Probiotics and Human Milk Differentially Influence the Gut Microbiome and NEC Incidence in Preterm Pigs
Abstract
1. Introduction
2. Materials and Methods
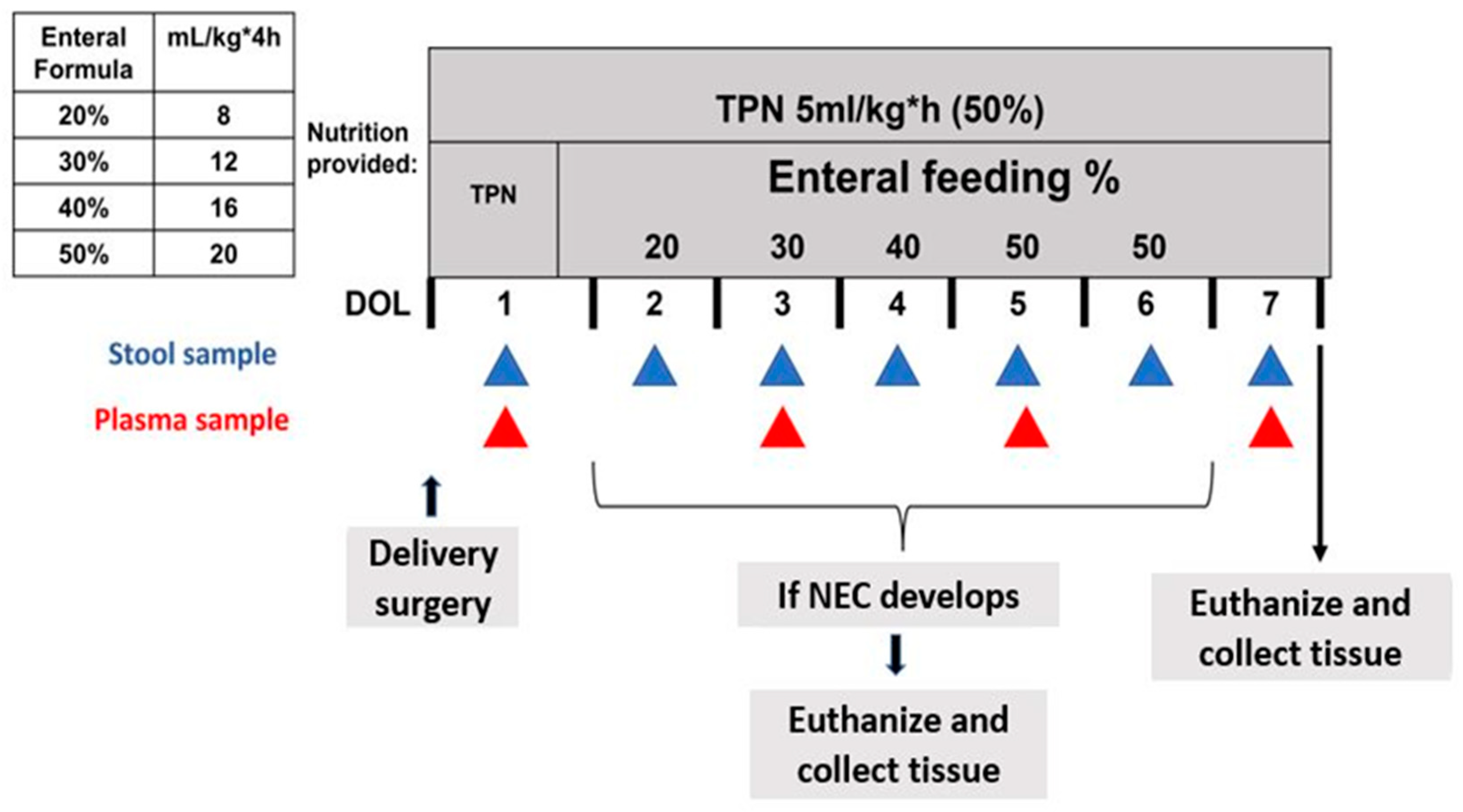
2.1. Delivery, Clinical Care, and Nutrition
2.2. NEC Monitoring
2.3. Sample Collection
2.4. Microbiome Analysis
2.5. Bifidobacterium Longum Quantification in Colon Content Samples
2.6. Evaluation of Pro-Inflammatory Cytokine Expression in Distal Ileum Tissue
2.7. Statistical Analysis
3. Results
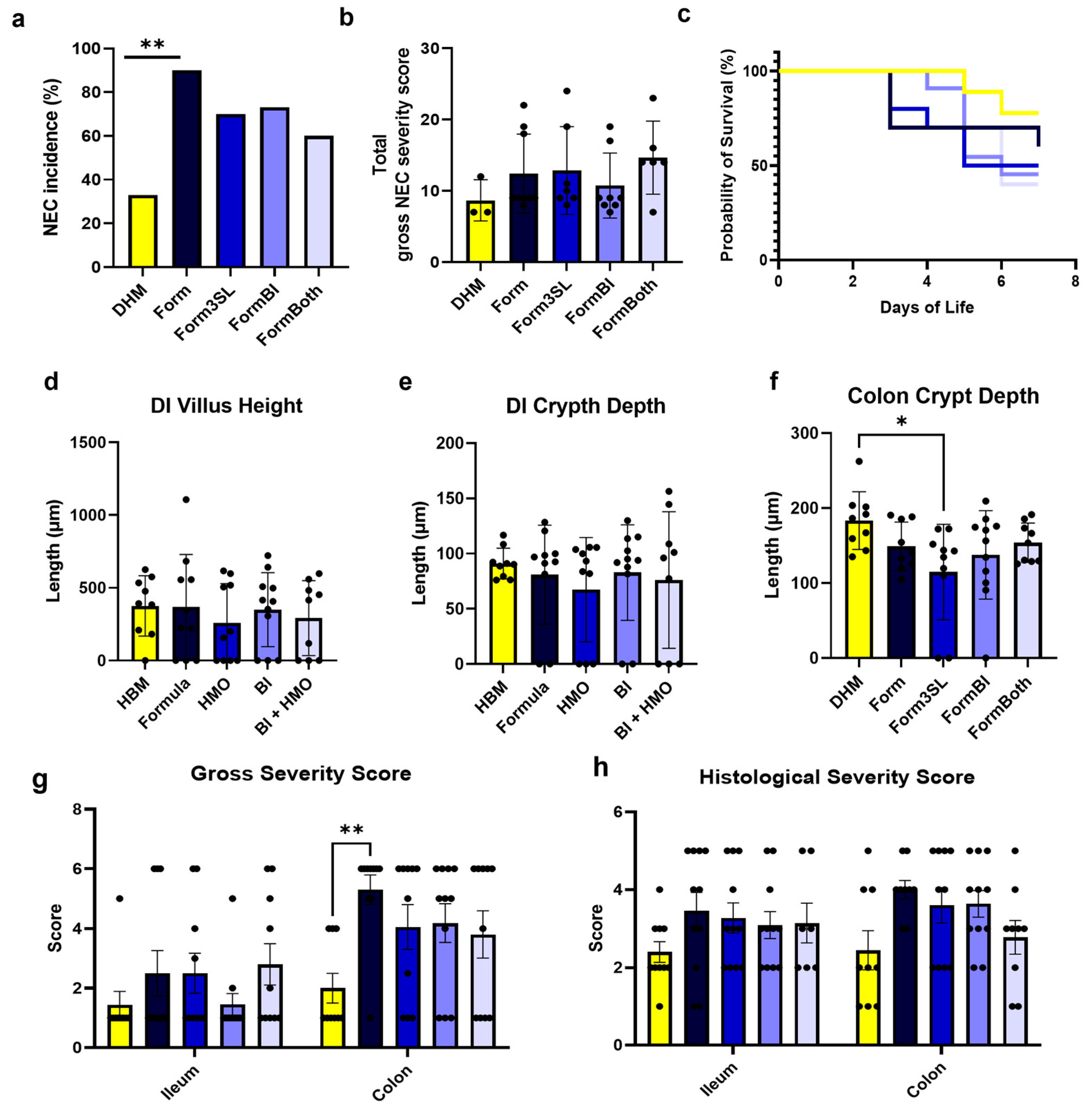
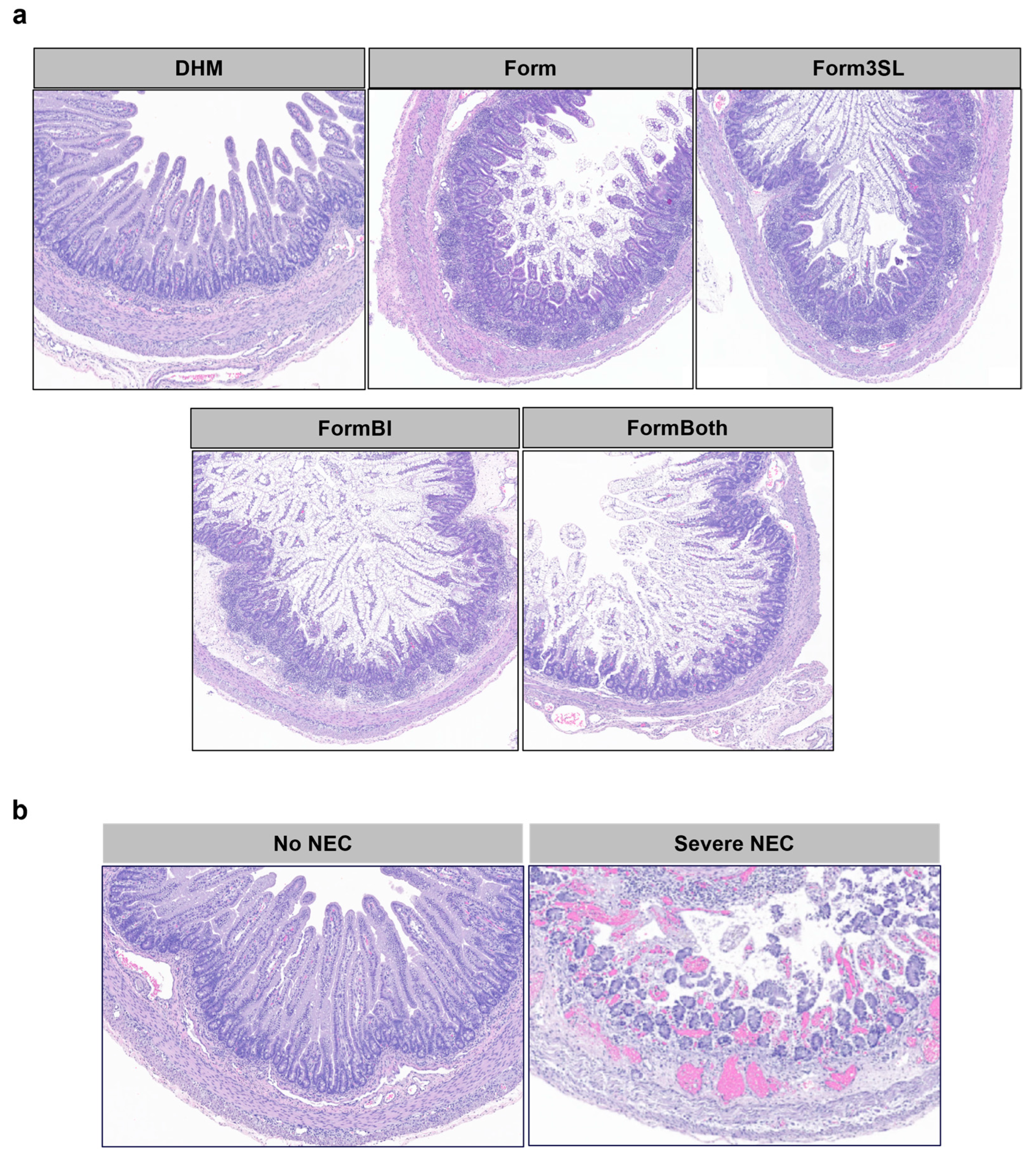
3.1. SL and BL. infantis Supplementation Did Not Confer Protection against NEC Incidence or Severity
3.2. Relative Abundance of Top 10 Taxa by Treatment and Diagnosis
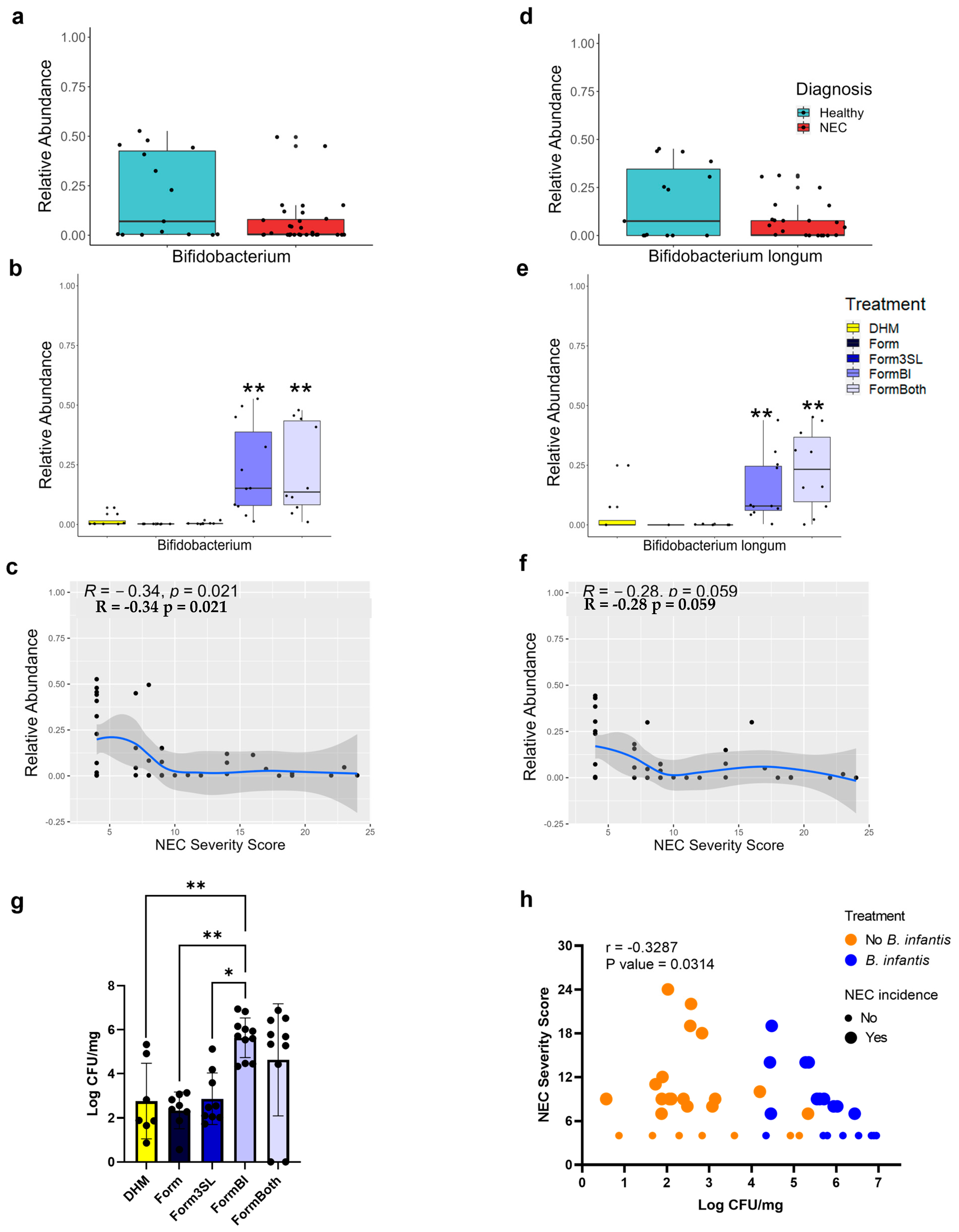
3.3. Bifidobacterium Genus and B. longum Are Significantly More Abundant in FormBI and FormBoth Groups and Negatively Correlate with Disease Severity
3.4. Escherichia-Shigella Is Significantly More Abundant in Healthy Piglets and Negatively Correlates with Disease Severity
3.5. Clostridium sensu stricto 1 Is Significantly More Abundant in Diseased Piglets and Correlates with Disease Severity
3.6. Enterococcus Is Significantly More Abundant in the Colon of Diseased Piglets and Correlates with Disease Severity
3.7. Donor Human Milk-Fed Piglets Exhibited Lower Abundance of Clostridium sensu stricto 1
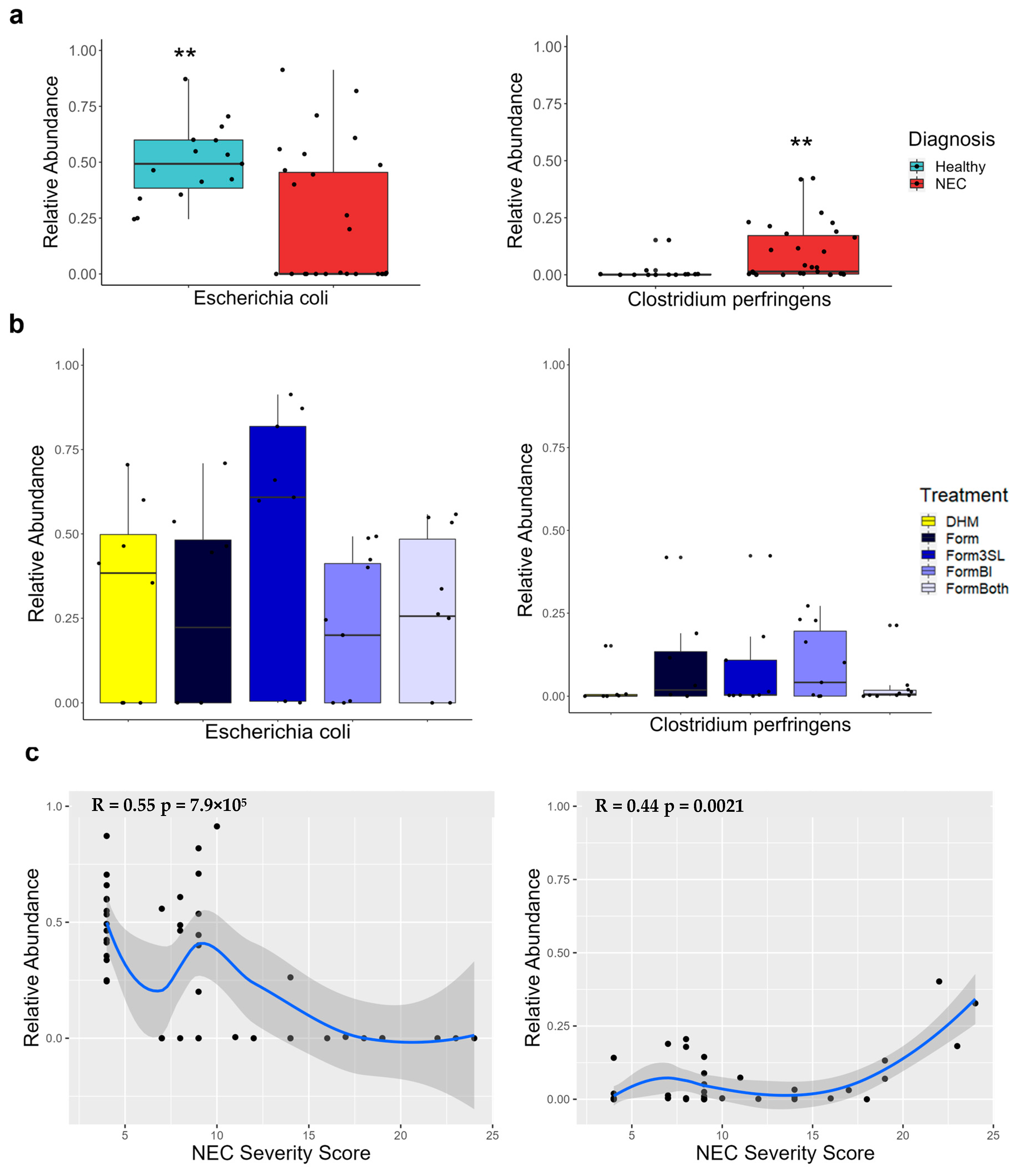
3.8. Clostridium Perfringens Is Significantly More Abundant in Piglets with NEC and Positively Correlates with Disease Severity
3.9. Escherichia coli Is Significantly More Abundant in Healthy Piglets and Negatively Correlates with Disease Severity
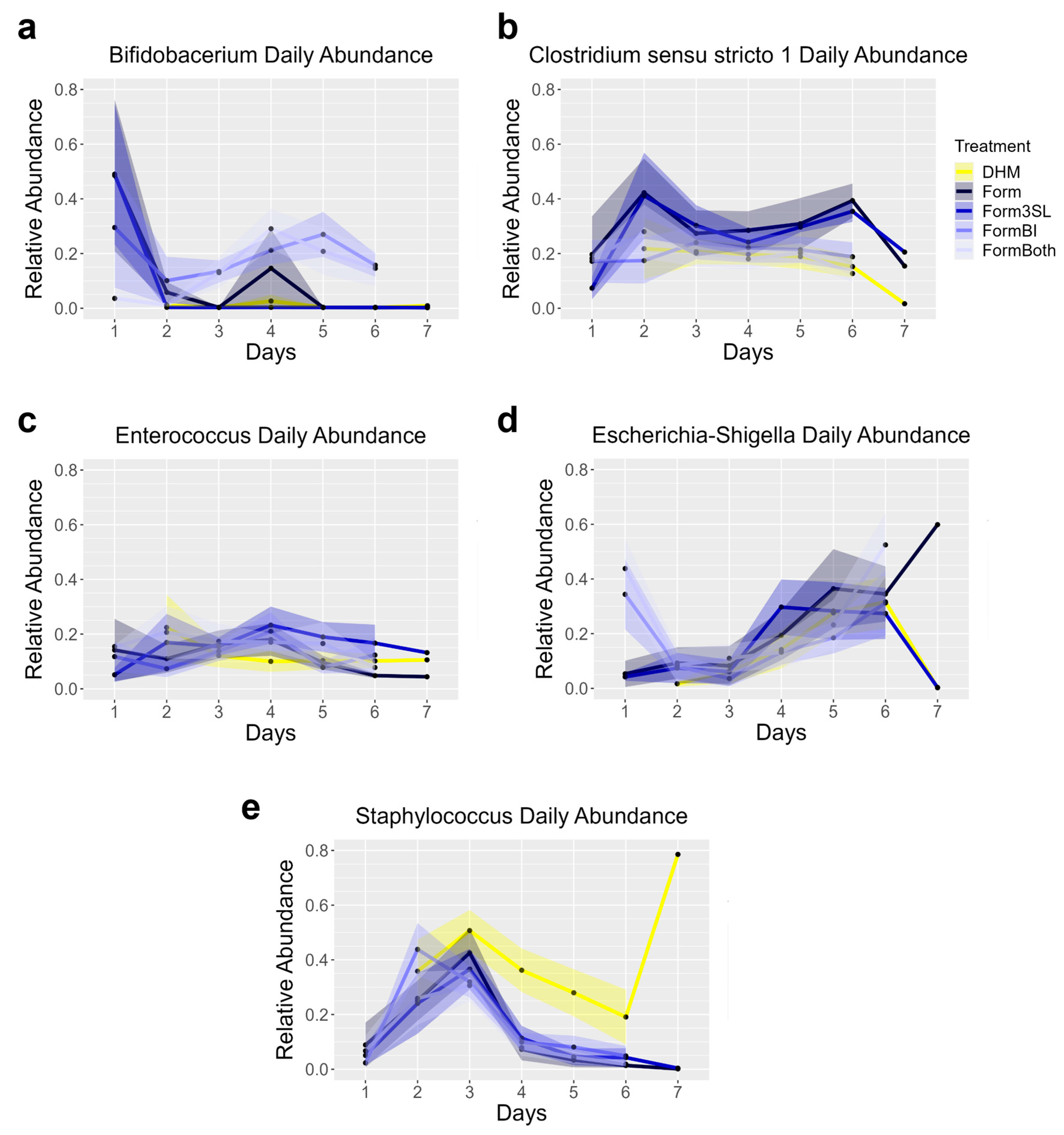
3.10. 16S Sequencing Shows Daily Patterns of Abundance of the Top 5 Genera in the Stool
