Genetic Preference for Sweet Taste in Mothers Associates with Mother-Child Preference and Intake
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Diet Recordings for the Mothers and the Children
2.3. Anthropometric Information for the Mothers and the Children
2.4. Saliva Collection
2.5. DNA Extraction
2.6. Selection of Taste- and Food Preference-Related SNPs and Sequencing
2.7. Caries Assessment
2.8. Data Handling and Statistical Analyses
3. Results
3.1. Participant Characteristics
3.2. SNP Genotyping
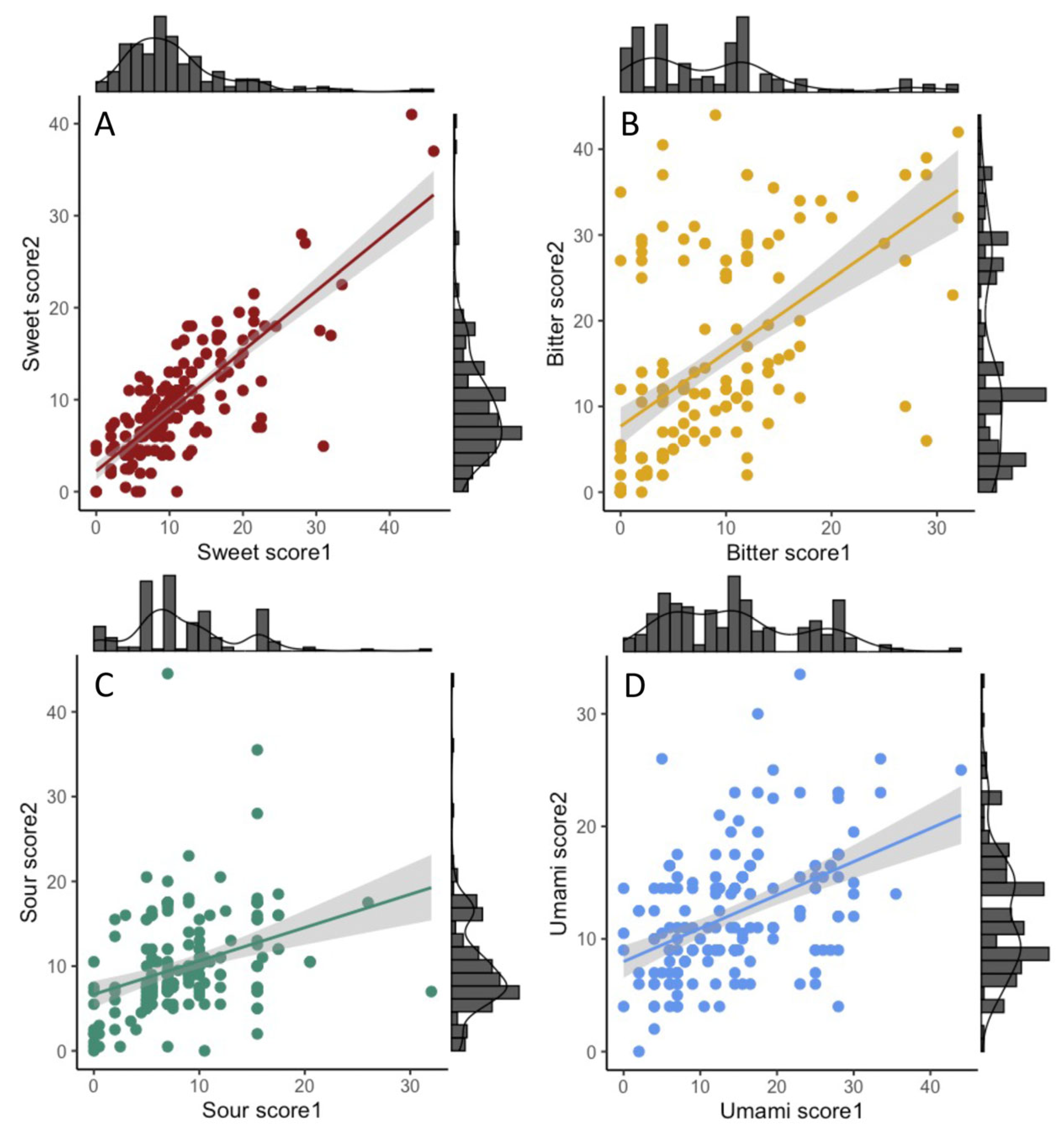
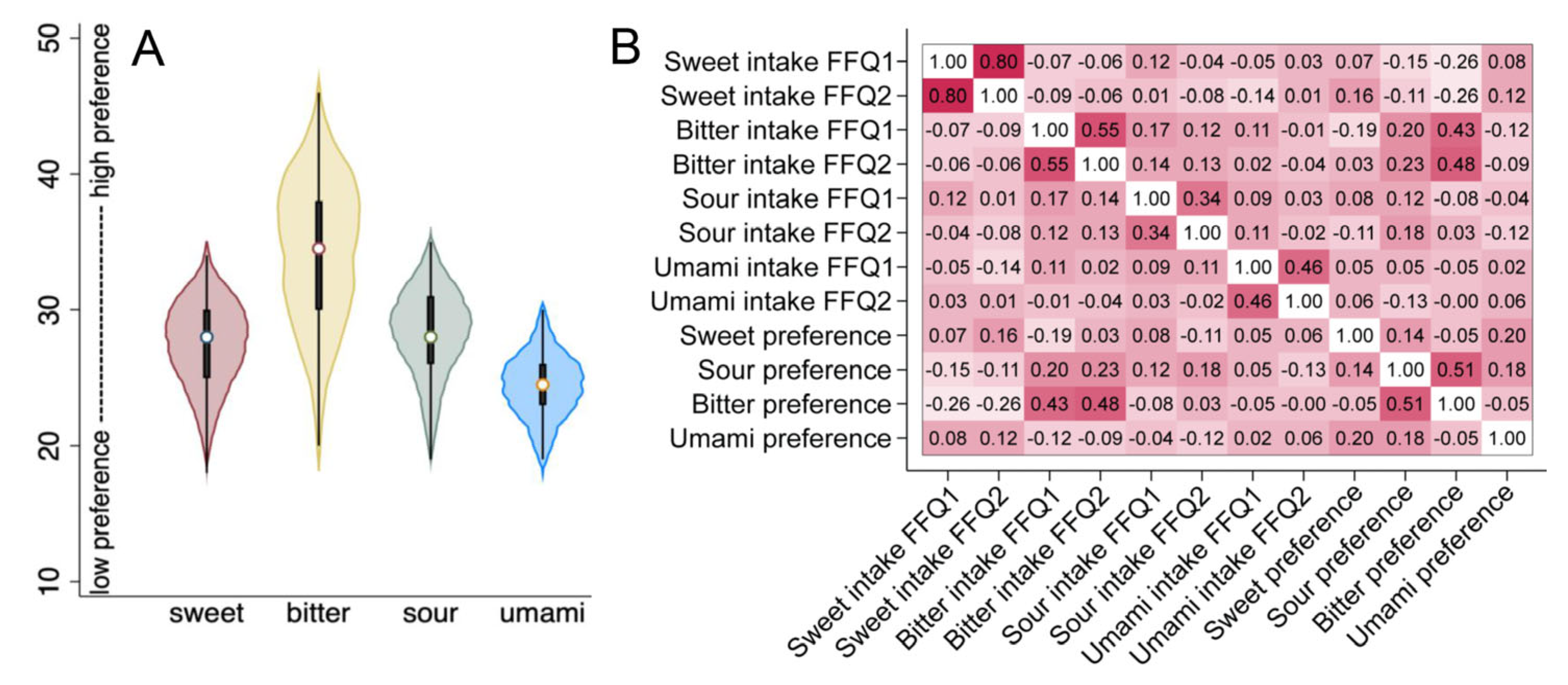
3.3. Mothers’ Reported Intake and Preferences for Sweet and Other Taste-Representing Foods
3.4. Associations between SNPs and Taste Preference and Food Intake in the Mothers
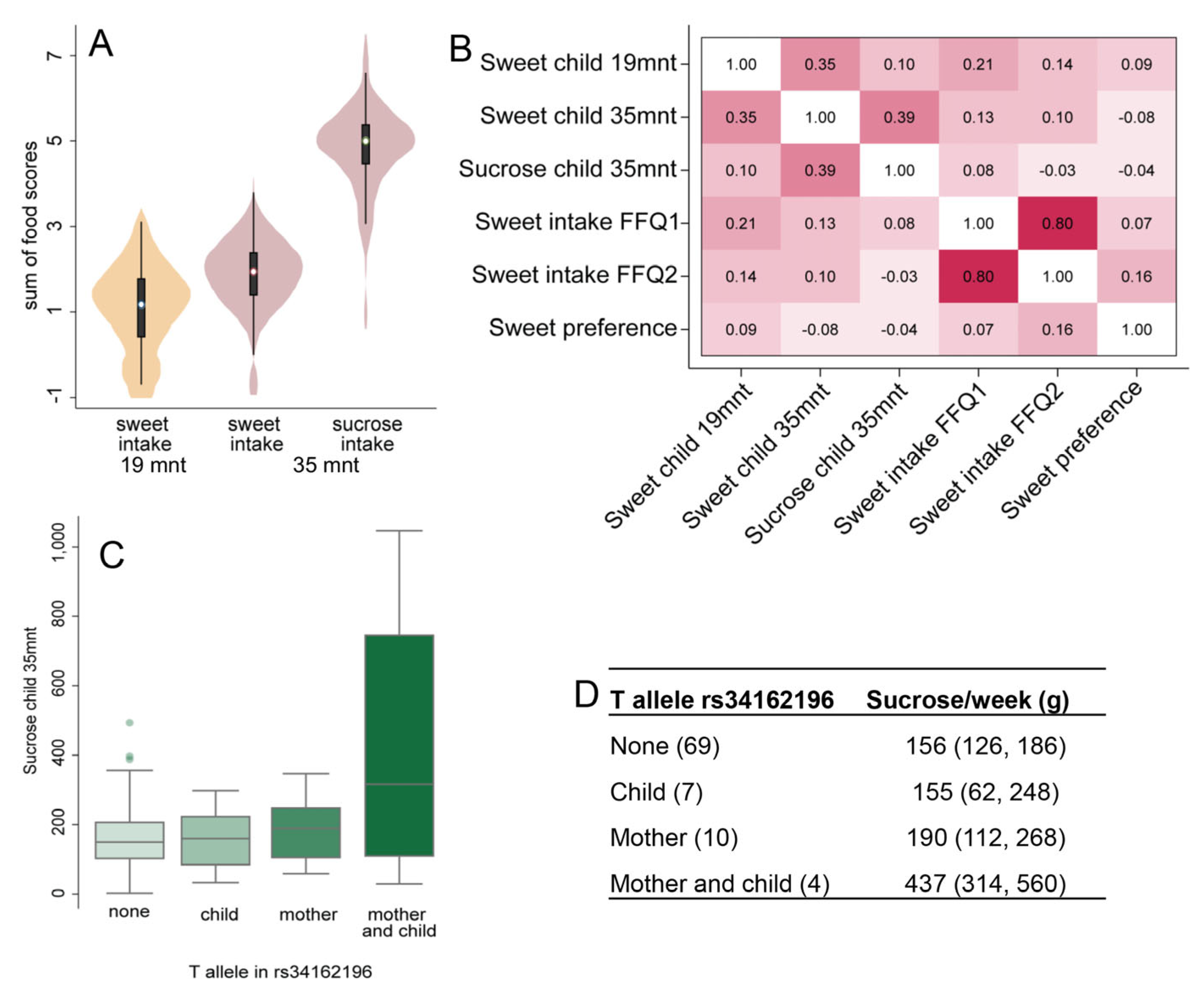
3.5. Child Feeding in Relation to Their Mothers’ Genetic Association with Sweet Preference
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collaborators GBDRF. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1223–1249. [Google Scholar] [CrossRef] [PubMed]
- Story, M.; Neumark-Sztainer, D.; French, S. Individual and environmental influences on adolescent eating behaviors. J. Am. Diet Assoc. 2002, 102, S40–S51. [Google Scholar] [CrossRef] [PubMed]
- Sanematsu, K.; Yoshida, R.; Shigemura, N.; Ninomiya, Y. Structure, function, and signaling of taste G-protein-coupled receptors. Curr. Pharm. Biotechnol. 2014, 15, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Diepeveen, J.; Moerdijk-Poortvliet, T.C.W.; van der Leij, F.R. Molecular insights into human taste perception and umami tastants: A review. J. Food Sci. 2022, 87, 1449–1465. [Google Scholar] [CrossRef]
- Roper, S.D. The taste of table salt. Pflug. Arch. 2015, 467, 457–463. [Google Scholar] [CrossRef]
- Liman, E.R.; Kinnamon, S.C. Sour taste: Receptors, cells and circuits. Curr. Opin. Physiol. 2021, 20, 8–15. [Google Scholar] [CrossRef]
