Hematopoietic Stem Cell: Regulation and Nutritional Intervention
Abstract
1. Introduction
2. Factors Regulating the Homeostatic Function of Hematopoietic Stem Cells
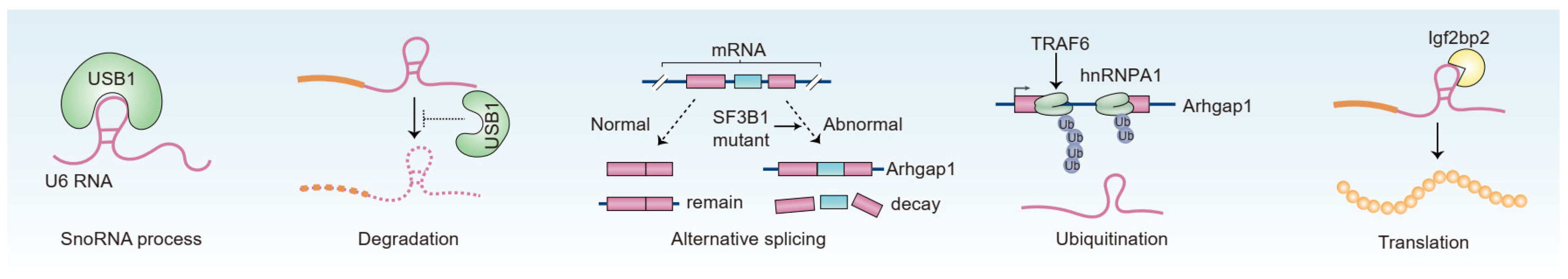
2.1. RNA-Binding Protein
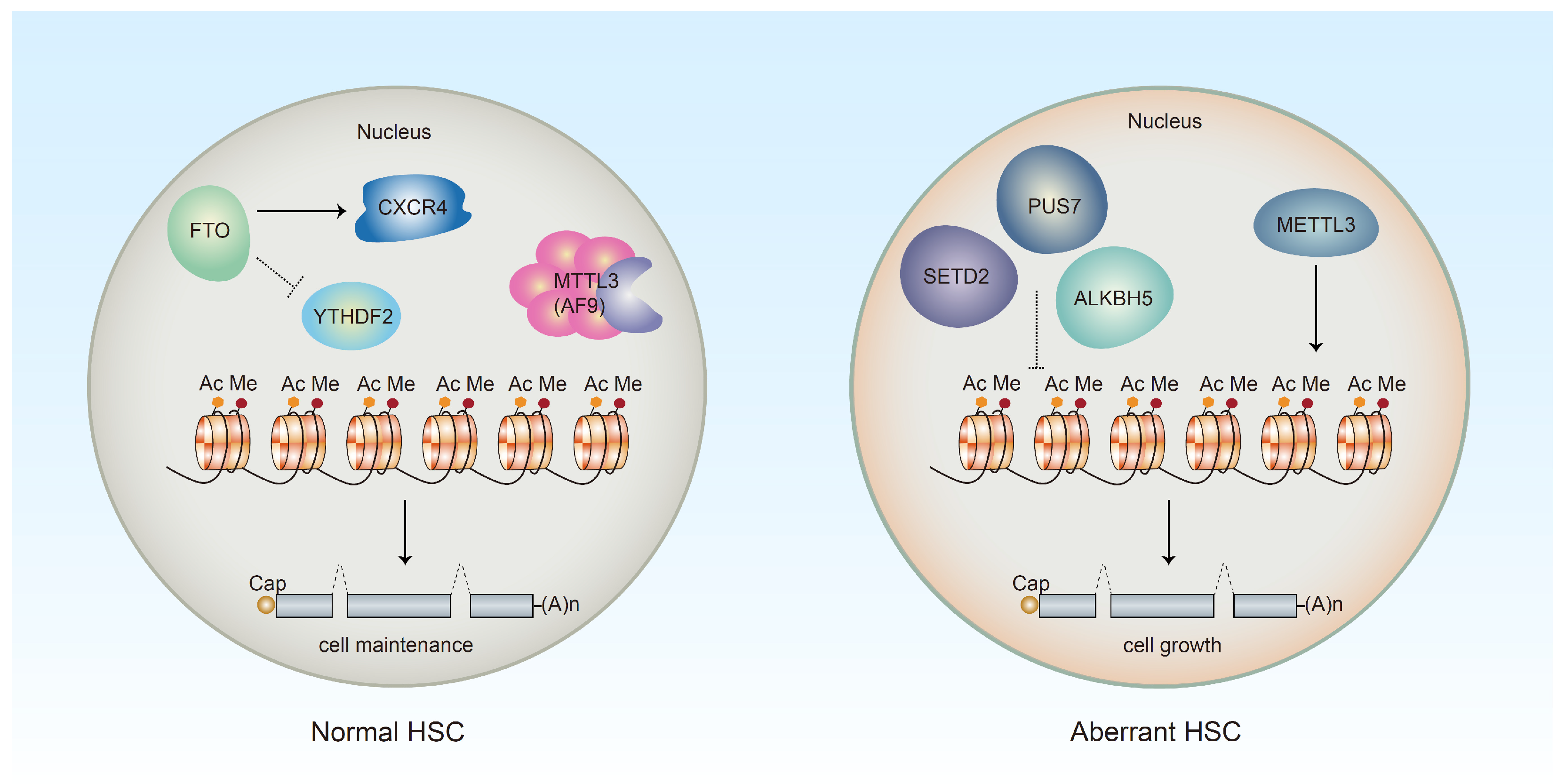
2.2. Epigenetic Regulation of HSC
2.3. Other Factors Regulating HSC Dormancy and Maintenance
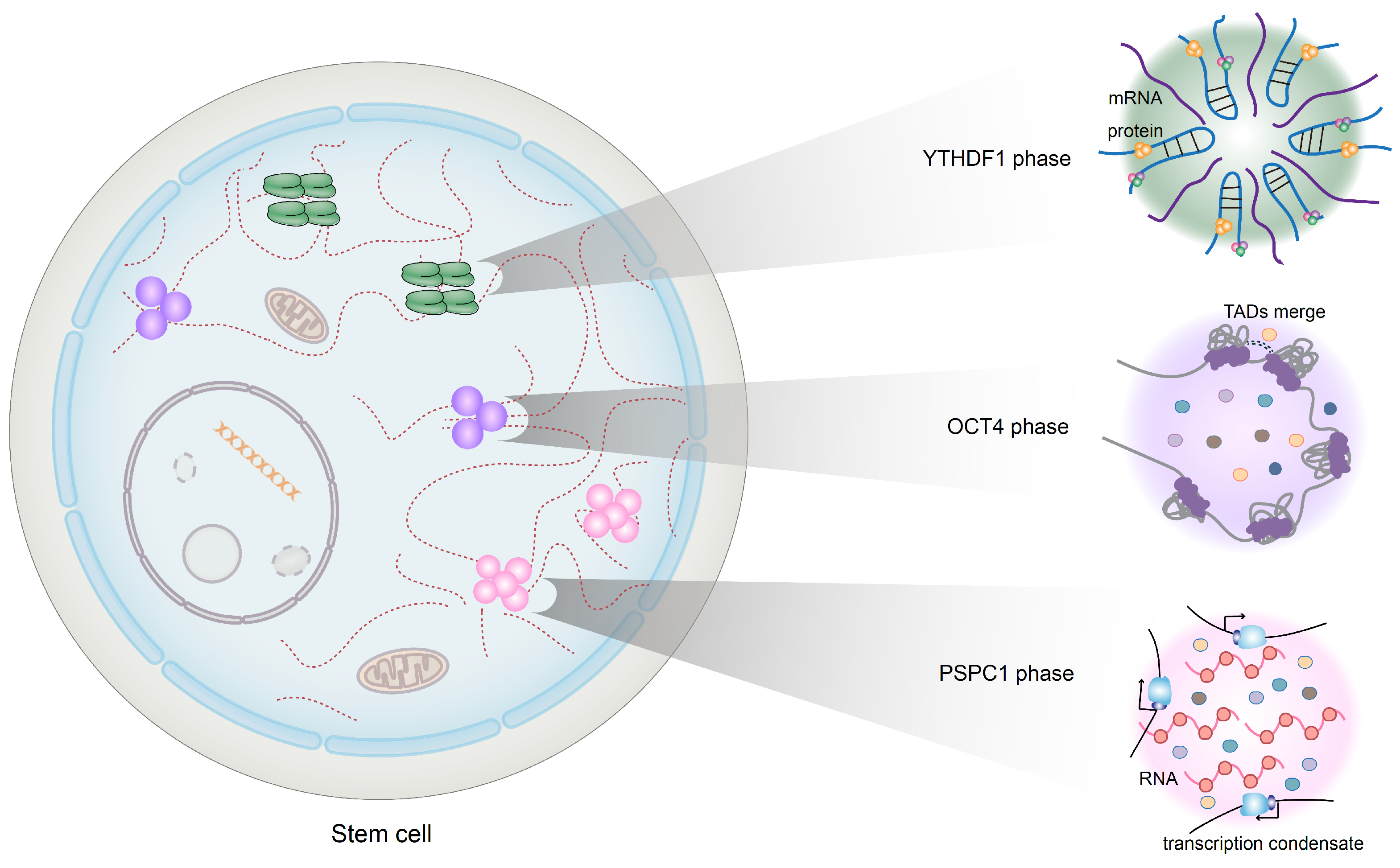
2.4. Phase Separation
3. Therapies for Bone Marrow Transplantation
4. The Relationship between HSCs and Autoimmune Diseases (ADs)
5. High-Fat Diet (HFD) Affects the Activity of HSCs
6. Specific Nutrients Essential to Maintain HSC
6.1. Vitamins
6.1.1. Vitamin A
6.1.2. Vitamin B3
6.1.3. Vitamin C
6.1.4. Vitamin D
6.2. Amino Acids
6.3. Probiotics and Prebiotics
7. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jacobson, L.O.; Simmons, E.L.; Marks, E.K.; Eldredge, J.H. Recovery from radiation injury. Science 1951, 113, 510–511. [Google Scholar] [CrossRef]
- Ford, C.E.; Hamerton, J.L.; Barnes, D.W.; Loutit, J.F. Cytological identification of radiation-chimaeras. Nature 1956, 177, 452–454. [Google Scholar] [CrossRef]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.; Jin, G.; Ma, Y.; Zhao, H.; Zhang, G.; Li, M.O.; Peng, M. SZT2 maintains hematopoietic stem cell homeostasis via nutrient-mediated mTORC1 regulation. J. Clin. Investig. 2022, 132, e146272. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, Y.; Hoshii, T.; Yamazaki, S.; Eto, K.; Ema, H.; Kobayashi, M.; Ueno, M.; Ohta, K.; Arai, Y.; Hara, E.; et al. Spred1 safeguards hematopoietic homeostasis against diet-induced systemic stress. Cell Stem Cell 2018, 22, 713–725.e718. [Google Scholar] [CrossRef] [PubMed]
- Hermetet, F.; Buffiere, A.; Aznague, A.; Pais de Barros, J.P.; Bastie, J.N.; Delva, L.; Quere, R. High-fat diet disturbs lipid raft/TGF-beta signaling-mediated maintenance of hematopoietic stem cells in mouse bone marrow. Nat. Commun. 2019, 10, 523. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, G.L.; Hannemann, N.; Ipseiz, N.; Kronke, G.; Bauerle, T.; Munos, L.; Wirtz, S.; Schett, G.; Bozec, A. Microbiota from obese mice regulate hematopoietic stem cell differentiation by altering the bone niche. Cell. Metab. 2015, 22, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Kamimae-Lanning, A.N.; Krasnow, S.M.; Goloviznina, N.A.; Zhu, X.; Roth-Carter, Q.R.; Levasseur, P.R.; Jeng, S.; McWeeney, S.K.; Kurre, P.; Marks, D.L. Maternal high-fat diet and obesity compromise fetal hematopoiesis. Mol. Metab. 2015, 4, 25–38. [Google Scholar] [CrossRef]
- Rzepecki, P.; Barzal, J.; Sarosiek, T.; Szczylik, C. Biochemical indices for the assessment of nutritional status during hematopoietic stem cell transplantation: Are they worth using? A single center experience. Bone Marrow Transpl. 2007, 40, 567–572. [Google Scholar] [CrossRef]
- Chanda, B.; Ditadi, A.; Iscove, N.N.; Keller, G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell 2013, 155, 215–227. [Google Scholar] [CrossRef]