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thanert, R.; Keen, E.C.; Dantas, G.; Warner, B.B.; Tarr, P.I. Necrotizing Enterocolitis and the Microbiome: Current Status and Future Directions. J. Infect. Dis. 2021, 223 (Suppl. S2), S257–S263. [Google Scholar] [CrossRef]
- Lucas, A.; Cole, T.J. Breast milk and neonatal necrotising enterocolitis. Lancet 1990, 336, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Meinzen-Derr, J.; Poindexter, B.; Wrage, L.; Morrow, A.L.; Stoll, B.; Donovan, E.F. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J. Perinatol. 2009, 29, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.T.; Patel, R.M. A critical analysis of risk factors for necrotizing enterocolitis. Semin. Fetal. Neonatal Med. 2018, 23, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.L.; Thymann, T.; Cilieborg, M.S.; Lykke, M.; Molbak, L.; Jensen, B.B.; Schmidt, M.; Kelly, D.; Mulder, I.; Burrin, D.G.; et al. Antibiotics modulate intestinal immunity and prevent necrotizing enterocolitis in preterm neonatal piglets. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G59–G71. [Google Scholar] [CrossRef]
- Warner, B.B.; Deych, E.; Zhou, Y.; Hall-Moore, C.; Weinstock, G.M.; Sodergren, E.; Shaikh, N.; Hoffmann, J.A.; Linneman, L.A.; Hamvas, A.; et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: A prospective case-control study. Lancet 2016, 387, 1928–1936. [Google Scholar] [CrossRef]
- Sangild, P.T.; Siggers, R.H.; Schmidt, M.; Elnif, J.; Bjornvad, C.R.; Thymann, T.; Grondahl, M.L.; Hansen, A.K.; Jensen, S.K.; Boye, M.; et al. Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology 2006, 130, 1776–1792. [Google Scholar] [CrossRef]
- Gopalakrishna, K.P.; Macadangdang, B.R.; Rogers, M.B.; Tometich, J.T.; Firek, B.A.; Baker, R.; Ji, J.; Burr, A.H.P.; Ma, C.; Good, M.; et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 2019, 25, 1110–1115. [Google Scholar] [CrossRef]
