Development of a Structural Equation Model to Examine the Relationships between Genetic Polymorphisms and Cardiovascular Risk Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample
2.2. DNA Extraction and Genotyping
2.3. Blood Pressure Measurements
2.4. Biochemical Measurements
2.5. Development of Latent Variables
2.6. Data Analysis and Validation (Structural Equation Modeling)
3. Results
Goodness-of-Fit of the Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amini, M.; Zayeri, F.; Salehi, M. Trend analysis of cardiovascular disease mortality, incidence, and mortality-to-incidence ratio: Results from global burden of disease study 2017. BMC Public Health 2021, 21, 401. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Todd, C.V.; Nisha, H. Calculating Risk vs. