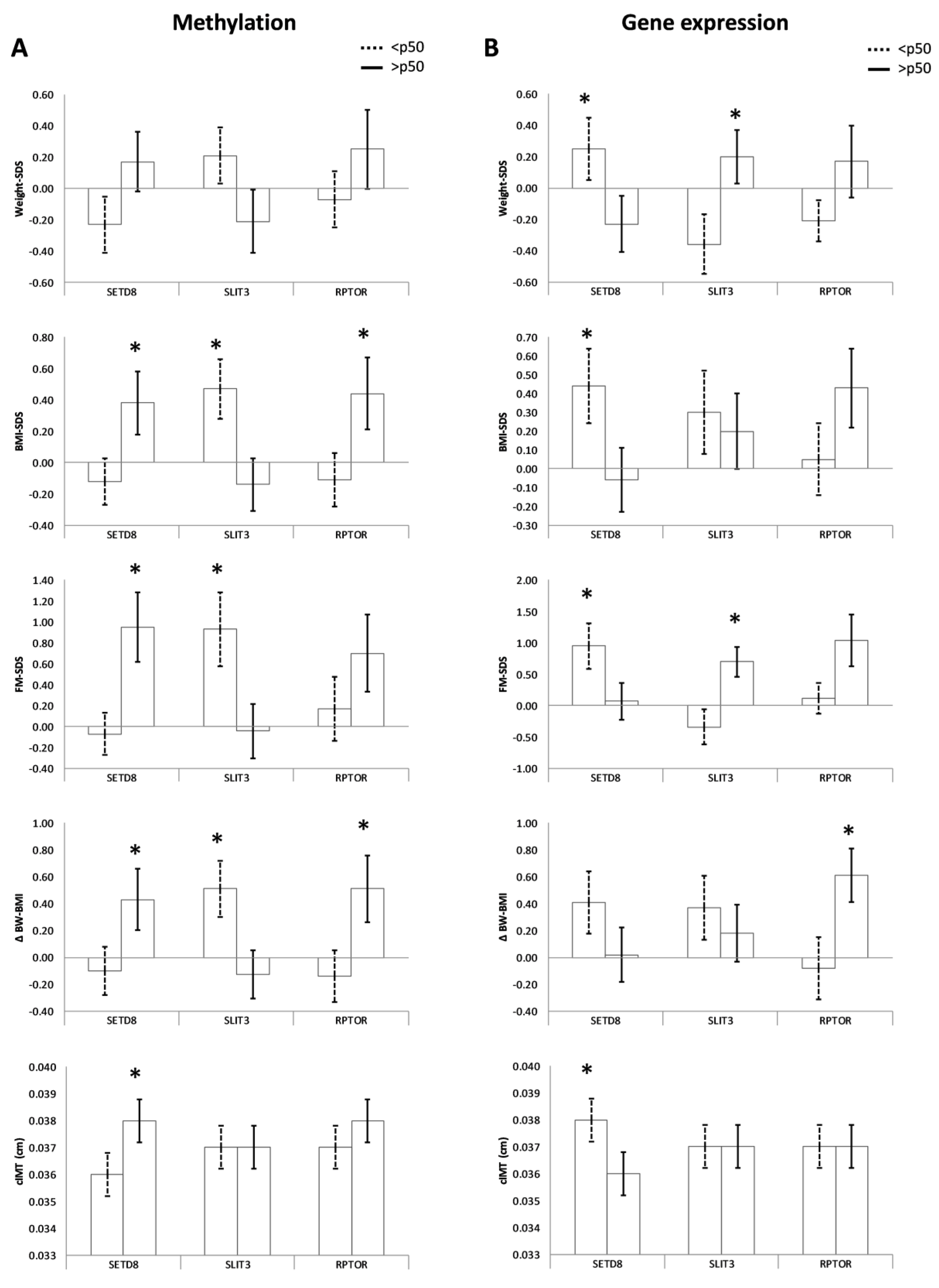
3.3. CpGs Methylation and Obesity-Related Parameters in the Offspring
3.4. Gene Expression and Obesity-Related Parameters in the Offspring
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agarwal, P.; Morriseau, T.S.; Kereliuk, S.M.; Doucette, C.A.; Wicklow, B.A.; Dolinsky, V.W. Maternal obesity, diabetes during pregnancy and epigenetic mechanisms that influence the developmental origins of cardiometabolic disease in the offspring. Crit. Rev. Clin. Lab. Sci. 2018, 55, 71–101. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Fraticelli, F.; Stuppia, L.; Vitacolonna, E. Nutrigenetics, epigenetics and gestational diabetes: Consequences in mother and child. Epigenetics 2019, 14, 215–235. [Google Scholar] [CrossRef]
- Boney, C.M.; Verma, A.; Tucker, R.; Vohr, B.R. Metabolic syndrome in childhood: Association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 2005, 115, e290–e296. [Google Scholar] [CrossRef] [PubMed]
- Fraser, A.; Tilling, K.; Macdonald-Wallis, C.; Sattar, N.; Brion, M.J.; Benfield, L.; Ness, A.; Deanfield, J.; Hingorani, A.; Nelson, S.M. Association of maternal weight gain in pregnancy with offspring obesity and metabolic and vascular traits in childhood. Circulation 2010, 121, 2557–2564. [Google Scholar] [CrossRef]
- Margerison-Zilko, C.E.; Shrimali, B.P.; Eskenazi, B.; Lahiff, M.; Lindquist, A.R.; Abrams, B.F. Trimester of maternal gestational weight gain and offspring body weight at birth and age five. Matern. Child Health J. 2012, 16, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, R.; Welten, M.; Oddy, W.H.; Beilin, L.J.; Mori, T.A.; Jaddoe, V.W.; Huang, R.C. Associations of maternal prepregnancy body mass index and gestational weight gain with cardio-metabolic risk factors in adolescent offspring: A prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2016, 123, 207–216. [Google Scholar] [CrossRef]
- Lillycrop, K.A.; Burdge, G.C. Epigenetic changes in early life and future risk of obesity. Int. J. Obes. 2011, 35, 72–83. [Google Scholar] [CrossRef]
- Chavira-Suárez, E.; Ramírez-Mendieta, A.J.; Martínez-Gutiérrez, S.; Zárate-Segura, P.; Beltrán-Montoya, J.; Espinosa-Maldonado, N.C.; de la Cerda-Ángeles, J.C.; Vadillo-Ortega, F. Influence of pre-pregnancy body mass index (p-BMI) and gestational weight gain (GWG) on DNA methylation and protein expression of obesogenic genes in umbilical vein. PLoS ONE 2019, 14, e0226010. [Google Scholar] [CrossRef]