- Heida, F.H.; van Zoonen, A.; Hulscher, J.B.F.; Te Kiefte, B.J.C.; Wessels, R.; Kooi, E.M.W.; Bos, A.F.; Harmsen, H.J.M.; de Goffau, M.C. A Necrotizing Enterocolitis-Associated Gut Microbiota Is Present in the Meconium: Results of a Prospective Study. Clin. Infect. Dis. 2016, 62, 863–870. [Google Scholar] [CrossRef]
- Kiu, R.; Shaw, A.; Sim, K.; Bedwell, H.; Cornwell, E.; Pickard, D.; Belteki, G.; Malsom, J.; Philips, S.; Young, G.R.; et al. Dissemination and pathogenesis of toxigenix Clostridium perfringens strains linked to neonatal intensive care units and Necrotising Enterocolitis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Autran, C.A.; Kellman, B.P.; Kim, J.H.; Asztalos, E.; Blood, A.B.; Spence, E.C.H.; Patel, A.L.; Hou, J.; Lewis, N.E.; Bode, L. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 2018, 67, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Wejryd, E.; Marti, M.; Marchini, G.; Werme, A.; Jonsson, B.; Landberg, E.; Abrahamsson, T.R. Low Diversity of Human Milk Oligosaccharides is Associated with Necrotising Enterocolitis in Extremely Low Birth Weight Infants. Nutrients 2018, 10, 1556. [Google Scholar] [CrossRef]
- Preidis, G.A.; Weizman, A.V.; Kashyap, P.C.; Morgan, R.L. AGA Technical Review on the Role of Probiotics in the Management of Gastrointestinal Disorders. Gastroenterology 2020, 159, 708–738.e4. [Google Scholar] [CrossRef]
- Autran, C.A.; Schoterman, M.H.; Jantscher-Krenn, E.; Kamerling, J.P.; Bode, L. Sialylated galacto-oligosaccharides and 2′-fucosyllactose reduce necrotising enterocolitis in neonatal rats. Br. J. Nutr. 2016, 116, 294–299. [Google Scholar] [CrossRef]
- Jantscher-Krenn, E.; Zherebtsov, M.; Nissan, C.; Goth, K.; Guner, Y.S.; Naidu, N.; Choudhury, B.; Grishin, A.V.; Ford, H.R.; Bode, L. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 2012, 61, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Cilieborg, M.S.; Sangild, P.T.; Jensen, M.L.; Ostergaard, M.V.; Christensen, L.; Rasmussen, S.O.; Morbak, A.L.; Jorgensen, C.B.; Bering, S.B. alpha1,2-Fucosyllactose Does Not Improve Intestinal Function or Prevent Escherichia coli F18 Diarrhea in Newborn Pigs. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.O.; Martin, L.; Ostergaard, M.V.; Rudloff, S.; Roggenbuck, M.; Nguyen, D.N.; Sangild, P.T.; Bering, S.B. Human milk oligosaccharide effects on intestinal function and inflammation after preterm birth in pigs. J. Nutr. Biochem. 2017, 40, 141–154. [Google Scholar] [CrossRef]
- Underwood, M.A. Bifidobacterium infantis, Necrotizing Enterocolitis, Death, and the Role of Parents in the Neonatal Intensive Care Unit. J. Pediatr. 2022, 244, 14–16. [Google Scholar] [CrossRef]
- Hoyos, A.B. Reduced incidence of necrotizing enterocolitis associated with enteral administration of Lactobacillus acidophilus and Bifidobacterium infantis to neonates in an intensive care unit. Int. J. Infect. Dis. 1999, 3, 197–202. [Google Scholar] [CrossRef]
- Robertson, C.; Savva, G.M.; Clapuci, R.; Jones, J.; Maimouni, H.; Brown, E.; Minocha, A.; Hall, L.J.; Clarke, P. Incidence of necrotising enterocolitis before and after introducing routine prophylactic Lactobacillus and Bifidobacterium probiotics. Arch. Dis. Child Fetal. Neonatal Ed. 2020, 105, 380–386. [Google Scholar] [CrossRef]
- Hartel, C.; Pagel, J.; Rupp, J.; Bendiks, M.; Guthmann, F.; Rieger-Fackeldey, E.; Heckmann, M.; Franz, A.; Schiffmann, J.H.; Zimmermann, B.; et al. Prophylactic use of Lactobacillus acidophilus/Bifidobacterium infantis probiotics and outcome in very low birth weight infants. J. Pediatr. 2014, 165, 285–289.e1. [Google Scholar] [CrossRef] [PubMed]
- Tobias, J.; Olyaei, A.; Laraway, B.; Jordan, B.K.; Dickinson, S.L.; Golzarri-Arroyo, L.; Fialkowski, E.; Owora, A.; Scottoline, B. Bifidobacteriumlongum subsp. infantis EVC001 Administration Is Associated with a Significant Reduction in the Incidence of Necrotizing Enterocolitis in Very Low Birth Weight Infants. J. Pediatr. 2022, 244, 64–71.e2. [Google Scholar] [CrossRef] [PubMed]
