Abstract
Background: Advanced glycation end products (AGEs) are involved in age-related diseases, but the interaction of gut microbiota with dietary AGEs (dAGEs) and tissue AGEs in the population is unknown. Objective: Our objective was to investigate the association of dietary and tissue AGEs with gut microbiota in the population-based Rotterdam Study, using skin AGEs as a marker for tissue accumulation and stool microbiota as a surrogate for gut microbiota. Design: Dietary intake of three AGEs (dAGEs), namely carboxymethyl-lysine (CML), N-(5-hydro-5-methyl-4-imidazolon-2-yl)-ornithine (MGH1), and carboxyethyl-lysine (CEL), was quantified at baseline from food frequency questionnaires. Following up after a median of 5.7 years, skin AGEs were measured using skin autofluorescence (SAF), and stool microbiota samples were sequenced (16S rRNA) to measure microbial composition (including alpha-diversity, beta-dissimilarity, and taxonomic abundances) as well as predict microbial metabolic pathways. Associations of both dAGEs and SAF with microbial measures were investigated using multiple linear regression models in 1052 and 718 participants, respectively. Results: dAGEs and SAF were not associated with either the alpha-diversity or beta-dissimilarity of the stool microbiota. After multiple-testing correction, dAGEs were not associated with any of the 188 genera tested, but were nominally inversely associated with the abundance of Barnesiella, Colidextribacter, Oscillospiraceae UCG-005, and Terrisporobacter, in addition to being positively associated with Coprococcus, Dorea, and Blautia. A higher abundance of Lactobacillus was associated with a higher SAF, along with several nominally significantly associated genera. dAGEs and SAF were nominally associated with several microbial pathways, but none were statistically significant after multiple-testing correction. Conclusions: Our findings did not solidify a link between habitual dAGEs, skin AGEs, and overall stool microbiota composition. Nominally significant associations with several genera and functional pathways suggested a potential interaction between gut microbiota and AGE metabolism, but validation is required. Future studies are warranted, to investigate whether gut microbiota modifies the potential impact of dAGEs on health.
1. Introduction
Advanced glycation end products (AGEs) are a group of molecules formed nonenzymatically, after the initial attachment of reducing sugars to amino groups of proteins, lipids, or nucleic acids [1]. Long-lived tissues are more prone to AGEs accumulation, and sustain damage due to the formation of cross-links, the modification of proteins, or from inflammation due to their binding to the receptor for AGEs (RAGE) [2,3,4]. Evidences are gathering for their possible involvement in aging [5] and age-related diseases, such as cardiovascular diseases, dementia, and diabetes [6].
AGEs are also generated from food processing, especially under high temperatures and low moisture [7]. Dietary AGEs (dAGEs) were associated with elevated inflammation, as well as incidences of type 2 diabetes [8] and pancreatic cancer [9], but these associations are still inconclusive, and their underlying mechanisms remain unclear. Aside from entering the circulation, it was estimated that a large fraction (20–50%) of dAGEs are excreted in feces [10]. Thus, an interaction with the commensal microbiota in the intestinal tract is likely. Gut microbiota composition and metabolic activities can vary in response to dietary factors, which in turn may influence host health through their involvement in inflammation, metabolic processes, immunoregulation, etc. [11,12].
Exposure to dAGEs was reported to increase colon permeability in rats [13]. Their cross-linking structures can also hinder microbe mobility [14]. dAGEs may activate downstream inflammatory responses once recognized by RAGE, which is highly expressed in the intestinal tract [15]. They may also influence microbial and host metabolism, such as by reducing the abundance of microbes that produce health-provoking short-chain fatty acids (SCFAs) and induce insulin resistance [16]. In contrast, some dAGEs were substrates for SCFA production [17,18]. Potential interactions of dAGEs and the gut microbiota have been summarized in Supplementary Figure S1. An intervention study of one month’s dAGE restriction was reported to already alter the gut microbiota of patients undergoing peritoneal dialysis, as measured in stool [19]. However, the way in which AGEs interact with the human gut microbiota is still largely unexplored, and data from large population studies are lacking [20,21]. Gut microbial metabolism and fermentation in turn provide a large panel of metabolites to the host, including AGEs and their precursors [22]. Gut microbiota may also indirectly influence AGE levels by contributing to systemic inflammation and oxidative stress.
To sum up, whether dietary AGEs modulate the gut microbiota or whether activities of gut microbiota influence the AGE burden in vivo may be relevant for host health but remains largely unknown. To explore the intricate connections between dietary AGEs, gut microbiota, and host AGEs accumulation, we obtained data from the population-based Rotterdam Study on stool microbiota as a surrogate for gut microbiota, estimating dAGEs from dietary information, and using skin AGEs as a reflection of AGE accumulation in long-lived tissues [23]. We examined the associations of dietary and skin AGEs with the overall diversity, dissimilarity, and the taxonomic and functional abundance of the stool microbiota.
2. Subjects and Methods
2.1. Study Population
The Rotterdam Study is a general population-based cohort consisting of three subcohorts of inhabitants from the Rotterdam suburb Ommoord who were invited to participate. All the participants of our study were from the third subcohort of the Rotterdam Study (RS III), which started in 2006, and the majority of whom were of European ancestry. In total, N = 3932 participants (≥45 years old) were included at baseline (RS III 1st visit) with follow-up examinations every 4–6 years. The design and objectives of the Rotterdam Study have been extensively described previously [24].
The dietary intakes of three types of AGEs, namely carboxymethyl-lysine (CML), N-(5-hydro-5-methyl-4-imidazolon-2-yl)-ornithine (MGH1), and carboxyethyl-lysine (CEL), were estimated based on a food frequency questionnaire (FFQ) at baseline (RS III 1st visit) for 2676 participants. Skin AGEs were measured (n = 1167), and stool samples for microbiota analyses were collected (n = 1420) during RS III 2nd visit (the median follow-up time being 5.7 years later).
A total of 1120 participants had data taken on both dAGEs and stool microbiome, after excluding 47 participants with implausible energy intake (<500 kcal/d or >5000 kcal/d). Of this 1120, a further 27 participants were excluded because the stool samples had been in ambient temperature for more than 6 days, or for an unknown duration. Another 37 participants were excluded either because they had used antibiotics less than 1 month before stool sample collection, or because their antibiotic usage was unknown. An additional 3 participants (n = 3) with remaining outlying values in dietary AGEs (>±6 SD) were additionally excluded (Details in Supplementary Figure S2). For the stool microbiota and SAF analysis, 752 participants had both measurements available. From this group, we excluded 2 participants with outlying values of SAF (>±4 SD), and a further 32 participants based on the samples’ time in ambient temperature and their antibiotic usage, leaving 718 participants for analysis (Supplementary Figure S3).
2.2. Estimation of Dietary AGEs and Other Dietary Characteristics
When visiting the RS research center, participants received a 389-item food frequency questionnaire (FFQ). This FFQ collected detailed information, including food types, frequencies, portions, and some preparation methods of 389 food and beverage items covering the preceding month. The FFQ has been validated to properly rank subjects for nutrient intake in other studies among Dutch adults [25,26].
Briefly, dAGEs were calculated from the FFQ using reference contents of three types of AGEs (CML, MGH1, and CEL) in 190 food items from a Dutch database [27], as well as from another reference database on CML contents in 257 food items from Northern Ireland [28]. Both databases reported the content of protein-bound AGEs, determined using ultraperformance liquid chromatography-tandem mass-spectrometry (UPLC-MS/MS). Details of the method were described elsewhere [29]. We used energy-adjusted AGE intake for our analyses in order to reduce reporting bias related to measurement error [30]. Dietary energy intake was calculated using the Dutch Food Composition Table (NEVO). Overall diet quality was approximated in a diet quality score (with a range of 0–14), reflecting adherence to the Dutch Dietary Guidelines [31].
2.3. Measurement of Skin AGEs as SAF
Skin AGEs were measured as SAF, using AGE Reader™ (DiagnOptics B.V., Groningen, The Netherlands) on the inner skin of the dominant forearm at the Rotterdam Study research center during the RS III 2nd visit. This assessed the overall level of AGEs in the skin, based on the fluorescent property of some AGEs and their correlations with other AGEs. Details of the measurement have been described elsewhere [32].
2.4. Stool Microbiota Profiling and Processing
Fecal samples were collected at home by the participants and sent to Erasmus MC by mail. Upon arrival, samples were recorded and stored at −20 °C. DNA was isolated in accordance with the manufacturer’s protocol (Arrow Stool DNA; Isogen Life Science, Utrecht, The Netherlands). The V3 and V4 variable regions of the 16S rRNA gene were amplified using the 309F-806R primer pair and sequenced on an Illumina MiSeq sequencer (MiSeq Reagent Kit v3, 2 × 300 bp) with an average depth of 50,000 reads per sample [33].
Raw reads from the MiSeq were demultiplexed using a custom script to separate sample FASTQ files based on the dual index. Primers, barcodes, and heterogeneity spacers were trimmed off using tagcleaner v0.16 [34]. Trimmed FASTQ files were loaded into R v4.0.0 with the DADA2 [35] package (version 1.18.0). Quality filtering was performed in DADA2 using the following criteria: trim = 0, maxEE = c(2,2), truncQ = 2, rm.phix = TRUE. Filtered reads were run through the DADA2 Amplicon Sequence Variant (ASV) assignment tool to denoise, cluster, and merge the reads. ASVs were assigned a taxonomy from the SILVA version 138.1 rRNA database [36] using the RDP naïve Bayesian classifier [37]. The resulting data tables were combined into a phyloseq object using phyloseq R package (version 1.38.0) [38] and a phylogenetic tree was generated using the phangorn R package (version 2.6.2) [39] based on the sequences of the ASVs, and then added to the phyloseq object.
ASVs with a read count of less than 0.05% of the total reads, as well as those observed in less than 1% of the total samples, were precluded from the analysis. Technical covariates included the season of stool production, number of total reads, time in the mail (assessing the days of the samples’ being in an ambient temperature), and batch (marking each run of DNA isolation and DNA sequencing).
2.5. Assessment of Covariates and Population Characteristics
Covariates were assessed at the RS III 2nd visit. Information on smoking, alcohol intake, physical activity, medications used during the past week, and medical history was collected during home interviews. Smoking status was categorized as never, past, or current smoker, based on cigarette, cigar, and pipe smoking information. Alcohol intake was assessed in glasses/day and harmonized to grams of alcohol per day. The use of antibiotics was provided by the participants when stool samples were collected and included no use of antibiotics, used in less than one month, 1–3 months, or 3 months–1 year before sample collection. Height and weight were measured at the research center and BMI (kg/m2) was calculated as weight divided by height squared. Fasting blood samples were obtained, and HDL cholesterol, total cholesterol, triglycerides, creatinine, and glucose were measured using routine techniques. LDL cholesterol (mmol/L) was calculated with the Friedewald formula. The estimated glomerular filtration rate (eGFR) was calculated using the CKD-EPI equation. Diabetes was defined as a fasting glucose of ≥7.0 mmol/L, or a nonfasting glucose of ≥11.0 mmol/L and/or the use of glucose-lowering medications, and was verified with medical records.
2.6. Statistical Analysis
All statistical analyses were performed in R Studio (Version 1.4.1717, R Version 4.1.0). The number of independent tests was calculated based on the correlation matrix between outcomes of interest using the method described by Li and Ji [40]. Bonferroni correction was used to maintain a low false positive rate in the presence of multiple testing. Results which had a p-value less than 0.05 but did not pass the p-value threshold after correction were regarded as nominally significant.
2.6.1. Characteristics of Microbial Composition
Three indexes were used to describe the overall alpha diversity of the stool microbiota, namely the Shannon Index, the Inverse Simpson Index, and the number of ASVs observed. The calculation of the former two indexes was carried out using the diversity function in vegan (R package) based on the abundance of all ASVs after initial quality control and filtering. Centered log-ratio (CLR) transformation of the abundance of the individual taxon was performed using the transform function in the Microbiome R package. Zeros in taxon abundance were replaced with a pseudo count of min (nonzero abundance)/2 before taking logs. Beta dissimilarity, which measures the between-participants dissimilarities of stool microbiota composition, was calculated as Aitchison distance, i.e., Euclidean distance, based on the CLR transformed abundance of all ASVs, using the vegdist function from the vegan package [41].
2.6.2. Microbial Pathway Prediction
We used the PICRUSt2 (v.2.5.0) tool to obtain predicted microbial functions based on the MetaCyc dataset and the ASV table [42,43]. In short, the prediction was performed using the default EPA-NG polygenetic replacement option and MinPath biological pathway reconstruction, based on Enzyme Commission (EC) numbers. ASVs with less than 0.05% of the total read count, or which were present in less than 1% of the samples, were precluded.
2.6.3. dAGEs and Microbiota Analyses
Descriptives of the lifestyle, clinical, dietary, and stool microbiota characteristics of the participants are presented in tertile groups of low, medium, and high CML intake. Results on CML were presented in the main text since it has been extensively studied in the literature, and the estimation in diet was more accurate based on reference values from more food items than it was for the other two types of AGEs.
- (1)
- dAGEs and alpha diversity
Associations between dAGEs and alpha diversity indexes were analyzed in multiple linear regression models, adjusting for factors that would potentially influence the level of AGEs in the diet or gut microbiota composition. Two linear regression models were constructed where the Shannon Index, Inverse Simpson Index, or the number of observed ASVs were the outcomes, respectively: model 1 was adjusted for age, sex, seasons of stool production, number of total reads, time in the mail, and batches of DNA isolation and sequencing. Model 2 was further adjusted for covariates that were reported to influence stool microbiota substantially or were associated with dAGEs, including the use of PPI and antibiotics, alcohol consumption, BMI, diabetes, diet quality score, energy intake, and smoking status.
- (2)
- dAGEs and beta dissimilarity
To visualize sample distances, we extracted the first two principal components (PC) that explained the most variation of stool microbiota composition based on CLR transformed ASV abundance. The samples were plotted in dAGE tertile groups with a two-dimensional ordination plot based on the first two PCs. We tested for differences in microbiota beta dissimilarity among dAGEs tertile groups using permutational multivariate analysis of variance (PERMANOVA) with 999 times permutations using the adonis2 function in vegan package. All covariates and categorical dAGEs were entered into PERMANOVA models sequentially.
- (3)
- dAGEs and abundance of individual genera
After aggregating all the ASVs to the genus level, the analysis was confined to 188 genera. We studied the association using linear regression, accounting for confounders in models described previously. To achieve a better model fit and reduce the chance of false positive discoveries due to the compositional nature of the microbiome data, the dependent variable (i.e., abundance of each genus) was CLR transformed. Heteroscedasticity and model diagnostics were inspected by plotting the linear regression residuals against the predicted outcome. All results in this study were derived from model 2 for genera presented in at least 30% of all participants, unless specified otherwise.
- (4)
- dAGEs and predicted microbial pathways
For further insight into the link between dAGEs and microbial metabolic potential, we studied the association between dAGEs and the putative microbial metabolic pathways in the linear regression models. adjusting for the covariates described previously. The abundances of predicted metabolic pathways were CLR transformed and used as the outcome in our models (360 pathways were analyzed).
2.6.4. Stool Microbiota and SAF
Sex-specific, age-adjusted SAF was obtained as the residuals of SAF regressed against age in women and men, separately. Participants were grouped into tertiles of sex-specific, age-adjusted SAF to obtain descriptive statistics, which then underwent beta dissimilarity testing after adjusting for other covariates. We examined the relationship between stool microbiota and SAF through the same serial analyses as those of the dAGEs study, but now with stool microbiota traits as exposures and SAF as the outcome (except for analysis on beta dissimilarity by SAF groups). Confounders were adjusted in two models: Model 1 was adjusted for age, sex, seasons of stool production, number of total reads, time in the mail, and DNA isolation and sequencing batches; Model 2 was additionally adjusted for the use of PPI and antibiotics, alcohol consumption, BMI, diabetes, eGFR, and smoking status. For all analyses, model 2 was considered to be the main model.
2.6.5. Sensitivity Analysis
We repeated the dAGEs analyses after further excluding dAGEs outside of the mean ± 4 SD range (n = 14) so as to exclude results driven by them, although the potential impact from dietary components is more likely to be observed with exceptional intake. We also repeated all the analyses after further excluding participants who had used antibiotics within 3 months prior to stool sample collection (n = 67). Only the overlapping results for genera presented in at least 30% of the population were discussed further, as we are more certain that these results were more robust to influences from outlying dAGE values, antibiotics, and the sparse presence of microbes in the population.
2.6.6. Imputation of Missing Values
Missing values in the use of antibiotics, smoking, alcohol, eGFR, and BMI were imputed through multiple imputation (m = 10) in R (MICE package, version 3.14.0).
3. Results
3.1. dAGEs and Stool Microbiota
3.1.1. Descriptive statistics
For the dAGEs and microbiome study (n = 1052), clinical and lifestyle characteristics of the total study population are shown in Table 1. The mean (SD) age of the participants was 62.5 (5.5) years, with 59% being female. Mean (SD) daily intake was 2.5 (0.9) mg for CML, 29.5 (8.1) mg for MGH1, and 2.5 (0.9) mg for CEL after energy adjustment. Diabetes mellitus was present in 107 (10%) participants (mostly type 2). Shannon Index, Inverse Simpson Index, and the number of observed ASVs were higher in the low CML group compared to the medium and high CML groups.

Table 1.
Characteristics of all participants included for dAGEs and stool microbiome analyses, presented in tertile groups of CML intake.
3.1.2. dAGEs and Overall Diversity and Dissimilarity of the Stool Microbiota
dAGEs were not associated with all three measures of microbial alpha diversity (see Supplementary Table S1). In PCA analysis, the first two PCs accounted for 4.5% and 2.5% of the total variance of the microbiota composition, respectively. We did not observe an obvious separation of individual microbiota for the dAGE tertile groups in the ordination plot based on the first two PCs (Supplementary Figure S4). No significant difference was observed in beta dissimilarity among dAGEs tertile groups in PERMANOVA (Supplementary Table S2). The results persisted after restricting the analysis to participants (n = 971) who had dAGEs within the mean ±4 SD range and had not used antibiotics for at least three months prior to sample collection.
3.1.3. dAGEs and Microbial Abundance
The three dAGEs showed 31 nominally significant associations (p < 0.05) with 188 genera, but no association passed the statistical significance threshold after Bonferroni correction (p < 0.0002 with 300 independent tests) (Supplementary Tables S3 and S4). Further exclusion of participants in sensitivity analyses did not change the results substantially (Supplementary Table S5). The associations of dAGEs with genera that were present in ≥30% of participants are summarized in Figure 1. Of them, a higher dAGE intake was associated with a lower abundance of genus Barnesiella of the Barnesiellaceae family, Colidextribacter and UCG-005 of the Oscillospiraceae family, and Terrisporobacter of the Peptostreptococcaceae family (and GCA_900066575 of the Lachnospiraceae family with a p < 0.1 in the sensitivity analysis), and a higher abundance of genera Coprococcus, Dorea, and Blautia of the Lachnospiraceae family, in both the main and sensitivity analyses.