- Cabezas-Wallscheid, N.; Buettner, F.; Sommerkamp, P.; Klimmeck, D.; Ladel, L.; Thalheimer, F.B.; Pastor-Flores, D.; Roma, L.P.; Renders, S.; Zeisberger, P.; et al. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 2017, 169, 807–823.e819. [Google Scholar] [CrossRef]
- Sun, X.; Cao, B.; Naval-Sanchez, M.; Pham, T.; Sun, Y.B.Y.; Williams, B.; Heazlewood, S.Y.; Deshpande, N.; Li, J.; Kraus, F.; et al. Nicotinamide riboside attenuates age-associated metabolic and functional changes in hematopoietic stem cells. Nat. Commun. 2021, 12, 2665. [Google Scholar] [CrossRef] [PubMed]
- Agathocleous, M.; Meacham, C.E.; Burgess, R.J.; Piskounova, E.; Zhao, Z.; Crane, G.M.; Cowin, B.L.; Bruner, E.; Murphy, M.M.; Chen, W.; et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 2017, 549, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Ghram, M.; Morris, G.; Culjkovic-Kraljacic, B.; Mars, J.C.; Gendron, P.; Skrabanek, L.; Revuelta, M.V.; Cerchietti, L.; Guzman, M.L.; Borden, K.L.B. The eukaryotic translation initiation factor eIF4E reprograms alternative splicing. EMBO J. 2023, 42, e110496. [Google Scholar] [CrossRef] [PubMed]
- McCarter, J.G.W.; Nemirovsky, D.; Famulare, C.A.; Farnoud, N.; Mohanty, A.S.; Stone-Molloy, Z.S.; Chervin, J.; Ball, B.J.; Epstein-Peterson, Z.D.; Arcila, M.E.; et al. Interaction between myelodysplasia-related gene mutations and ontogeny in acute myeloid leukemia. Blood Adv. 2023. [Google Scholar] [CrossRef] [PubMed]
- Willekens, C.; Laplane, L.; Dagher, T.; Benlabiod, C.; Papadopoulos, N.; Lacout, C.; Rameau, P.; Catelain, C.; Alfaro, A.; Edmond, V.; et al. SRSF2-P95H decreases JAK/STAT signaling in hematopoietic cells and delays myelofibrosis development in mice. Leukemia 2023. [Google Scholar] [CrossRef]
- Wang, Y.H.; Lin, C.C.; Yao, C.Y.; Amaral, F.; Yu, S.C.; Kao, C.J.; Shih, P.T.; Hou, H.A.; Chou, W.C.; Tien, H.F. High BM plasma S100A8/A9 is associated with a perturbed microenvironment and poor prognosis in myelodysplastic syndromes. Blood Adv. 2023. [Google Scholar] [CrossRef]
- Mortera-Blanco, T.; Dimitriou, M.; Woll, P.S.; Karimi, M.; Elvarsdottir, E.; Conte, S.; Tobiasson, M.; Jansson, M.; Douagi, I.; Moarii, M.; et al. SF3B1-initiating mutations in MDS-RSs target lymphomyeloid hematopoietic stem cells. Blood 2017, 130, 881–890. [Google Scholar] [CrossRef]
- Obeng, E.A.; Chappell, R.J.; Seiler, M.; Chen, M.C.; Campagna, D.R.; Schmidt, P.J.; Schneider, R.K.; Lord, A.M.; Wang, L.; Gambe, R.G.; et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 2016, 30, 404–417. [Google Scholar] [CrossRef]
- Lieu, Y.K.; Liu, Z.; Ali, A.M.; Wei, X.; Penson, A.; Zhang, J.; An, X.; Rabadan, R.; Raza, A.; Manley, J.L.; et al. SF3B1 mutant-induced missplicing of MAP3K7 causes anemia in myelodysplastic syndromes. Proc. Natl. Acad. Sci. USA 2022, 119, e2111703119. [Google Scholar] [CrossRef]