- Hichami, A.; Khan, A.S.; Khan, N.A. Cellular and Molecular Mechanisms of Fat Taste Perception. Handb. Exp. Pharmacol. 2022, 275, 247–270. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Esberg, A.; Haworth, S.; Holgerson, P.L.; Johansson, I. Allelic Variation in Taste Genes Is Associated with Taste and Diet Preferences and Dental Caries. Nutrients 2019, 11, 1491. [Google Scholar] [CrossRef] [PubMed]
- Patrikainen, M.; Pan, P.; Kulesskaya, N.; Voikar, V.; Parkkila, S. The role of carbonic anhydrase VI in bitter taste perception: Evidence from the Car6(-)/(-) mouse model. J. Biomed. Sci. 2014, 21, 82. [Google Scholar] [CrossRef]
- Garcia-Bailo, B.; Toguri, C.; Eny, K.M.; El-Sohemy, A. Genetic variation in taste and its influence on food selection. OMICS 2009, 13, 69–80. [Google Scholar] [CrossRef]
- Cole, J.B.; Florez, J.C.; Hirschhorn, J.N. Comprehensive genomic analysis of dietary habits in UK Biobank identifies hundreds of genetic associations. Nat. Commun. 2020, 11, 1467. [Google Scholar] [CrossRef]
- Liszt, K.I.; Wang, Q.; Farhadipour, M.; Segers, A.; Thijs, T.; Nys, L.; Deleus, E.; Van der Schueren, B.; Gerner, C.; Neuditschko, B.; et al. Human intestinal bitter taste receptors regulate innate immune responses and metabolic regulators in obesity. J. Clin. Investig. 2022, 132, e144828. [Google Scholar] [CrossRef] [PubMed]
- Ki, S.Y.; Jeong, Y.T. Taste Receptors beyond Taste Buds. Int. J. Mol. Sci. 2022, 23, 9677. [Google Scholar] [CrossRef] [PubMed]
- Orlova, E.; Dudding, T.; Chernus, J.M.; Alotaibi, R.N.; Haworth, S.; Crout, R.J.; Lee, M.K.; Mukhopadhyay, N.; Feingold, E.; Levy, S.M.; et al. Association of Early Childhood Caries with Bitter Taste Receptors: A Meta-Analysis of Genome-Wide Association Studies and Transcriptome-Wide Association Study. Genes 2022, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Godfrey, K.M.; Poston, L.; Szajewska, H.; van Goudoever, J.B.; de Waard, M.; Brands, B.; Grivell, R.M.; Deussen, A.R.; Dodd, J.M.; et al. Nutrition During Pregnancy, Lactation and Early Childhood and its Implications for Maternal and Long-Term Child Health: The Early Nutrition Project Recommendations. Ann. Nutr. Metab. 