- Dolinoy, D.C.; Weidman, J.R.; Jirtle, R.L. Epigenetic gene regulation: Linking early developmental environment to adult disease. Reprod. Toxicol. 2007, 23, 297–307. [Google Scholar] [CrossRef]
- Sakurai, K.; Shioda, K.; Eguchi, A.; Watanabe, M.; Miyaso, H.; Mori, C.; Shioda, T. DNA methylome of human neonatal umbilical cord: Enrichment of differentially methylated regions compared to umbilical cord blood DNA at transcription factor genes involved in body patterning and effects of maternal folate deficiency or children’s sex. PLoS ONE 2019, 14, e0214307. [Google Scholar] [CrossRef]
- Godfrey, K.M.; Sheppard, A.; Gluckman, P.D.; Lillycrop, K.A.; Burdge, G.C.; McLean, C.; Rodford, J.; Slater-Jefferies, J.L.; Garratt, E.; Crozier, S.R. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes 2011, 60, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.G. The Epigenomic Analysis of Human Obesity. Obesity 2017, 25, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Sharp, G.C.; Salas, L.A.; Monnereau, C.; Allard, C.; Yousefi, P.; Everson, T.M.; Bohlin, J.; Xu, Z.; Huang, R.C.; Reese, S.E. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: Findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum. Mol. Genet. 2017, 26, 4067–4085. [Google Scholar] [CrossRef]
- Martin, C.L.; Jima, D.; Sharp, G.C.; McCullough, L.E.; Park, S.S.; Gowdy, K.M.; Skaar, D.; Cowley, M.; Maguire, R.L.; Fuemmeler, B. Maternal pre-pregnancy obesity, offspring cord blood DNA methylation, and offspring cardiometabolic health in early childhood: An epigenome-wide association study. Epigenetics 2019, 14, 325–340. [Google Scholar] [CrossRef]
- Yeung, E.H.; Guan, W.; Mumford, S.L.; Silver, R.M.; Zhang, C.; Tsai, M.Y.; Schisterman, E.F. Measured maternal prepregnancy anthropometry and newborn DNA methylation. Epigenomics 2019, 11, 187–198. [Google Scholar] [CrossRef]
- Opsahl, J.O.; Moen, G.H.; Qvigstad, E.; Böttcher, Y.; Birkeland, K.I.; Sommer, C. Epigenetic signatures associated with maternal body mass index or gestational weight gain: A systematic review. J. Dev. Orig. Health Dis. 2020, 12, 373–383. [Google Scholar] [CrossRef]
- Lesseur, C.; Armstrong, D.A.; Paquette, A.G.; Li, Z.; Padbury, J.F.; Marsit, C.J. Maternal obesity and gestational diabetes are associated with placental leptin DNA methylation. Am. J. Obstet. Gynecol. 2014, 211, 654.e1–654.e9. [Google Scholar] [CrossRef]