- Lee, S.C.; North, K.; Kim, E.; Jang, E.; Obeng, E.; Lu, S.X.; Liu, B.; Inoue, D.; Yoshimi, A.; Ki, M.; et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 2018, 34, 225–241.e228. [Google Scholar] [CrossRef]
- Kon, A.; Yamazaki, S.; Nannya, Y.; Kataoka, K.; Ota, Y.; Nakagawa, M.M.; Yoshida, K.; Shiozawa, Y.; Morita, M.; Yoshizato, T.; et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood 2018, 131, 621–635. [Google Scholar] [CrossRef]
- Fang, J.; Bolanos, L.C.; Choi, K.; Liu, X.; Christie, S.; Akunuru, S.; Kumar, R.; Wang, D.; Chen, X.; Greis, K.D.; et al. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 2017, 18, 236–245. [Google Scholar] [CrossRef]
- Jeong, H.C.; Shukla, S.; Fok, W.C.; Huynh, T.N.; Batista, L.F.Z.; Parker, R. USB1 is a miRNA deadenylase that regulates hematopoietic development. Science 2023, 379, 901–907. [Google Scholar] [CrossRef]
- Suo, M.; Rommelfanger, M.K.; Chen, Y.; Amro, E.M.; Han, B.; Chen, Z.; Szafranski, K.; Chakkarappan, S.R.; Boehm, B.O.; MacLean, A.L.; et al. Age-dependent effects of Igf2bp2 on gene regulation, function, and aging of hematopoietic stem cells in mice. Blood 2022, 139, 2653–2665. [Google Scholar] [CrossRef] [PubMed]
- De Andrés-Aguayo, L.; Varas, F.; Kallin, E.M.; Infante, J.F.; Wurst, W.; Floss, T.; Graf, T. Musashi 2 is a regulator of the HSC compartment identified by a retroviral insertion screen and knockout mice. Blood 2011, 118, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Rentas, S.; Holzapfel, N.; Belew, M.S.; Pratt, G.; Voisin, V.; Wilhelm, B.T.; Bader, G.D.; Yeo, G.W.; Hope, K.J. Musashi-2 attenuates AHR signalling to expand human haematopoietic stem cells. Nature 2016, 532, 508–511. [Google Scholar] [CrossRef]
- Stumpo, D.J.; Broxmeyer, H.E.; Ward, T.; Cooper, S.; Hangoc, G.; Chung, Y.J.; Shelley, W.C.; Richfield, E.K.; Ray, M.K.; Yoder, M.C.; et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 2009, 114, 2401–2410. [Google Scholar] [CrossRef]
- Shen, C.; Sheng, Y.; Zhu, A.C.; Robinson, S.; Jiang, X.; Dong, L.; Chen, H.; Su, R.; Yin, Z.; Li, W.; et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 2020, 27, 64–80.e69. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.; Morgan, M.; Campos, J.; Spencer, G.J.; Shmakova, A.; Ivanova, I.; Mapperley, C.; Lawson, H.; Wotherspoon, D.A.; Sepulveda, C.; et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 2019, 25, 137–148.e136. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Wang, X.; Cooper, S.; Broxmeyer, H.E.; Kapur, R. Transient regulation of RNA methylation in human hematopoietic stem cells promotes their homing and engraftment. Leukemia 2023, 37, 453–464. [Google Scholar] [CrossRef]
- Mapperley, C.; van de Lagemaat, L.N.; Lawson, H.; Tavosanis, A.; Paris, J.; Campos, J.; Wotherspoon, D.; Durko, J.; Sarapuu, A.; Choe, J.; et al. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J. Exp. Med. 2021, 218, e20200829. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qian, P.; Shao, W.; Shi, H.; He, X.C.; Gogol, M.; Yu, Z.; Wang, Y.; Qi, M.; Zhu, Y.; et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018, 28, 904–917. [Google Scholar] [CrossRef] [PubMed]