- Ghoneim, N.; Bauchart-Thevret, C.; Oosterloo, B.; Stoll, B.; Kulkarni, M.; de Pipaon, M.S.; Zamora, I.J.; Olutoye, O.O.; Berg, B.; Wittke, A.; et al. Delayed initiation but not gradual advancement of enteral formula feeding reduces the incidence of necrotizing enterocolitis (NEC) in preterm pigs. PLoS ONE 2014, 9, e106888. [Google Scholar] [CrossRef] [PubMed]
- Call, L.; Stoll, B.; Oosterloo, B.; Ajami, N.; Sheikh, F.; Wittke, A.; Waworuntu, R.; Berg, B.; Petrosino, J.; Olutoye, O.; et al. Metabolomic signatures distinguish the impact of formula carbohydrates on disease outcome in a preterm piglet model of NEC. Microbiome 2018, 6, 111. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool microbiota and vaccine responses of infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Liu, J.; Taft, D.H.; Maldonado-Gomez, M.X.; Johnson, D.; Treiber, M.L.; Lemay, D.G.; DePeters, E.J.; Mills, D.A. The fecal resistome of dairy cattle is associated with diet during nursing. Nat. Commun. 2019, 10, 4406. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Burrin, D.; Sangild, P.T.; Stoll, B.; Thymann, T.; Buddington, R.; Marini, J.; Olutoye, O.; Shulman, R.J. Translational Advances in Pediatric Nutrition and Gastroenterology: New Insights from Pig Models. Annu. Rev. Anim. Biosci. 2020, 8, 321–354. [Google Scholar] [CrossRef]
- Oosterloo, B.C.; Premkumar, M.; Stoll, B.; Olutoye, O.; Thymann, T.; Sangild, P.T.; Burrin, D.G. Dual purpose use of preterm piglets as a model of pediatric GI disease. Vet. Immunol. Immunopathol. 2014, 159, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Sangild, P.T. Gut responses to enteral nutrition in preterm infants and animals. Exp. Biol. Med. 2006, 231, 1695–1711. [Google Scholar] [CrossRef]