- Lesseur, C.; Armstrong, D.A.; Paquette, A.G.; Koestler, D.C.; Padbury, J.F.; Marsit, C.J. Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol. Cell. Endocrinol. 2013, 381, 160–167. [Google Scholar] [CrossRef]
- Breton, E.; Gagné-Ouellet, V.; Thibeault, K.; Guérin, R.; Van Lieshout, R.; Perron, P.; Hivert, M.; Bouchard, L. Placental NEGR1 DNA methylation is associated with BMI and neurodevelopment in preschool-age children. Epigenetics 2020, 15, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, D.; Ouidir, M.; Wirkalemahu, T.; Zeng, X.; Tekola-Ayele, F. Placental DNA methylation changes associated with maternal pre-pregnancy BMI and gestational weight gain. Int. J. Obes. 2020, 44, 1406–1416. [Google Scholar] [CrossRef]
- Kawai, T.; Yamada, T.; Abe, K.; Okamura, K.; Kamura, H.; Akaishi, R.; Minakami, H.; Nakabayashi, K.; Hata, K. Increased epigenetic alterations at the promoters of transcriptional regulators following inadequate maternal gestational weight gain. Sci. Rep. 2015, 5, 14224. [Google Scholar] [CrossRef] [PubMed]
- Thakali, K.M.; Faske, J.B.; Ishwar, A.; Alfaro, M.P.; Cleves, M.A.; Badger, T.M.; Andres, A.; Shankar, K. Maternal obesity and gestational weight gain are modestly associated with umbilical cord DNA methylation. Placenta 2017, 57, 194–203. [Google Scholar] [CrossRef]
- Koskinen, A.; Lehtoranta, L.; Laiho, A.; Laine, J.; Kääpä, P.; Soukka, H. Maternal diabetes induces changes in the umbilical cord gene expression. Placenta 2015, 36, 767–774. [Google Scholar] [CrossRef]
- Mas-Parés, B.; Xargay-Torrent, S.; Bonmati, A.; Lizarraga-Mollinedo, E.; Martínez-Calcerrada, J.M.; Carreras-Badosa, G.; Prats-Puig, A.; de Zegher, F.; Ibanez, L.; Lopez-Bermejo, A. Umbilical Cord miRNAs in Small-for-Gestational-Age Children and Association with Catch-Up Growth: A Pilot Study. J. Clin. Endocrinol. Metab. 2019, 104, 5285–5298. [Google Scholar] [CrossRef] [PubMed]
- Lezcano, A.C.; García, J.F.; Ramos, C.F.; Longás, Á.F.; Siguero, J.P.L. Estudio transversal español de crecimiento 2008. Parte II: Valores de talla, peso e índice de masa corporal desde el nacimiento a la talla adulta. An. Pediatr. 2008, 68, 552–569. [Google Scholar] [CrossRef] [PubMed]
- Treuth, M.S.; Butte, N.F.; Wong, W.W.; Ellis, K.J. Body composition in prepubertal girls: Comparison of six methods. Int. J. Obes. 2001, 25, 1352–1359. [Google Scholar] [CrossRef]