- Ciesla, M.; Ngoc, P.C.T.; Muthukumar, S.; Todisco, G.; Madej, M.; Fritz, H.; Dimitriou, M.; Incarnato, D.; Hellstrom-Lindberg, E.; Bellodi, C. m(6)A-driven SF3B1 translation control steers splicing to direct genome integrity and leukemogenesis. Mol. Cell 2023, 83, 1165–1179.e11. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.Y.; Song, J.; Hu, C.L.; Chen, S.B.; Zhang, Q.; Xu, C.H.; Wu, J.C.; Hou, D.; Sun, M.; Zhang, Y.L.; et al. SETD2 deficiency accelerates MDS-associated leukemogenesis via S100a9 in NHD13 mice and predicts poor prognosis in MDS. Blood 2020, 135, 2271–2285. [Google Scholar] [CrossRef]
- Guzzi, N.; Cieśla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimková, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 2018, 173, 1204–1216.e1226. [Google Scholar] [CrossRef]
- Calvanese, V.; Nguyen, A.T.; Bolan, T.J.; Vavilina, A.; Su, T.; Lee, L.K.; Wang, Y.; Lay, F.D.; Magnusson, M.; Crooks, G.M.; et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 2019, 576, 281–286. [Google Scholar] [CrossRef]
- Mohrin, M.; Shin, J.; Liu, Y.; Brown, K.; Luo, H.; Xi, Y.; Haynes, C.M.; Chen, D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015, 347, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zeng, H.; Chen, S.; Xu, Y.; Wang, S.; Tang, Y.; Wang, X.; Du, C.; Shen, M.; Chen, F.; et al. SRC-3 is involved in maintaining hematopoietic stem cell quiescence by regulation of mitochondrial metabolism in mice. Blood 2018, 132, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; He, J.; Liu, S.; Gao, A.; Yang, L.; Sun, G.; Ding, W.; Li, C.Y.; Gou, F.; He, M.; et al. A comprehensive RNA editome reveals that edited Azin1 partners with DDX1 to enable hematopoietic stem cell differentiation. Blood 2021, 138, 1939–1952. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Kong, W.X.; Tang, X.Y.; Xu, M.; Wang, Q.H.; Zhang, B.; Hu, L.D.; Chen, H. The transcription factor Zfp90 regulates the self-renewal and differentiation of hematopoietic stem cells. Cell Death Dis. 2018, 9, 677. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.T.; Warr, M.R.; Adelman, E.R.; Lansinger, O.M.; Flach, J.; Verovskaya, E.V.; Figueroa, M.E.; Passegué, E. Autophagy maintains the metabolism and function of young and old stem cells. Nature 2017, 543, 205–210. [Google Scholar] [CrossRef]
- Arai, F.; Hirao, A.; Ohmura, M.; Sato, H.; Matsuoka, S.; Takubo, K.; Ito, K.; Koh, G.Y.; Suda, T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 2004, 118, 149–161. [Google Scholar] [CrossRef]
- Ito, K.; Turcotte, R.; Cui, J.; Zimmerman, S.E.; Pinho, S.; Mizoguchi, T.; Arai, F.; Runnels, J.M.; Alt, C.; Teruya-Feldstein, J.; et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 2016, 354, 1156–1160. [Google Scholar] [CrossRef]