- Sangild, P.T.; Thymann, T.; Schmidt, M.; Stoll, B.; Burrin, D.G.; Buddington, R.K. Invited review: The preterm pig as a model in pediatric gastroenterology. J. Anim. Sci. 2013, 91, 4713–4729. [Google Scholar] [CrossRef] [PubMed]
- Lennon, D.; Zanganeh, T.; Borum, P.R. Development of the piglet neonatal intensive care unit for translational research. Lab. Anim. 2011, 40, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.L.; Sangild, P.T.; Lykke, M.; Schmidt, M.; Boye, M.; Jensen, B.B.; Thymann, T. Similar efficacy of human banked milk and bovine colostrum to decrease incidence of necrotizing enterocolitis in preterm piglets. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R4–R12. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.R.; Chow, J.M.; Buck, R.H. Multifunctional Benefits of Prevalent HMOs: Implications for Infant Health. Nutrients 2021, 13, 3364. [Google Scholar] [CrossRef]
- Quinn, E.M.; Slattery, H.; Walsh, D.; Joshi, L.; Hickey, R.M. Bifidobacterium longum subsp. infantis ATCC 15697 and Goat Milk Oligosaccharides Show Synergism In Vitro as Anti-Infectives against Campylobacter jejuni. Foods 2020, 9, 348. [Google Scholar] [CrossRef]
- Kang, L.J.; Oh, E.; Cho, C.; Kwon, H.; Lee, C.G.; Jeon, J.; Lee, H.; Choi, S.; Han, S.J.; Nam, J.; et al. 3′-Sialyllactose prebiotics prevents skin inflammation via regulatory T cell differentiation in atopic dermatitis mouse models. Sci. Rep. 2020, 10, 5603. [Google Scholar] [CrossRef]
- Kavanaugh, D.W.; O’Callaghan, J.; Butto, L.F.; Slattery, H.; Lane, J.; Clyne, M.; Kane, M.; Joshi, L.; Hickey, R.M. Exposure of Bifidobacterium longum subsp. infantis to Milk Oligosaccharides Increases Adhesion to Epithelial Cells and Induces a Substantial Transcriptional Response. PLoS ONE 2013, 8, e67224. [Google Scholar] [CrossRef]