- Bassols, J.; Martínez-Calcerrada, J.M.; Prats-Puig, A.; Carreras-Badosa, G.; Xargay-Torrent, S.; Lizarraga-Mollinedo, E.; Feliu-Alsina, M.; Riera-Pérez, E.; Osiniri, I.; de Zegher, F. Perirenal fat is related to carotid intima-media thickness in children. Int. J. Obes. 2018, 42, 641–647. [Google Scholar] [CrossRef]
- Lorente-Pozo, S.; Parra-Llorca, A.; Núñez-Ramiro, A.; Cernada, M.; Hervás, D.; Boronat, N.; Sandoval, J.; Vento, M. The Oxygen Load Supplied during Delivery Room Stabilization of Preterm Infants Modifies the DNA Methylation Profile. J. Pediatr. 2018, 202, 70–76.e2. [Google Scholar] [CrossRef]
- Rosenbloom, K.R.; Sloan, C.A.; Malladi, V.S.; Dreszer, T.R.; Learned, K.; Kirkup, V.M.; Wong, M.C.; Maddren, M.; Fang, R.; Heitner, S.G. ENCODE Data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013, 41, D56–D63. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.S.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef]
- Yajnik, C.S. Transmission of obesity-adiposity and related disorders from the mother to the Baby. Ann. Nutr. Metab. 2014, 64 (Suppl. 1), 8–17. [Google Scholar] [CrossRef] [PubMed]
- Piao, L.; Feng, Y.; Che, N.; Li, M.; Li, X.; Jin, Y.; Xuan, Y. SETD8 is a prognostic biomarker that contributes to stem-like cell properties in non-small cell lung cancer. Pathol. Res. Pract. 2020, 216, 153258. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin 2018, 11, 37. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Okamura, M.; Tsutsumi, S.; Nishikawa, N.S.; Tanaka, T.; Sakakibara, I.; Kitakami, J.I.; Ihara, S.; Hashimoto, Y.; Hamakubo, T. The Peroxisome Proliferator-Activated Receptor γ/Retinoid X Receptor α Heterodimer Targets the Histone Modification Enzyme PR-Set7/Setd8 Gene and Regulates Adipogenesis through a Positive Feedback Loop. Mol. Cell. Biol. 2009, 29, 3544–3555. [Google Scholar] [CrossRef] [PubMed]
- Milite, C.; Feoli, A.; Viviano, M.; Rescigno, D.; Cianciulli, A.; Balzano, A.L.; Mai, A.; Castellano, S.; Sbardella, G. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin. Epigenetics 2016, 8, 102. [Google Scholar] [CrossRef]
- Li, Z.; Nie, F.; Wang, S.; Li, L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc. Natl. Acad. Sci. USA 2011, 108, 3116–3123. [Google Scholar] [CrossRef]