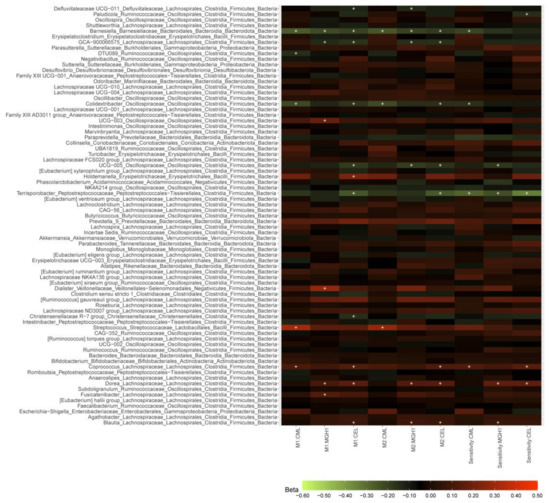
Figure 1.
dAGEs-microbial taxa associations in the total population and after further excluding individuals in sensitivity analyses. M1, M2: results from model 1 and model 2 in the total study population (N = 1052). Sensitivity: sensitivity analysis results from model 2 after further excluding participants who had used antibiotics in less than three months before stool sample collection and whose dAGEs intake exceeding the mean ± 4 SD range (N = 971). Associations between dAGEs and the abundance of 188 genera were derived from linear regression model 2 in 1052 and 973 participants, respectively. The color of the boxes represents the direction of the association, with red being positive and green being negative and color lightness implies the magnitude of beta coefficients obtained from linear regression analyses. Beta coefficients are adjusted differences of the centered log-ratio transformed genera abundance associated with one SD difference of dAGEs. “+” denotes p < 0.05. The associations were adjusted for age, sex, season of stool production, number of total reads, batches of DNA isolation and sequencing, and time in the mail in model 1, and in model 2 further for the use of PPI, the use of antibiotics, diabetes, BMI, diet quality score, energy intake, alcohol intake, and smoking status. Source data in Supplementary Tables S3–S5.
3.1.4. dAGEs and Microbial Pathways
We tested the associations of three types of dAGEs with 360 predicted microbial metabolic pathways. A total of 49 nominally significant associations were observed (p < 0.05) but none of them reached statistical significance (p < 0.0003) after Bonferroni correction (number of independent tests: 148). Among the nominally significantly associated pathways that presented in at least 30% of the samples, inversely associated pathways included the superpathway of N-acetylneuraminate degradation, pyruvate fermentation to acetone, acetyl-CoA fermentation to butanoate II, the superpathway of UDP-N-acetylglucosamine-derived O-antigen building block biosynthesis, biotin biosynthesis II, L-glutamate degradation V (via hydroxyglutarate), adenosylcobalamin biosynthesis I (early cobalt insertion), the superpathway of polyamine biosynthesis II, and L-lysine fermentation to acetate and butanoate. Positively associated pathways included inosine-5′-phosphate biosynthesis III, L-lysine biosynthesis II, the superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate), peptidoglycan biosynthesis II (staphylococci), mevalonate pathway I, and the superpathway of 2,3-butanediol biosynthesis. (Details in Supplementary Table S6).
3.2. Stool Microbiota and SAF
Characteristics of all the participants included in the SAF study and arranged in sex-specific, age-adjusted SAF tertile groups are shown in Supplementary Table S7. The low SAF group showed slightly higher alpha diversity (Supplementary Table S8), but no association was observed in multiple linear regression models (Supplementary Tables S9 and S10). In the PCA analysis, the first two PCs accounted for 4.6% and 2.5% of the total variance of the microbiota, respectively. We also did not observe an apparent separation of the SAF tertile groups based on the first two PCs (Supplementary Figure S5). No significant difference in Aitchison distance was observed among sex-specific, age-adjusted SAF tertiles in PERMANOVA after adjusting for covariates in model 2 (Details in Supplementary Table S11).
Regarding individual taxa, a higher abundance of genus Lactobacillus from the Lactobacillaceae family was associated with a higher SAF after Bonferroni correction (p-value cut-off: p < 0.0007) but microbes of this genus only presented in a small fraction (23%) of samples (Supplementary Table S10). Among genera that presented in more than 30% of the studied population, SAF was inversely associated with the abundance of genera [Eubacterium] ventriosum group, Anaerostipes, Roseburia, Lachnospiraceae NK4A136 group, [Eubacterium] eligens group, and Lachnospiraceae UCG-001 from the Lachnospiraceae family, Oscillospiraceae UCG-005 from the Oscillospiraceae family, and genus Sutterella from the Sutterellaceae family. SAF was nominally positively associated with the abundance of the genus Negativibacillus from the Ruminococcaceae family.
We observed three pathways, present in at least 30% of the participants, that were nominally (p < 0.05) associated with SAF: positive associations with the superpathway of L-aspartate and L-asparagine biosynthesis, pyrimidine deoxyribonucleosides salvage, and L-histidine degradation I. However, none of them were statistically significant after Bonferroni correction (p < 0.0007, based on 74 independent tests) (Supplementary Table S12).
4. Discussion
We investigated dietary and skin AGEs in relation to gut microbiota composition in a general population cohort using stool microbiota as a surrogate. dAGEs were not associated with microbial alpha diversity or beta dissimilarity. We observed nominally significant associations between dAGEs and genera abundance, but none of them survived multiple testing correction. Despite a higher alpha diversity being observed in the lowest SAF tertile, neither alpha diversity nor beta dissimilarity was associated with SAF, but the abundance of the genus Lactobacillus was positively associated with SAF.
Limited studies were available on dAGEs and microbiota concerning intervention in humans and animals and in vitro fermentation [13,16,19,44,45,46,47,48], and no general population-based data were available. Glycated products were often suggested to reduce the alpha diversity and abundance of SCFA producing microbes in stool microbiota [49], but some studies reported elevated SCFA production and indicated potential health benefits [46,50,51]. One mice study showed a high dAGEs diet led to inflammation and altered gut microbiota composition and this was reversed following low dAGEs intake [52]. However, our study did not show any apparent dissimilarity of stool microbiota in the dAGE groups, paralleling a 4-week dAGEs intervention study in obese individuals that did not observe apparent different microbial composition [53]. This could be explained by the fact that our study population may be rather homogenous in dietary AGEs, while the stool microbiota varies drastically with factors such as age, diet, and comorbidities. Further, we studied the habitual diet that the gut microbiota might have adapted to [54].
For the analysis of dAGEs and genera abundance, with a sample size of 1052, we only have 50.8% power to demonstrate the largest observed difference (beta = 0.28, alpha = 0.003), suggesting our study is underpowered to detect any smaller differences after Bonferroni correction. Despite that, a higher level of dAGEs was nominally associated with a lower abundance of Barnesiella, Colidextribacter, Oscillospiraceae UCG-005, Terrisporobacter, and Lachnospiraceae GCA_900066575, and a higher abundance of Coprococcus, Dorea, and Blautia. The differentially abundant genera found in our study differ from those found in other studies, which could be partially attributed to differences in study duration, amount of AGEs, possible human-specific adaptations of the microbiota to thermally processed food [55], and absence of interaction with host physiology in in vitro studies.