- Dong, S.; Wang, Q.; Kao, Y.R.; Diaz, A.; Tasset, I.; Kaushik, S.; Thiruthuvanathan, V.; Zintiridou, A.; Nieves, E.; Dzieciatkowska, M.; et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 2021, 591, 117–123. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Fang, Q.; Tian, G.G.; Wang, Q.; Liu, M.; He, L.; Li, S.; Wu, J. YTHDF1 phase separation triggers the fate transition of spermatogonial stem cells by activating the IkappaB-NF-kappaB-CCND1 axis. Cell Rep. 2023, 42, 112403. [Google Scholar] [CrossRef]
- Shao, W.; Bi, X.; Pan, Y.; Gao, B.; Wu, J.; Yin, Y.; Liu, Z.; Peng, M.; Zhang, W.; Jiang, X.; et al. Phase separation of RNA-binding protein promotes polymerase binding and transcription. Nat. Chem. Biol. 2022, 18, 70–80. [Google Scholar] [CrossRef]
- Shao, X.; Chen, Y.; Xu, A.; Xiang, D.; Wang, W.; Du, W.; Huang, Y.; Zhang, X.; Cai, M.; Xia, Z.; et al. Deneddylation of PML/RARalpha reconstructs functional PML nuclear bodies via orchestrating phase separation to eradicate APL. Cell Death Differ. 2022, 29, 1654–1668. [Google Scholar] [CrossRef]
- Xiao, C.; Wu, G.; Chen, P.; Gao, L.; Chen, G.; Zhang, H. Phase separation in epigenetics and cancer stem cells. Front. Oncol. 2022, 12, 922604. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Ma, Q.; Zeng, P.; Wu, D.; Hou, Y.; Liu, X.; Jia, L.; Sun, J.; Chen, Y.; et al. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell 2021, 28, 1868–1883.e1811. [Google Scholar] [CrossRef] [PubMed]
- Maneix, L.; Iakova, P.; Moree, S.; Sweeney, M.; Moka, N.; Yellapragada, S.V.; Catic, A. Protein phase separation in hematopoietic stem cell aging. Blood 2019, 134, 5003. [Google Scholar] [CrossRef]
- Christopherson, K.W., II; Hangoc, G.; Mantel, C.R.; Broxmeyer, H.E. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science 2004, 305, 1000–1003. [Google Scholar] [CrossRef]
- Czechowicz, A.; Kraft, D.; Weissman, I.L.; Bhattacharya, D. Efficient transplantation via antibody-based clearance of hematopoietic stem cell niches. Science 2007, 318, 1296–1299. [Google Scholar] [CrossRef]
- McDermott, D.H.; Gao, J.L.; Liu, Q.; Siwicki, M.; Martens, C.; Jacobs, P.; Velez, D.; Yim, E.; Bryke, C.R.; Hsu, N.; et al. Chromothriptic cure of WHIM syndrome. Cell 2015, 160, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Fares, I.; Chagraoui, J.; Gareau, Y.; Gingras, S.; Ruel, R.; Mayotte, N.; Csaszar, E.; Knapp, D.J.; Miller, P.; Ngom, M.; et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 2014, 345, 1509–1512. [Google Scholar] [CrossRef]
- Aiuti, A.; Biasco, L.; Scaramuzza, S.; Ferrua, F.; Cicalese, M.P.; Baricordi, C.; Dionisio, F.; Calabria, A.; Giannelli, S.; Castiello, M.C.; et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 2013, 341, 1233151. [Google Scholar] [CrossRef] [PubMed]
- Cartier, N.; Hacein-Bey-Abina, S.; Bartholomae, C.C.; Veres, G.; Schmidt, M.; Kutschera, I.; Vidaud, M.; Abel, U.; Dal-Cortivo, L.; Caccavelli, L.; et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009, 326, 818–823. [Google Scholar] [CrossRef]