- Rousseaux, A.; Brosseau, C.; Le Gall, S.; Piloquet, H.; Barbarot, S.; Bodinier, M. Human Milk Oligosaccharides: Their Effects on the Host and Their Potential as Therapeutic Agents. Front. Immunol. 2021, 12, 680911. [Google Scholar] [CrossRef]
- Angeloni, S.; Ridet, J.L.; Kusy, N.; Gao, H.; Crevoisier, F.; Guinchard, S.; Kochhar, S.; Sigrist, H.; Sprenger, N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 2005, 15, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.V.; Scholz, M.; Zolfo, M.; Taft, D.H.; Schibler, K.R.; Tett, A.; Segata, N.; Morrow, A.L. Metagenomic Sequencing with Strain-Level Resolution Implicates Uropathogenic E. coli in Necrotizing Enterocolitis and Mortality in Preterm Infants. Cell Rep. 2016, 14, 2912–2924. [Google Scholar] [CrossRef] [PubMed]
- Olm, M.R.; Bhattacharya, N.; Crits-Christoph, A.; Firek, B.A.; Baker, R.; Song, Y.S.; Morowitz, M.J.; Banfield, J.F. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci. Adv. 2019, 5, eaax5727. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishna, K.P.; Hand, T.W. Influence of Maternal Milk on the Neonatal Intestinal Microbiome. Nutrients 2020, 12, 823. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.L.; Aldape, M.J.; Bryant, A.E. Life-threatening clostridial infections. Anaerobe 2012, 18, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Sim, K.; Shaw, A.G.; Randell, P.; Cox, M.J.; McClure, Z.E.; Li, M.S.; Haddad, M.; Langford, P.R.; Cookson, W.O.; Moffatt, M.F.; et al. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin. Infect. Dis. 2015, 60, 389–397. [Google Scholar] [CrossRef]
- Shaw, A.G.; Cornwell, E.; Sim, K.; Thrower, H.; Scott, H.; Brown, J.C.S.; Dixon, R.A.; Kroll, J.S. Dynamics of toxigenic Clostridium perfringens colonisation in a cohort of prematurely born neonatal infants. BMC Pediatr. 2020, 20, 75. [Google Scholar] [CrossRef]