Disbiome, an online dataset documenting published associations between microbes and diseases, gives hints of potential health relevance [56]. A reduced Barnesiella was seen in chronic kidney disease, autism, inflammatory bowel disease, and obesity, among others. A reduced Terrisporobacter abundance was seen in obesity, autism, and type 2 diabetes. Lachnospiraceae GCA_900066575 was inversely associated with BMI [57]. A higher Dorea abundance was reported in atrial fibrillation, autism, irritable bowel syndrome, and non-alcoholic fatty liver disease. A higher Blautia was seen in atrial fibrillation, major depressive disorder, and type 1 diabetes. A higher abundance of Coprococcus was also associated with diets rich in sugars but also linked to health-promoting traits, such as the maintenance of microbial homeostasis. In addition, Coprococcus and Blautia are potential SCFA producers [58,59]. It is worth noting that both higher and lower abundances of some genera were seen in multiple diseases, suggesting that the patterns of disturbance in the entire microbiota might be more informative than variations of a single genus.
We also examined dAGEs in association with microbial pathways, which are thought to be more relevant to certain health risks than compositional variation alone, such as type 1 diabetes in infants [60]. Our results point to the hypothesis that exposing the gut microbiota to a higher dAGE intake may (1) result in lower SCFAs, which are substrates for the positively associated pathways, such as the “superpathway of 2,3-butanediol biosynthesis”, as well as products for the inversely associated pathways such as “N-acetylneuraminate degradation”, “acetyl-CoA fermentation to butanoate II”, and “L-lysine fermentation to acetate and butanoate”; (2) modulate cholesterol levels through the mevalonate pathway I; and (3) modulate the redox status by influencing the pathways that affect the NAD+/NADH ratio and L-glutamate. However, further research is necessary as none of these associations remained statistically significant after multiple testing correction.
As for skin AGEs, both alpha diversity and beta dissimilarity were not associated with SAF. It is worth noting that some covariates also act as intermediates in the association (e.g., diabetes), which could have led to overadjustment. We did not observe a consistent pattern of associations for phylogenetically related taxa; thus, it seems unlikely that tissue AGEs originate from a group of related microbes. This is supported by the lack of association between microbial functions and SAF, although we also lack the statistical power to detect these associations. These results imply that overall gut microbial diversity may not be a major contributor to long-term AGE accumulation in distant tissues, such as the skin.
On the genus level, the abundance of Lactobacillus (present in 23% of participants) was associated with a higher SAF. Although Lactobacillus is present in fermented foods and is considered a probiotic, a higher abundance in stool microbiota was also seen in obesity and type 2 diabetes, two conditions that increase AGEs. Among all the nominally significant associations observed, a lower abundance of potential SCFA producers [61], Eubacterium ventriosum group, Anaerostipes, Roseburia, Lachnospiraceae NK4A136 group, Lachnospiraceae UCG-001, and Oscillospiraceae UCG-005, was nominally associated with a higher SAF. Reduced abundance of the inversely associated genera in the stool microbiota was also observed in multiple diseases, such as: autism, colorectal cancer, Crohn’s disease for Eubacterium ventriosum group; inflammatory bowel disease and diabetes for Anaerostipes; chronic kidney disease, hypertension, Parkinson’s disease for Roseburia; dementia and obesity for Lachnospiraceae NK4A136; and diabetes and primary biliary cholangitis for Sutterella. The abundance of the associated genera could further be influenced by diet. For instance, [Eubacterium] eligens group, which was nominally inversely associated with SAF, was also inversely associated with the intake of AGE-forming fructose [62]. Negativibacillus, which was nominally positively associated with SAF, was also associated with ultra-processed foods [63]. Our findings imply that gut microbes may influence SCFAs levels and host health, or participate in the metabolism of AGE-related compounds in the diet and further affect tissue AGEs.
The strengths of this study include a relatively large sample size and a deeply phenotyped population that allows for the control of many potential confounders. We also focused on three representative protein-bound AGEs from the habitual diet. They are more likely to enter the gut microbiota habitat than free AGEs. The dAGEs estimation was based on a detailed 389-item FFQ. Additionally, SAF can reflect tissue AGE accumulation over the long term.
Limitations included that our findings need to be replicated, to reduce the possibility of chance findings. Moreover, our results should be interpreted in light of limited power, imperfect quantification of dAGEs, and potential residual confounding, especially from the diet. The results were restricted to the elderly Dutch population. We also acknowledge limitations inherent to microbiota analysis, including that linear regression might not be the best method to analyze compositional microbiome data.
To gain further insight into the interaction of AGEs and gut microbiota, larger sample sizes are needed to increase the statistical power. Dietary intervention studies and longitudinal studies combined with microbial functional and metabolic data are also warranted if we are to understand the causality of any associations observed. Studies in earlier life stages are needed to understand the health relevance across the lifespan. The link of AGEs with intestinal pathophysiology should also be elucidated, as a recent mice study revealed a critical role of CML in brain aging, and a microbiota-dependent increase in intestinal permeability was key to this process [64]. Studies may also explore the role of the gut microbiome in modifying the relationship between dAGEs and health outcomes. For instance, some microbes such as Collinsella intestinalis can degrade dAGEs to innocuous products [65]. This may help explain the inconsistent observations on dAGEs and health risks.
In conclusion, our human population study showed only nominally significant associations between dAGEs and stool microbiota, unlike intervention studies in animals. However, this does not mean that reducing dAGEs will not be beneficial as we assessed the cross-sectional associations and prospective intervention studies are needed to confirm the observations. A higher abundance of Lactobacillus was associated with a higher skin AGEs. We await other independent studies to verify this finding. Further research is also required to investigate the causality and health relevance of the associations we and others observed between AGEs and the stool microbiota.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nu15112567/s1, Figure S1: Summary of existing literature information on the mechanisms; Figure S2: Population inclusion and exclusion flow diagram for dAGEs and stool microbiome analyses; Figure S3: Population inclusion and exclusion flow diagram for stool microbiome and SAF analyses; Figure S4: Ordination plot of microbiota beta dissimilarity among dAGE tertile groups; Figure S5: Ordination plot of microbiota beta dissimilarity among SAF tertile groups; Table S1: Associations between dietary AGEs intake and measures of alpha diversity in the total population; Table S2: Summary of PERMANOVA of beta dissimilarity by dAGEs tertile groups; Tables S3–S5: dAGEs-taxa associations in the total population; Table S6: dAGEs-pathway associations in the total population; Table S7: Characteristics of participants included for stool microbiome and skin autofluorescence analyses and by tertile groups of skin autofluorescence; Table S8: The association between measures of alpha diversity and SAF; Tables S9 and S10: The associations between abundance of taxonomical units and SAF in the total population; Table S11: Summary of PERMANOVA of beta dissimilarity by SAF tertile groups; Table S12: The associations between microbial metabolic (MetaCyc) pathways and SAF in the total population.