- Biffi, A.; Montini, E.; Lorioli, L.; Cesani, M.; Fumagalli, F.; Plati, T.; Baldoli, C.; Martino, S.; Calabria, A.; Canale, S.; et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013, 341, 1233158. [Google Scholar] [CrossRef] [PubMed]
- Goodnow, C.C. Multistep pathogenesis of autoimmune disease. Cell 2007, 130, 25–35. [Google Scholar] [CrossRef]
- Gravano, D.M.; Al-Kuhlani, M.; Davini, D.; Sanders, P.D.; Manilay, J.O.; Hoyer, K.K. CD8(+) T cells drive autoimmune hematopoietic stem cell dysfunction and bone marrow failure. J. Autoimmun. 2016, 75, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, A.; Madley, R.; Danzl, N.; Borsotti, C.; Marharlooei, M.K.; Li, H.W.; Nauman, G.; Ding, X.; Ho, S.H.; Fousteri, G.; et al. T1D patient-derived hematopoietic stem cells are programmed to generate Tph, Tfh, and autoimmunity-associated B cell subsets in human immune system mice. Clin. Immunol. 2022, 240, 109048. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Chang, X.; Lu, Q.; Liu, Y. Cytopenia and autoimmune diseases: A vicious cycle fueled by mTOR dysregulation in hematopoietic stem cells. J. Autoimmun. 2013, 41, 182–187. [Google Scholar] [CrossRef]
- Snowden, J.A.; Badoglio, M.; Labopin, M.; Giebel, S.; McGrath, E.; Marjanovic, Z.; Burman, J.; Moore, J.; Rovira, M.; Wulffraat, N.M.; et al. Evolution, trends, outcomes, and economics of hematopoietic stem cell transplantation in severe autoimmune diseases. Blood Adv. 2017, 1, 2742–2755. [Google Scholar] [CrossRef]
- Alexander, T.; Farge, D.; Badoglio, M.; Lindsay, J.O.; Muraro, P.A.; Snowden, J.A.; Autoimmune Diseases Working Party (ADWP) of the European Society for Blood and Marrow Transplantation (EBMT); Marrow, T. Hematopoietic stem cell therapy for autoimmune diseases—Clinical experience and mechanisms. J. Autoimmun. 2018, 92, 35–46. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Katagi, M.; Terashima, T.; Okano, J.; Urabe, H.; Nakae, Y.; Ogawa, N.; Udagawa, J.; Maegawa, H.; Matsumura, K.; Chan, L.; et al. Hyperglycemia induces abnormal gene expression in hematopoietic stem cells and their progeny in diabetic neuropathy. FEBS Lett. 2014, 588, 1080–1086. [Google Scholar] [CrossRef]
- Yamakawa, T.; Ohigashi, H.; Hashimoto, D.; Hayase, E.; Takahashi, S.; Miyazaki, M.; Minomi, K.; Onozawa, M.; Niitsu, Y.; Teshima, T. Vitamin A-coupled liposomes containing siRNA against HSP47 ameliorate skin fibrosis in chronic graft-versus-host disease. Blood 2018, 131, 1476–1485. [Google Scholar] [CrossRef]
- Vannini, N.; Campos, V.; Girotra, M.; Trachsel, V.; Rojas-Sutterlin, S.; Tratwal, J.; Ragusa, S.; Stefanidis, E.; Ryu, D.; Rainer, P.Y.; et al. The NAD-booster nicotinamide riboside potently stimulates hematopoiesis through increased mitochondrial clearance. Cell Stem Cell 2019, 24, 405–418.e407. [Google Scholar] [CrossRef]