2019, 74, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Netting, M.J.; Middleton, P.F.; Makrides, M. Does maternal diet during pregnancy and lactation affect outcomes in offspring? A systematic review of food-based approaches. Nutrition 2014, 30, 1225–1241. [Google Scholar] [CrossRef]
- Ventura, A.K.; Worobey, J. Early influences on the development of food preferences. Curr. Biol. 2013, 23, 401–408. [Google Scholar] [CrossRef]
- Fisk, C.M.; Crozier, S.R.; Inskip, H.M.; Godfrey, K.M.; Cooper, C.; Robinson, S.M. Influences on the quality of young children’s diets: The importance of maternal food choices. Br. J. Nutr. 2011, 105, 287–296. [Google Scholar] [CrossRef]
- Ventura, A.K.; Phelan, S.; Silva Garcia, K. Maternal Diet During Pregnancy and Lactation and Child Food Preferences, Dietary Patterns, and Weight Outcomes: A Review of Recent Research. Curr. Nutr. Rep. 2021, 10, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Delahunt, A.; Conway, M.C.; Callaghan, S.L.; O’Brien, E.C.; Geraghty, A.A.; O’Reilly, S.L.; McDonnell, C.M.; Mehegan, J.; McAuliffe, F.M. Maternal dietary quality during pregnancy and child appetitive traits at 5-years-old: Findings from the ROLO longitudinal birth cohort study. Appetite 2022, 179, 106291. [Google Scholar] [CrossRef]
- Noakes, P.; Taylor, A.; Hale, J.; Breckler, L.; Richmond, P.; Devadason, S.G.; Prescott, S.L. The effects of maternal smoking on early mucosal immunity and sensitization at 12 months of age. Pediatr. Allergy Immunol. 2007, 18, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Esberg, A.; Eriksson, L.; Johansson, I. Site- and Time-Dependent Compositional Shifts in Oral Microbiota Communities. Front. Oral. Health 2022, 3, 826996. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.M.; Boston, E.S.M.; Finarelli, J.A.; Murphy, W.J.; Higgins, D.G.; Teeling, E.C. The Birth and Death of Olfactory Receptor Gene Families in Mammalian Niche Adaptation. Mol. Biol. Evol. 2018, 35, 1390–1406. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Schulze, M.B.; Hu, F.B. Intake of sugar-sweetened beverages and weight gain: A systematic review. Am. J. Clin. Nutr. 2006, 84, 274–288. [Google Scholar] [CrossRef]
- Patterson, E.; Ryan, P.M.; Cryan, J.F.; Dinan, T.G.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. Gut microbiota, obesity and diabetes. Postgrad. Med. J. 2016, 92, 286–300. [Google Scholar] [CrossRef]
- Krasse, B. The Vipeholm Dental Caries Study: Recollections and reflections 50 years later. J. Dent. Res. 2001, 80, 1785–1788. [Google Scholar] [CrossRef]
- Haworth, S.; Esberg, A.; Lif Holgerson, P.; Ruja-Hakola, R.; Timpson, N.J.; Magnusson, P.K.E.; Franks, P.W.; Johansson, I. Heritability of Caries Scores, Trajectories, and Disease Subtypes. J. Dent. Res. 2000, 99, 264–270. [Google Scholar] [CrossRef]
- Willett, W.C.; Howe, G.R.; Kushi, L.H. Adjustment for total energy intake in epidemiologic studies. Am. J. Clin. Nutr. 1997, 65, 1220S–1228S. [Google Scholar] [CrossRef]
- Kusuhara, Y.; Yoshida, R.; Ohkuri, T.; Yasumatsu, K.; Voigt, A.; Hubner, S.; Maeda, K.; Boehm, U.; Meyerhof, W.; Ninomiya, Y. Taste responses in mice lacking taste receptor subunit T1R1. J. Physiol. 2013, 591, 1967–1985. [Google Scholar] [CrossRef]
- Hartley, I.E.; Liem, D.G.; Keast, R. Umami as an ‘Alimentary’ Taste. A New Perspective on Taste Classification. Nutrients 2019, 11, 182. [Google Scholar] [CrossRef]
- Lundberg, R. Valideringsstudie av Matfrekvensformulär Besvarade av Gravida i NorthPop-Studien. Master’s Thesis, Umeå University, Umeå, Sweden, 2022. [Google Scholar]
- Johansson, I.; Hallmans, G.; Wikman, A.; Biessy, C.; Riboli, E.; Kaaks, R. Validation and calibration of food-frequency questionnaire measurements in the Northern Sweden Health and Disease cohort. Public Health Nutr. 2002, 5, 487–496. [Google Scholar] [CrossRef]
| Variable | Measurement |
|---|---|
| Age (years mean, 95% CI; min-max) 1 | 31.1 (30.5, 31.6; 19.45) |
| BMI (kg/m2, mean, 95% CI) | 24.3 (23.8, 24.8) |
| Underweight (BMI < 20 kg/m2), % | 8.1 |
| Normal weight (BMI 20 ≤ 25 kg/m2), % | 54.3 |
| Overweight (BMI 25 ≤ 29 kg/m2), % | 24.8 |
| Obese (BMI ≥ 30 kg/m2), % | 10.1 |
| University or college education, % | 68.2 |
| Maternal FFQ1 in the 3rd trimester, n (%) | 241 (96.0) |
| Maternal FFQ2 when the child is 24 months old, n (%) | 188 (74.9) |
| Taste preference questionnaire, n (%) | 142 (56.7) |
| Saliva sample for DNA extraction, n (%) | 187 (74.5) |
| Oral examination, n | 208 |
| Number of teeth (mean, 95% CI; min–max) | 27.3 (27.1, 27.5; 15–28) |
| Caries prevalence (mean, 95% CI; min–max) 2 | 11.2 (9.8, 12.6; 0–59) |
| Major Allele CC (n = 148) | Minor Allele CT/TC/TT (n = 36) | p- Value | |
|---|---|---|---|
| Diet, mean (95% CI) 1 | |||
| Sweet preference score | 26.9 (26.3, 27.5) | 29.5 (28.3, 30.7) | <0.001 |
| Bitter preference score | 34.4 (33.3, 35.4) | 32.4 (30.2, 34.7) | 0.128 |
| Sour preference score | 29.1 (27.5, 28.7) | 28.7 (27.4, 30.0) | 0.395 |
| Umami preference score | 24.2 (23.8, 24.6) | 24.9 (24.0, 25.9) | 0.156 |
| Sweet food intake/week (FFQ1) | 10.4 (9.2, 11.7) | 11.5 (8.9, 14.2) | 0.489 |
| Sweet food intake/week (FFQ2) | 9.1 (8.1, 10.2) | 11.8 (9.4, 14.1) | 0.044 |
| Bitter food intake/week (FFQ1) | 8.8 (7.6, 9.9) | 7.7 (5.2, 10.0) | 0.413 |
| Bitter food intake/week (FFQ2) | 15.2 (13.0, 17.3) | 14.7 (9.8, 19.5) | 0.854 |
| Sour food intake/week (FFQ1) | 8.4 (7.6, 9.2) | 7.6 (5.9, 9.3) | 0.380 |
| Sour food intake/week (FFQ2) | 10.3 (9.3, 11.4) | 8.3 (5.9, 10.7) | 0.123 |
| Umami food intake/week (FFQ1) | 13.5 (12.1, 15.0) | 17.0 (13.9, 20.0) | 0.045 |
| Umami food intake/week (FFQ2) | 12.1 (11.1, 13.2) | 12.0 (9.7, 14.3) | 0.918 |
| Secondary outcomes, mean (95% CI) | |||
| BMI, kg/m2 | 24.1 (23.4, 24.8) | 25.6 (24.2, 27.0) | 0.049 |
| Proportion overweight, % | 21.9 | 25.7 | 0.314 2 |
| Proportion obese, % | 11.6 | 20.0 | |
| DMFS 3 | 10.7 (8.9, 12.6) | 12.6 (9.0, 16.1) | 0.364 |
| Major Allele TT (n = 156) | Minor Allele TG/GT/GG (n = 31) | p- Value | |
|---|---|---|---|
| Diet, mean (95% CI) 1 | |||
| Sweet preference score | 27.5 (26.9, 28.1) | 26.8 (25.5, 28.1) | 0.343 |
| Bitter preference score | 34.3 (33.2, 35.3) | 32.8 (30.5, 35.1) | 0.252 |
| Sour preference score | 28.3 (27.7, 28.9) | 27.9 (26.5, 29.2) | 0.536 |
| Umami preference score | 24.4 (24.0, 24.8) | 24.0 (23.1, 24.9) | 0.451 |
| Sweet food intake/week (FFQ1) | 10.1 (8.9, 11.3) | 13.2 (10.4, 16.0) | 0.046 |
| Sweet food intake/week (FFQ2) | 9.0 (7.9, 10.0) | 11.8 (9.5, 14.1) | 0.031 |
| Bitter food intake/week (FFQ1) | 8.7 (7.6, 9.8) | 7.5 (5.0, 10.1) | 0.414 |
| Bitter food intake/week (FFQ2) | 15.8 (13.7, 17.9) | 11.5 (6.7, 16.2) | 0.102 |
| Sour food intake/week (FFQ1) | 8.4 (7.7, 9.1) | 7.9 (6.1, 9.7) | 0.642 |
| Sour food intake/week (FFQ2) | 10.2 (9.1, 11.3) | 9.2 (6.7, 11.6) | 0.448 |
| Umami food intake/week (FFQ1) | 13.9 (12.5, 15.3) | 15.8 (12.6, 19.0) | 0.277 |
| Umami food intake/week (FFQ2) | 12.0 (11.0, 13.0) | 13.3 (11.0, 15.6) | 0.302 |
| Secondary outcomes, mean (95% CI) | |||
| BMI kg/m2 | 24.6 (23.9, 25.2) | 23.3 (21.8, 24.7) | 0.112 |
| Proportion overweight, % | 24.0 | 16.7 | 0.516 2 |
| Proportion obese, % | 13.6 | 10.0 | |
| DMFS 3 | 11.3 (9.5, 13.1) | 10.8 (6.8, 14.7) | 0.808 |
| Variable | Measurement |
|---|---|
| Child FFQ at 19 months, n (%) | 203 (80.9) |
| Child FFQ at 35 months, n (%) | 161 (64.1) |
| Child BMI percentile at 35 months (mean, 95% CI) | 60.3 (56.2, 64.3) |
| Underweight at 35 months (BMI < 5% percentile), % | 3.1 |
| Normal weight at 35 months (BMI 6 ≤ 85 percentile), % | 71.5 |
| Overweight at 35 months (BMI 85 ≤ 95 percentile), % | 11.1 |
| Obese at 35 months (BMI ≥ 95% percentile), % | 14.2 |