Author Contributions
Conceptualization, J.C., D.R., R.K. and M.C.Z.; Methodology, J.C., D.R., C.M.-G. and M.C.Z.; Software, J.C., D.R.; Validation, D.R.; Formal analysis, J.C.; Investigation, J.C., D.R., R.K. and M.C.Z.; Resources, C.M.-G., T.V., J.B.J.v.M., M.A.I., A.G.U., R.K. and M.C.Z.; Data curation, C.M.-G., J.B.J.v.M., R.K. and M.C.Z.; Writing—original draft, J.C.; Writing—review & editing, J.C., D.R., C.M.-G., T.V., J.B.J.v.M., M.A.I., A.G.U., R.K. and M.C.Z.; Visualization, J.C.; Supervision, A.G.U., R.K. and M.C.Z.; Project administration, R.K. and M.C.Z.; Funding acquisition, A.G.U., R.K. and M.C.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The Rotterdam Study is supported by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Netherlands Genomics Initiative, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. Djawad Radjabzadeh was funded by an Erasmus MC mRACE grant “Profiling of the human gut microbiome”. Jinluan Chen was supported by the China Scholarship Council for PhD fellowship (No. 201606170110).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (Medical Ethics Committee) of Erasmus Medical Center and by the review board of The Netherlands Ministry of Health, Welfare and Sports.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the management team of the Rotterdam Study (secretariat.epi@erasmusmc.nl). The data are not publicly available due to restrictions based on privacy regulations and informed consent of the participants.
Acknowledgments
The generation and management of the 16S microbiome data for The Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, University Medical Center Rotterdam, the Netherlands. We thank Nahid El Faquir and Jolande Verkroost-Van Heemst for their help in sample collection and registration and Kamal Arabe, Hedayat Razawy, Karan Singh Asra, Pelle van der Wal, Sergio Chavez, and Djawad Radjabzadeh for their help in DNA isolation and sequencing, and Joost Verlouw, Constanza Vallerga, and Marijn Verkerk for their help with the bioinformatic analyses. We thank Ruolin Li, Cindy Boer, Robert Kraaij, Carolina Medina-Gomez, and Joyce van Meurs for overseeing the quality control of the generated datasets. In addition, Ruolin Li generated the data on predicted MetaCyc pathways. The contribution of the inhabitants, general practitioners, and pharmacists of the Ommoord district to the Rotterdam Study is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Henle, T. Protein-bound advanced glycation endproducts (AGEs) as bioactive amino acid derivatives in foods. Amino Acids. Dec. 2005, 29, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Verzijl, N.; DeGroot, J.; Thorpe, S.R.; Bank, R.A.; Shaw, J.N.; Lyons, T.J.; Bijlsma, J.W.; Lafeber, F.P.; Baynes, J.W.; TeKoppele, J.M. Effect of collagen turnover on the accumulation of advanced glycation end products. J. Biol. Chem. 2000, 275, 39027–39031. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Barden, A.; Mori, T.; Beilin, L. Advanced glycation end-products: A review. Diabetologia 2001, 44, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Peng, Y.; Shen, Y.; Zhang, Y.; Liu, L.; Yang, X. Dietary polyphenols: Regulate the advanced glycation end products-RAGE axis and the microbiota-gut-brain axis to prevent neurodegenerative diseases. Crit. Rev. Food Sci. Nutr. 2022, 1–27. [Google Scholar] [CrossRef]
- Semba, R.D.; Nicklett, E.J.; Ferrucci, L. Does Accumulation of Advanced Glycation End Products Contribute to the Aging Phenotype? J. Gerontol. A Biol. 2010, 65, 963–975. [Google Scholar] [CrossRef]
- Twarda-Clapa, A.; Olczak, A.; Bialkowska, A.M.; Koziolkiewicz, M. Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs. Cells 2022, 11, 1312. [Google Scholar] [CrossRef]
- Birlouez-Aragon, I.; Saavedra, G.; Tessier, F.J.; Galinier, A.; Ait-Ameur, L.; Lacoste, F.; Niamba, C.N.; Alt, N.; Somoza, V.; Lecerf, J.M. A diet based on high-heat-treated foods promotes risk factors for diabetes mellitus and cardiovascular diseases. Am. J. Clin. Nutr. 2010, 91, 1220–1226. [Google Scholar] [CrossRef]
- Luevano-Contreras, C.; Gomez-Ojeda, A.; Macias-Cervantes, M.H.; Garay-Sevilla, M.E. Dietary Advanced Glycation End Products and Cardiometabolic Risk. Curr. Diabetes Rep. 2017, 17, 63. [Google Scholar] [CrossRef]
- Jiao, L.; Stolzenberg-Solomon, R.; Zimmerman, T.P.; Duan, Z.; Chen, L.; Kahle, L.; Risch, A.; Subar, A.F.; Cross, A.J.; Hollenbeck, A.; et al. Dietary consumption of advanced glycation end products and pancreatic cancer in the prospective NIH-AARP Diet and Health Study. Am. J. Clin. Nutr. 2015, 101, 126–134. [Google Scholar] [CrossRef]
- Delgado-Andrade, C.; Tessier, F.J.; Niquet-Leridon, C.; Seiquer, I.; Pilar Navarro, M. Study of the urinary and faecal excretion of Nepsilon-carboxymethyllysine in young human volunteers. Amino Acids 2012, 43, 595–602. [Google Scholar] [CrossRef]
- Vojinovic, D.; Radjabzadeh, D.; Kurilshikov, A.; Amin, N.; Wijmenga, C.; Franke, L.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat. Commun. 2019, 10, 5813. [Google Scholar] [CrossRef] [PubMed]
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Yuan, X.; Zhao, J.; Zhang, Y.; Hu, J.; Wang, J.; Li, J. Dietary advanced glycation end products modify gut microbial composition and partially increase colon permeability in rats. Mol. Nutr. Food Res. 2017, 61, 1700118. [Google Scholar] [CrossRef]
- Zhao, D.; Le, T.T.; Larsen, L.B.; Li, L.; Qin, D.; Su, G.; Li, B. Effect of glycation derived from alpha-dicarbonyl compounds on the in vitro digestibility of beta-casein and beta-lactoglobulin: A model study with glyoxal, methylglyoxal and butanedione. Food Res. Int. 2017, 102, 313–322. [Google Scholar] [CrossRef]