- Chen, N.; Wang, J. Wnt/β-Catenin signaling and obesity. Front. Physiol. 2018, 9, 792. [Google Scholar] [CrossRef]
- Blockus, H.; Chédotal, A. Slit-robo signaling. Dev. 2016, 143, 3037–3044. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Kang, Y.J.; Bak, H.J.; Kim, M.S.; Lee, H.J.; Kwak, D.W.; Han, Y.J.; Kim, M.Y.; Boo, H.; Kim, S.Y. Epigenome-wide DNA methylation profiling of preeclamptic placenta according to severe features. Clin. Epigenetics 2020, 12, 128. [Google Scholar] [CrossRef]
- Lim, R.; Barker, G.; Lappas, M. SLIT3 is Increased in Supracervical Human Foetal Membranes and in Labouring Myometrium and Regulates Pro-Inflammatory Mediators. Am. J. Reprod. Immunol. 2014, 71, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Ma, L.; Ma, G.H.; Ren, H. Genome-wide Analysis Reveals DNA Methylation Alterations in Obesity Associated with High Risk of Colorectal Cancer. Sci. Rep. 2019, 9, 5100. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Sabatini, D.M. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 2005, 280, 39505–39509. [Google Scholar] [CrossRef]
- Berndt, S.I.; Gustafsson, S.; Mägi, R.; Ganna, A.; Wheeler, E.; Feitosa, M.F.; Justice, A.E.; Monda, K.L.; Croteau-Chonka, D.C.; Day, F.R. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013, 45, 501–512. [Google Scholar] [CrossRef]
- Kvaløy, K.; Page, C.M.; Holmen, T.L. Epigenome-wide methylation differences in a group of lean and obese women—A HUNT Study. Sci. Rep. 2018, 8, 16330. [Google Scholar] [CrossRef]
| Gene | Estimate Coefficient | OR | Chromosome | Position | Relation to Gene | Relation to CpG Island | |
|---|---|---|---|---|---|---|---|
| SETD8 | CpG 1 | −0.10000675 | 0.90483131 | 12 | 123868662 | TSS200 | Island |
| CpG 2 | −0.04700338 | 0.95408418 | 12 | 123868665 | TSS200 | Island | |
| TMEM214 | CpG 1 | −0.05779695 | 0.94384157 | 2 | 27255615 | TSS200 | Island |
| CpG 2 | −0.08774743 | 0.9159922 | 2 | 27255618 | TSS200 | Island | |
| SLIT3 | CpG 1 | 0.07056568 | 1.07311505 | 5 | 168271855 | Body | NA |
| CpG 2 | 0.10201274 | 1.10739758 | 5 | 168271859 | Body | NA | |
| RPTOR | CpG 1 | 0.08914043 | 1.09323417 | 17 | 78915842 | Body | Island |
| CpG 2 | 0.07583486 | 1.07878441 | 17 | 78915881 | Body | Island | |
| HOXC8 | CpG 1 | 0.0872253 | 1.09114248 | 12 | 54402697 | TSS200 | Island |
| CpG 2 | 0.121894 | 1.12963435 | 12 | 54402699 | TSS200 | Island | |
| CpG 3 | 0.11646746 | 1.12352096 | 12 | 54402714 | TSS200 | Island | |