| Diet in Children (Mean (95% CI) 1) by Maternal rs34162196 SNP Polymorphism | Major Allele CC (n = 148) | Minor Allele CT/TC/TT (n = 36) | p-Value |
|---|---|---|---|
| Sucrose intake/week, 35 months old | 154 (127, 181) | 238 (181, 295) | 0.010 |
| Sweet food intake/week, 19 months old | 3.6 (2.8, 4.3) | 5.4 (3.8, 7.1) | 0.046 |
| Sweet food intake/week, 35 months old | 7.5 (6.3, 8.7) | 11.0 (8.3, 13.6) | 0.022 |
| Diet in Children (Mean (95% CI) 1) by Maternal rs7513755 SNP Polymorphism | Major allele TT (n = 156) | Minor allele TG/GT/GG (n = 31) | p- Value |
| Sucrose intake/week, 35 months old | 175 (148, 201) | 132 (74, 191) | 0.196 |
| Sweet food intake/week, 19 months old | 3.6 (2.8, 4.4) | 5.1 (3.3, 6.8) | 0.135 |
| Sweet food intake/week, 35 months old | 8.2 (6.9, 9.4) | 7.3 (4.6, 10.0) | 0.568 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lif Holgerson, P.; Hasslöf, P.; Esberg, A.; Haworth, S.; Domellöf, M.; West, C.E.; Johansson, I. Genetic Preference for Sweet Taste in Mothers Associates with Mother-Child Preference and Intake. Nutrients 2023, 15, 2565. https://doi.org/10.3390/nu15112565
Lif Holgerson P, Hasslöf P, Esberg A, Haworth S, Domellöf M, West CE, Johansson I. Genetic Preference for Sweet Taste in Mothers Associates with Mother-Child Preference and Intake. Nutrients. 2023; 15(11):2565. https://doi.org/10.3390/nu15112565
Chicago/Turabian StyleLif Holgerson, Pernilla, Pamela Hasslöf, Anders Esberg, Simon Haworth, Magnus Domellöf, Christina E. West, and Ingegerd Johansson. 2023. "Genetic Preference for Sweet Taste in Mothers Associates with Mother-Child Preference and Intake" Nutrients 15, no. 11: 2565. https://doi.org/10.3390/nu15112565
APA StyleLif Holgerson, P., Hasslöf, P., Esberg, A., Haworth, S., Domellöf, M., West, C. E., & Johansson, I. (2023). Genetic Preference for Sweet Taste in Mothers Associates with Mother-Child Preference and Intake. Nutrients, 15(11), 2565. https://doi.org/10.3390/nu15112565