- Tadie, J.M.; Bae, H.B.; Banerjee, S.; Zmijewski, J.W.; Abraham, E. Differential activation of RAGE by HMGB1 modulates neutrophil-associated NADPH oxidase activity and bacterial killing. Am. J. Physiol. -Cell. Physiol. 2012, 302, C249–C256. [Google Scholar] [CrossRef]
- Wang, J.; Cai, W.; Yu, J.; Liu, H.; He, S.; Zhu, L.; Xu, J. Dietary Advanced Glycation End Products Shift the Gut Microbiota Composition and Induce Insulin Resistance in Mice. Diabetes Metab. Syndr. Obes. 2022, 15, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.P.N.; Ritari, J.; Boeren, S.; de Waard, P.; Plugge, C.M.; de Vos, W.M. Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat. Commun. 2015, 6, 10062. [Google Scholar] [CrossRef]
- Hellwig, M.; Auerbach, C.; Müller, N.; Samuel, P.; Kammann, S.; Beer, F.; Gunzer, F.; Henle, T. Metabolization of the Advanced Glycation End Product N-epsilon-Carboxymethyllysine (CML) by Different Probiotic E. coli Strains. J. Agric. Food Chem. 2019, 67, 1963–1972. [Google Scholar] [CrossRef]
- Yacoub, R.; Nugent, M.; Cai, W.; Nadkarni, G.N.; Chaves, L.D.; Abyad, S.; Honan, A.M.; Thomas, S.A.; Zheng, W.; Valiyaparambil, S.A.; et al. Advanced glycation end products dietary restriction effects on bacterial gut microbiota in peritoneal dialysis patients; a randomized open label controlled trial. PLoS ONE 2017, 12, e0184789. [Google Scholar] [CrossRef]
- Nesreen, A.L.; Carbonero, F. Impact of Maillard reaction products on nutrition and health: Current knowledge and need to understand their fate in the human digestive system. Crit. Rev. Food Sci. Nutr. 2019, 59, 474–487. [Google Scholar]
- Phuong-Nguyen, K.; McNeill, B.A.; Aston-Mourney, K.; Rivera, L.R. Advanced Glycation End-Products and Their Effects on Gut Health. Nutrients 2023, 15, 405. [Google Scholar] [CrossRef]
- Cohen-Or, I.; Katz, C.; Ron, E.Z. AGEs Secreted by Bacteria Are Involved in the Inflammatory Response. PLoS ONE 2011, 6, e17974. [Google Scholar] [CrossRef]
- Rajaobelina, K.; Cougnard-Gregoire, A.; Delcourt, C.; Gin, H.; Barberger-Gateau, P.; Rigalleau, V. Autofluorescence of Skin Advanced Glycation End Products: Marker of Metabolic Memory in Elderly Population. J. Gerontol. A Biol. 2015, 70, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Ikram, M.A.; Brusselle, G.; Ghanbari, M.; Goedegebure, A.; Ikram, M.K.; Kavousi, M.; Kieboom, B.C.; Klaver, C.C.; de Knegt, R.J.; Luik, A.I.; et al. Objectives, design and main findings until 2020 from the Rotterdam Study. Eur. J. Epidemiol. 2020, 35, 483–517. [Google Scholar] [CrossRef] [PubMed]
- Feunekes, G.I.J.; Vanstaveren, W.A.; Devries, J.H.M.; Burema, J.; Hautvast, J.G.A.J. Relative and Biomarker-Based Validity of a Food-Frequency Questionnaire Estimating Intake of Fats and Cholesterol. Am. J. Clin. Nutr. 1993, 58, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Goldbohm, R.A.; Vandenbrandt, P.A.; Brants, H.A.; van’t Veer, P.; Al, M.; Sturmans, F.; Hermus, R.J. Validation of a Dietary Questionnaire Used in a Large-Scale Prospective Cohort Study on Diet and Cancer. Eur. J. Clin. Nutr. 1994, 48, 253–265. [Google Scholar] [PubMed]
- Scheijen, J.L.; Clevers, E.; Engelen, L.; Dagnelie, P.C.; Brouns, F.; Stehouwer, C.D.; Schalkwijk, C.G. Analysis of advanced glycation endproducts in selected food items by ultra-performance liquid chromatography tandem mass spectrometry: Presentation of a dietary AGE database. Food Chem. 2016, 190, 1145–1150. [Google Scholar] [CrossRef]
- Hull, G.L.J.; Woodside, J.V.; Ames, J.M.; Cuskelly, G.J. N-epsilon-(carboxymethyl)lysine content of foods commonly consumed in a Western style diet. Food Chem. 2012, 131, 170–174. [Google Scholar] [CrossRef]
- Chen, J.; Waqas, K.; Tan, R.C.; Voortman, T.; Ikram, M.A.; Nijsten, T.E.; De Groot, L.C.; Uitterlinden, A.G.; Zillikens, M.C. The association between dietary and skin advanced glycation end products: The Rotterdam Study. Am. J. Clin. Nutr. 2020, 112, 129–137. [Google Scholar] [CrossRef]
- Willett, W.C.; Howe, G.R.; Kushi, L.H. Adjustment for total energy intake in epidemiologic studies. Am. J. Clin. Nutr. 1997, 65 (Suppl. 4), 1220S–1228S, discussion 1229S–1231S. [Google Scholar] [CrossRef]
- Voortman, T.; Kiefte-de Jong, J.C.; Ikram, M.A.; Stricker, B.H.; van Rooij, F.J.; Lahousse, L.; Tiemeier, H.; Brusselle, G.G.; Franco, O.H.; Schoufour, J.D. Adherence to the 2015 Dutch dietary guidelines and risk of non-communicable diseases and mortality in the Rotterdam Study. Eur. J. Epidemiol. 2017, 32, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; van der Duin, D.; Campos-Obando, N.; Ikram, M.A.; Nijsten, T.E.; Uitterlinden, A.G.; Zillikens, M.C. Serum 25-hydroxyvitamin D-3 is associated with advanced glycation end products (AGEs) measured as skin autofluorescence: The Rotterdam Study. Eur. J. Epidemiol. 2019, 34, 67–77. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Boer, C.G.; Beth, S.A.; van der Wal, P.; Kiefte-De Jong, J.C.; Jansen, M.A.; Konstantinov, S.R.; Peppelenbosch, M.P.; Hays, J.P.; Jaddoe, V.W.; et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci. Rep. 2020, 10, 1040. [Google Scholar] [CrossRef]
- Schmieder, R.; Lim, Y.W.; Rohwer, F.; Edwards, R. TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinform. 2010, 11, 341. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Li, J.; Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity Sep. 2005, 95, 221–227. [Google Scholar] [CrossRef]
- Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 23 May 2023).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Seiquer, I.; Rubio, L.A.; Peinado, M.J.; Delgado-Andrade, C.; Navarro, M.P. Maillard reaction products modulate gut microbiota composition in adolescents. Mol. Nutr. Food Res. 2014, 58, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Mills, D.J.; Tuohy, K.M.; Booth, J.; Buck, M.; Crabbe, M.J.; Gibson, G.R.; Ames, J.M. Dietary glycated protein modulates the colonic microbiota towards a more detrimental composition in ulcerative colitis patients and non-ulcerative colitis subjects. J. Appl. Microbiol. 2008, 105, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Yao, Y.; Dong, S.; Jin, S.; Xiao, H.; Wu, H.; Zeng, M. Chemical characterization of the glycated myofibrillar proteins from grass carp (Ctenopharyngodon idella) and their impacts on the human gut microbiota in vitro fermentation. Food Funct. 2017, 8, 1184–1194. [Google Scholar] [CrossRef]
- Zhao, Q.; Ou, J.; Huang, C.; Qiu, R.; Wang, Y.; Liu, F.; Zheng, J.; Ou, S. Absorption of 1-Dicysteinethioacetal-5-Hydroxymethylfurfural in Rats and Its Effect on Oxidative Stress and Gut Microbiota. J. Agr. Food Chem. 2018, 66, 11451–11458. [Google Scholar] [CrossRef]
- Qu, W.; Nie, C.; Zhao, J.; Ou, X.; Zhang, Y.; Yang, S.; Bai, X.; Wang, Y.; Wang, J.; Li, J. Microbiome-Metabolomics Analysis of the Impacts of Long-Term Dietary Advanced-Glycation-End-Product Consumption on C57BL/6 Mouse Fecal Microbiota and Metabolites. J. Agr. Food Chem. 2018, 66, 8864–8875. [Google Scholar] [CrossRef]
- Dong, L.; Li, Y.; Chen, Q.; Liu, Y.; Qiao, Z.; Sang, S.; Zhang, J.; Zhan, S.; Wu, Z.; Liu, L. Research advances of advanced glycation end products in milk and dairy products: Formation, determination, control strategy and immunometabolism via gut microbiota. Food Chem. 2023, 417, 135861. [Google Scholar] [CrossRef]
- Delgado-Andrade, C.; Pastoriza de la Cueva, S.; Peinado, M.J.; Rufian-Henares, J.A.; Navarro, M.P.; Rubio, L.A. Modifications in bacterial groups and short chain fatty acid production in the gut of healthy adult rats after long-term consumption of dietary Maillard reaction products. Food Res. Int. 2017, 100 Pt 1, 134–142. [Google Scholar] [CrossRef]
- Swiatecka, D.; Narbad, A.; Ridgway, K.P.; Kostyra, H. The study on the impact of glycated pea proteins on human intestinal bacteria. Int. J. Food Microbiol. 2011, 145, 267–272. [Google Scholar]
- van Dongen, K.C.; Linkens, A.M.; Wetzels, S.M.; Wouters, K.; Vanmierlo, T.; van de Waarenburg, M.P.; Scheijen, J.L.; de Vos, W.M.; Belzer, C.; Schalkwijk, C.G. Dietary advanced glycation endproducts (AGEs) increase their concentration in plasma and tissues, result in inflammation and modulate gut microbial composition in mice; evidence for reversibility. Food Res. Int. 2021, 147, 110547. [Google Scholar] [CrossRef] [PubMed]
- Linkens, A.M.; van Best, N.; Niessen, P.M.; Wijckmans, N.E.; de Goei, E.E.; Scheijen, J.L.; van Dongen, M.C.; van Gool, C.C.; de Vos, W.M.; Houben, A.J.; et al. A 4-Week Diet Low or High in Advanced Glycation Endproducts Has Limited Impact on Gut Microbial Composition in Abdominally Obese Individuals: The deAGEing Trial. Int. J. Mol. Sci. 2022, 23, 5328. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.P.N.; Troise, A.D.; Fogliano, V.; de Vos, W.M. Anaerobic Degradation of N-epsilon-Carboxymethyllysine, a Major Glycation End-Product, by Human Intestinal Bacteria. J. Agric. Food Chem. 2019, 67, 6594–6602. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D. Thermal processing of food reduces gut microbiota diversity of the host and triggers adaptation of the microbiota: Evidence from two vertebrates. Microbiome 2018, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Janssens, Y.; Nielandt, J.; Bronselaer, A.; Debunne, N.; Verbeke, F.; Wynendaele, E.; Van Immerseel, F.; Vandewynckel, Y.P.; De Tré, G.; De Spiegeleer, B. Disbiome database: Linking the microbiome to disease. BMC Microbiol. 2018, 18, 50. [Google Scholar] [CrossRef] [PubMed]
- Bailén, M.; Bressa, C.; Martínez-López, S.; González-Soltero, R.; Montalvo Lominchar, M.G.; San Juan, C.; Larrosa, M. Microbiota Features Associated With a High-Fat/Low-Fiber Diet in Healthy Adults. Front. Nutr. 2020, 7, 583608. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef]
- Chinda, D.; Nakaji, S.; Fukuda, S.; Sakamoto, J.; Shimoyama, T.; Nakamura, T.; Fujisawa, T.; Terada, A.; Sugawara, K. The fermentation of different dietary fibers is associated with fecal clostridia levels in men. J. Nutr. 2004, 134, 1881–1886. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell. Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Jones, R.B.; Alderete, T.L.; Kim, J.S.; Millstein, J.; Gilliland, F.D.; Goran, M.I. High intake of dietary fructose in overweight/obese teenagers associated with depletion of Eubacterium and Streptococcus in gut microbiome. Gut Microbes 2019, 10, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Atzeni, A.; Martínez, M.Á.; Babio, N.; Konstanti, P.; Tinahones, F.J.; Vioque, J.; Corella, D.; Fitó, M.; Vidal, J.; Moreno-Indias, I.; et al. Association between ultra-processed food consumption and gut microbiota in senior subjects with overweight/obesity and metabolic syndrome. Original Research. Front. Nutr. 2022, 9, 976547. [Google Scholar] [CrossRef] [PubMed]
- Mossad, O.; Batut, B.; Yilmaz, B.; Dokalis, N.; Mezö, C.; Nent, E.; Nabavi, L.S.; Mayer, M.; Maron, F.J.M.; Buescher, J.M.; et al. Gut microbiota drives age-related oxidative stress and mitochondrial damage in microglia via the metabolite N6-carboxymethyllysine. Nat. Neurosci. 2022, 25, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.R.; Wesener, D.A.; Cheng, J.; Houston-Ludlam, A.N.; Beller, Z.W.; Hibberd, M.C.; Giannone, R.J.; Peters, S.L.; Hettich, R.L.; Leyn, S.A.; et al. Bioremediation of a Common Product of Food Processing by a Human Gut Bacterium. Cell. Host Microbe 2019, 26, 463–477.e8. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).