| CpG 4 | 0.08129304 | 1.0846887 | 12 | 54402717 | TSS200 | Island |
| Methylation of SETD8 | Methylation of SLIT3 | Methylation of RPTOR | ||||
|---|---|---|---|---|---|---|
| <50th Centile | >50th Centile | <50th Centile | >50th Centile | <50th Centile | >50th Centile | |
| Mother | ||||||
| Age (years) | 30.53 ± 0.65 | 31.07 ± 0.56 | 30.87 ± 0.55 | 30.71 ± 0.80 | 30.21 ± 0.70 | 30.97 ± 0.63 |
| Pregestational BMI | 24.33 ± 0.66 | 24.61 ± 0.64 | 24.44 ± 0.62 | 24.43 ± 0.74 | 24.54 ± 0.70 | 24.29 ± 0.47 |
| 1st-trimester BMI | 24.74 ± 0.66 | 25.54 ± 0.62 | 25.16 ± 0.64 | 25.01 ± 0.71 | 24.94 ± 0.70 | 25.08 ± 0.64 |
| 2nd-trimester BMI | 26.61 ± 0.59 | 27.83 ± 0.62 | 27.33 ± 0.62 | 27.44 ± 0.66 | 26.95 ± 0.61 | 27.14 ± 0.68 |
| 3rd-trimester BMI | 29.03 ± 0.63 | 29.35 ± 0.60 | 28.91 ± 0.65 | 29.42 ± 0.67 | 29.10 ± 0.66 | 29.05 ± 0.66 |
| 1st-trimester GWG (kg) | 1.24 ± 0.25 | 2.01 ± 0.53 | 1.47 ± 0.31 | 1.95 ± 0.56 | 0.84 ± 0.24 | 2.28 ± 0.59 * |
| 2nd-trimester GWG (kg) | 6.04 ± 0.45 | 6.17 ± 0.50 | 6.19 ± 0.47 | 6.14 ± 0.53 | 6.08 ± 0.50 | 5.88 ± 0.44 |
| 3rd-trimester GWG (kg) | 5.75 ± 0.48 | 4.09 ± 0.27 * | 4.18 ± 0.30 | 5.20 ± 0.40 * | 5.07 ± 0.44 | 5.02 ± 0.46 |
| Total GWG (kg) | 14.40 ± 0.73 | 14.23 ± 0.87 | 13.51 ± 0.73 | 14.74 ± 0.85 | 13.45 ± 0.75 | 14.91 ± 0.86 |
| Newborn | ||||||
| Gender (%F) | 47 | 53 | 47 | 52 | 50 | 50 |
| GA (wk) | 39.82 ± 0.16 | 39.76 ± 0.16 | 39.79 ± 0.15 | 39.87 ± 0.18 | 39.67 ± 0.16 | 39.87 ± 0.18 |
| Placental weight (kg) | 5.86 ± 0.16 | 6.09 ± 0.18 | 5.74 ± 0.14 | 6.19 ± 0.21 | 5.82 ± 0.14 | 6.05 ± 0.21 |
| Birth weight-SDS (z-score) | −0.01 ± 0.09 | −0.01 ± 0.09 | −0.06 ± 0.09 | 0.01 ± 0.10 | −0.03 ± 0.10 | −0.04 ± 0.09 |
| Birth length-SDS (z-score) | −0.25 ± 0.11 | −0.18 ± 0.16 | −0.14 ± 0.13 | −0.34 ± 0.16 | −0.25 ± 0.14 | −0.31 ± 0.14 |
| Child | ||||||
| Gender (%F) | 42 | 58 | 58 | 42 | 44 | 56 |
| Age (years) | 5.87 ± 0.17 | 5.79 ± 0.18 | 5.80 ± 0.18 | 5.76 ± 0.19 | 5.71 ± 0.19 | 5.78 ± 0.18 |
| Weight-SDS (z-score) | −0.23 ± 0.18 | 0.17 ± 0.19 | 0.21 ± 0.18 | −0.21 ± 0.20 | −0.07 ± 0.18 | 0.25 ± 0.25 |
| Height-SDS (z-score) | −0.08 ± 0.20 | −0.17 ± 0.24 | −0.04 ± 0.19 | −0.24 ± 0.28 | 0.05 ± 0.24 | −0.21 ± 0.25 |
| BMI-SDS (z-score) | −0.12 ± 0.15 | 0.38 ± 0.20 * | 0.47 ± 0.19 | −0.14 ± 0.17 * | −0.11 ± 0.17 | 0.44 ± 0.23 * |