- Carr, A.C.; Vlasiuk, E.; Zawari, M.; Meijer, N.; Lauren, C.; MacPherson, S.; Williman, J.; Chambers, S.T. Supplementation with oral Vitamin C prior to and during myeloablative chemotherapy and autologous haematopoietic stem cell transplantation: A pilot study. Antioxidants 2022, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Bickford, P.C.; Kaneko, Y.; Grimmig, B.; Pappas, C.; Small, B.; Sanberg, C.D.; Sanberg, P.R.; Tan, J.; Douglas Shytle, R. Nutraceutical intervention reverses the negative effects of blood from aged rats on stem cells. Age 2015, 37, 103. [Google Scholar] [CrossRef] [PubMed]
- Mikirova, N.A.; Jackson, J.A.; Hunninghake, R.; Kenyon, J.; Chan, K.W.; Swindlehurst, C.A.; Minev, B.; Patel, A.N.; Murphy, M.P.; Smith, L.; et al. Nutraceutical augmentation of circulating endothelial progenitor cells and hematopoietic stem cells in human subjects. J. Transl. Med. 2010, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.G.; Wang, C.J.; Zhao, G.; Li, G.Y. Role of vitamin D in regulating the neural stem cells of mouse model with multiple sclerosis. Eur. Rev. Med. Pharm. Sci. 2015, 19, 4004–4011. [Google Scholar]
- Taya, Y.; Ota, Y.; Wilkinson, A.C.; Kanazawa, A.; Watarai, H.; Kasai, M.; Nakauchi, H.; Yamazaki, S. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science 2016, 354, 1152–1155. [Google Scholar] [CrossRef]
- Wilkinson, A.C.; Morita, M.; Nakauchi, H.; Yamazaki, S. Branched-chain amino acid depletion conditions bone marrow for hematopoietic stem cell transplantation avoiding amino acid imbalance-associated toxicity. Exp. Hematol. 2018, 63, 12–16.e11. [Google Scholar] [CrossRef]
- Li, C.; Wu, B.; Li, Y.; Chen, J.; Ye, Z.; Tian, X.; Wang, J.; Xu, X.; Pan, S.; Zheng, Y.; et al. Amino acid catabolism regulates hematopoietic stem cell proteostasis via a GCN2-eIF2alpha axis. Cell Stem Cell 2022, 29, 1119–1134.e1117. [Google Scholar] [CrossRef]
- Brevi, A.; Cogrossi, L.L.; Lorenzoni, M.; Mattorre, B.; Bellone, M. The insider: Impact of the gut microbiota on cancer immunity and response to therapies in multiple myeloma. Front. Immunol. 2022, 13, 845422. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, C.; Zhang, A. Gut microbiota in acute leukemia: Current evidence and future directions. Front. Microbiol. 2022, 13, 1045497. [Google Scholar] [CrossRef]
- Muratore, E.; Leardini, D.; Baccelli, F.; Venturelli, F.; Prete, A.; Masetti, R. Nutritional modulation of the gut microbiome in allogeneic hematopoietic stem cell transplantation recipients. Front. Nutr. 2022, 9, 993668. [Google Scholar] [CrossRef] [PubMed]
- Shono, Y.; van den Brink, M.R.M. Gut microbiota injury in allogeneic haematopoietic stem cell transplantation. Nat. Rev. Cancer 2018, 18, 283–295. [Google Scholar] [CrossRef]
- Yoshifuji, K.; Inamoto, K.; Kiridoshi, Y.; Takeshita, K.; Sasajima, S.; Shiraishi, Y.; Yamashita, Y.; Nisaka, Y.; Ogura, Y.; Takeuchi, R.; et al. Prebiotics protect against acute graft-versus-host disease and preserve the gut microbiota in stem cell transplantation. Blood Adv. 2020, 4, 4607–4617. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef]