- Schonherr-Hellec, S.; Aires, J. Clostridia and necrotizing enterocolitis in preterm neonates. Anaerobe 2019, 58, 6–12. [Google Scholar] [CrossRef]
- de la Cochetiere, M.F.; Piloquet, H.; des Robert, C.; Darmaun, D.; Galmiche, J.P.; Roze, J.C. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: The putative role of Clostridium. Pediatr. Res. 2004, 56, 366–370. [Google Scholar] [CrossRef]



| Direction | Organism | Taxa Level | Gene Target | Amplicon Size | Sequence (5′-3′) |
|---|---|---|---|---|---|
| Forward | Bacteria | B. longum group | 16S rRNA | 106 | CAGTTGATCGCATGGTCTT |
| Reverse | Bacteria | B. longum group | 16S rRNA | 106 | TACCCGTCGAAGCCAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melendez Hebib, V.; Taft, D.H.; Stoll, B.; Liu, J.; Call, L.; Guthrie, G.; Jensen, N.; Hair, A.B.; Mills, D.A.; Burrin, D.G. Probiotics and Human Milk Differentially Influence the Gut Microbiome and NEC Incidence in Preterm Pigs. Nutrients 2023, 15, 2585. https://doi.org/10.3390/nu15112585
Melendez Hebib V, Taft DH, Stoll B, Liu J, Call L, Guthrie G, Jensen N, Hair AB, Mills DA, Burrin DG. Probiotics and Human Milk Differentially Influence the Gut Microbiome and NEC Incidence in Preterm Pigs. Nutrients. 2023; 15(11):2585. https://doi.org/10.3390/nu15112585
Chicago/Turabian StyleMelendez Hebib, Valeria, Diana H. Taft, Barbara Stoll, Jinxin Liu, Lee Call, Gregory Guthrie, Nick Jensen, Amy B. Hair, David A. Mills, and Douglas G. Burrin. 2023. "Probiotics and Human Milk Differentially Influence the Gut Microbiome and NEC Incidence in Preterm Pigs" Nutrients 15, no. 11: 2585. https://doi.org/10.3390/nu15112585
APA StyleMelendez Hebib, V., Taft, D. H., Stoll, B., Liu, J., Call, L., Guthrie, G., Jensen, N., Hair, A. B., Mills, D. A., & Burrin, D. G. (2023). Probiotics and Human Milk Differentially Influence the Gut Microbiome and NEC Incidence in Preterm Pigs. Nutrients, 15(11), 2585. https://doi.org/10.3390/nu15112585