| FM-SDS (z-score) | −0.07 ± 0.20 | 0.95 ± 0.33 * | 0.93 ± 0.35 | −0.04 ± 0.26 * | 0.17 ± 0.31 | 0.70 ± 0.37 |
| ∆ BW − BMI (z-score) | −0.10 ± 0.18 | 0.43 ± 0.23 * | 0.51 ± 0.21 | −0.13 ± 0.18 * | −0.14 ± 0.19 | 0.51 ± 0.25 * |
| Waist (cm) | 56.32 ± 1.15 | 57.48 ± 1.53 | 58.00 ± 1.36 | 55.65 ± 1.52 | 56.08 ± 1.27 | 56.64 ± 1.39 |
| cIMT (cm) | 0.036 ± 0.01 | 0.038 ± 0.01 * | 0.037 ± 0.001 | 0.037 ± 0.001 | 0.037 ± 0.01 | 0.038 ± 0.01 |
| Relative Expression SETD8 | Relative Expression SLIT3 | Relative Expression RPTOR | ||||
|---|---|---|---|---|---|---|
| <50th Centile | >50th Centile | <50th Centile | >50th Centile | <50th Centile | >50th Centile | |
| Mother | ||||||
| Age (yrs) | 31.39 ± 0.61 | 30.19 ± 0.60 | 30.55 ± 0.72 | 31.05 ± 0.48 | 29.93 ± 0.63 | 31.67 ± 0.56 * |
| Pregestational BMI | 24.90 ± 0.60 | 24.01 ± 0.68 | 24.56 ± 0.66 | 24.37 ± 0.63 | 24.61 ± 0.62 | 24.31 ± 0.67 |
| 1st trimester BMI | 25.68 ± 0.61 | 24.55 ± 0.67 | 25.22 ± 0.64 | 25.04 ± 0.65 | 25.19 ± 0.63 | 25.06 ± 0.67 |
| 2nd trimester BMI | 28.18 ± 0.61 | 26.23 ± 0.58 * | 27.32 ± 0.61 | 27.09 ± 0.62 | 26.97 ± 0.58 | 27.45 ± 0.64 |
| 3rd trimester BMI | 30.09 ± 0.59 | 28.24 ± 0.61 * | 29.25 ± 0.60 | 29.13 ± 0.63 | 29.30 ± 0.58 | 29.08 ± 0.65 |
| 1st-trimester GWG (kg) | 1.61 ± 0.26 | 1.62 ± 0.53 | 1.90 ± 0.51 | 1.33 ± 0.27 | 1.37 ± 0.24 | 1.88 ± 0.55 |
| 2nd-trimester GWG (kg) | 6.70 ± 0.52 | 5.47 ± 0.39 | 5.95 ± 0.51 | 6.26 ± 0.44 | 5.93 ± 0.48 | 6.28 ± 0.47 |
| 3rd-trimester GWG (kg) | 5.16 ± 0.48 | 4.71 ± 0.33 | 5.19 ± 0.40 | 4.67 ± 0.42 | 5.51 ± 0.45 | 4.3 ± 0.35 * |
| Total GWG (kg) | 15.63 ± 0.82 | 12.97 ± 0.72 * | 14.17 ± 0.78 | 14.45 ± 0.82 | 14.50 ± 0.76 | 14.12 ± 0.84 |
| Newborn | ||||||
| Gender (%F) | 37 | 63 * | 50 | 50 | 55 | 45 |
| GA (wk) | 39.73 ± 0.18 | 39.86 ± 0.14 | 39.95 ± 0.15 | 39.63 ± 0.16 | 39.95 ± 0.16 | 39.63 ± 0.15 |
| Placental weight (kg) | 6.02 ± 0.16 | 5.93 ± 0.18 | 6.08 ± 17.31 | 5.86 ± 17.86 | 6.10 ± 0.16 | 5.83 ± 0.19 |
| Weight-SDS (z-score) | 0.07 ± 0.07 | −0.10 ± 0.10 | −0.02 ± 0.09 | −0.01 ± 0.09 | 0.14 ± 0.08 | −0.17 ± 0.09 * |
| Length-SDS (z-score) | −0.26 ± 0.13 | −0.17 ± 0.14 | −0.31 ± 0.14 | −0.12 ± 0.13 | −0.09 ± 0.13 | −0.35 ± 0.14 |
| Child | ||||||
| Gender (%F) | 41 | 59 | 41 | 59 | 56 | 44 |
| Age (yrs) | 5.88 ± 0.17 | 5.77 ± 0.17 | 5.80 ± 0.18 | 5.82 ± 0.16 | 5.61 ± 0.16 | 6.01 ± 0.16 |
| Weight-SDS (z-score) | 0.25 ± 0.20 | −0.23 ± 0.18 * | −0.36 ± 0.19 | 0.20 ± 0.17 * | −0.21 ± 0.13 | 0.17 ± 0.23 |