- Mizutani, Y.; Kawamoto, S.; Takahashi, M.; Doi, H.; Wakida, K.; Tabuchi, S.; Tanda, M.; Soga, A.; Chijiki, R.; Takakura, H.; et al. Efficacy and Safety of Synbiotics in Patients Undergoing Autologous Hematopoietic Stem Cell Transplantation: A Randomized, Double-blinded, Placebo-controlled Pilot Study. Intern. Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.C.; Ishida, R.; Kikuchi, M.; Sudo, K.; Morita, M.; Crisostomo, R.V.; Yamamoto, R.; Loh, K.M.; Nakamura, Y.; Watanabe, M.; et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 2019, 571, 117–121. [Google Scholar] [CrossRef]
- Olsson, A.; Venkatasubramanian, M.; Chaudhri, V.K.; Aronow, B.J.; Salomonis, N.; Singh, H.; Grimes, H.L. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016, 537, 698–702. [Google Scholar] [CrossRef]
- Hu, L.; Cheng, H.; Gao, Y.; Shi, M.; Liu, Y.; Hu, Z.; Xu, J.; Qiu, L.; Yuan, W.; Leung, A.Y.; et al. Antioxidant N-acetyl-L-cysteine increases engraftment of human hematopoietic stem cells in immune-deficient mice. Blood 2014, 124, e45–e48. [Google Scholar] [CrossRef]
| Nutrients | Functional Changes | Regulatory Mechanism | Ref. |
|---|---|---|---|
| Vitamin A | Improves fibrosis, facilitates HSC development | Downregulates HSP47 expression, suppresses level of ROS | [55,56,57,58] |
| Vitamin B3 | Stimulates hematopoiesis, attenuates age-associated metabolic and functional changes in HSC | A precursor to NAD+; increases mitophagy and reduces mitochondrial metabolism | [59,60] |
| Vitamin C | Improves acute myeloid leukemia condition, slackens leukemogenesis | Removes ROS, combines with Flt3 internal tandem duplication (Flt3ITD) | [61,62] |
| Vitamin D | Rescues aging stem cells, improves neural function | - | [63,64,65] |
| Amino acids | Maintains HSCs, reduces iatrogenic complications in HSC transplantation | Activates the GCN2-eIF2α axis, inhibits Src-mediated AKT activation | [66,67,68] |
| Probiotics, prebiotics and synbiotics | Prevents acute graft-versus-host disease (aGVHD), improves mucosal injury, ameliorates chemotherapy-induced mucosal damage improve diarrhea and anorexia after engraftment | Maintains butyrate-producing bacterial population, reduces gastrointestinal toxicity | [69,70,71,72,73,74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Han, Y.; Lei, Y.; Yu, Y.; Dong, Y.; Chen, J. Hematopoietic Stem Cell: Regulation and Nutritional Intervention. Nutrients 2023, 15, 2605. https://doi.org/10.3390/nu15112605
Sun S, Han Y, Lei Y, Yu Y, Dong Y, Chen J. Hematopoietic Stem Cell: Regulation and Nutritional Intervention. Nutrients. 2023; 15(11):2605. https://doi.org/10.3390/nu15112605
Chicago/Turabian StyleSun, Siyuan, Yingxue Han, Yumei Lei, Yifei Yu, Yanbin Dong, and Juan Chen. 2023. "Hematopoietic Stem Cell: Regulation and Nutritional Intervention" Nutrients 15, no. 11: 2605. https://doi.org/10.3390/nu15112605
APA StyleSun, S., Han, Y., Lei, Y., Yu, Y., Dong, Y., & Chen, J. (2023). Hematopoietic Stem Cell: Regulation and Nutritional Intervention. Nutrients, 15(11), 2605. https://doi.org/10.3390/nu15112605