| Height-SDS (z-score) | 0.05 ± 0.21 | −0.36 ± 0.22 | −0.43 ± 0.27 | 0.12 ± 0.16 * | −0.12 ± 0.20 | −0.09 ± 0.22 |
| BMI-SDS (z-score) | 0.44 ± 0.20 | −0.06 ± 0.17 * | 0.30 ± 0.22 | 0.20 ± 0.20 | 0.05 ± 0.19 | 0.43 ± 0.21 |
| FM-SDS (z-score) | 0.95 ± 0.36 | 0.07 ± 0.29 * | −0.34 ± 0.28 | 0.70 ± 0.24 * | 0.12 ± 0.24 | 1.04 ± 0.41 |
| ∆ BW-BMI (z-score) | 0.41 ± 0.23 | 0.02 ± 0.20 | 0.37 ± 0.24 | 0.18 ± 0.21 | −0.08 ± 0.23 | 0.61 ± 0.20 * |
| Waist (cm) | 59.46 ± 1.40 | 55.25 ± 1.25 * | 56.56 ± 1.20 | 57.75 ± 1.40 | 55.42 ± 1.03 | 59.11 ± 1.54 * |
| cIMT (cm) | 0.038 ± 0.001 | 0.036 ± 0.001 * | 0.037 ± 0.001 | 0.037 ± 0.001 | 0.037 ± 0.001 | 0.037 ± 0.001 |
| Umbilical cord | ||||||
| Methylation (%) | 0.66 ± 0.14 | 0.39 ± 0.07 * | 65.18 ± 1.43 | 58.80 ± 1.36 * | 38.92 ± 1.49 | 37.49 ± 1.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mas-Parés, B.; Xargay-Torrent, S.; Gómez-Vilarrubla, A.; Carreras-Badosa, G.; Prats-Puig, A.; De Zegher, F.; Ibáñez, L.; Bassols, J.; López-Bermejo, A. Gestational Weight Gain Relates to DNA Methylation in Umbilical Cord, Which, In Turn, Associates with Offspring Obesity-Related Parameters. Nutrients 2023, 15, 3175. https://doi.org/10.3390/nu15143175
Mas-Parés B, Xargay-Torrent S, Gómez-Vilarrubla A, Carreras-Badosa G, Prats-Puig A, De Zegher F, Ibáñez L, Bassols J, López-Bermejo A. Gestational Weight Gain Relates to DNA Methylation in Umbilical Cord, Which, In Turn, Associates with Offspring Obesity-Related Parameters. Nutrients. 2023; 15(14):3175. https://doi.org/10.3390/nu15143175
Chicago/Turabian StyleMas-Parés, Berta, Sílvia Xargay-Torrent, Ariadna Gómez-Vilarrubla, Gemma Carreras-Badosa, Anna Prats-Puig, Francis De Zegher, Lourdes Ibáñez, Judit Bassols, and Abel López-Bermejo. 2023. "Gestational Weight Gain Relates to DNA Methylation in Umbilical Cord, Which, In Turn, Associates with Offspring Obesity-Related Parameters" Nutrients 15, no. 14: 3175. https://doi.org/10.3390/nu15143175
APA StyleMas-Parés, B., Xargay-Torrent, S., Gómez-Vilarrubla, A., Carreras-Badosa, G., Prats-Puig, A., De Zegher, F., Ibáñez, L., Bassols, J., & López-Bermejo, A. (2023). Gestational Weight Gain Relates to DNA Methylation in Umbilical Cord, Which, In Turn, Associates with Offspring Obesity-Related Parameters. Nutrients, 15(14), 3175. https://doi.org/10.3390/nu15143175






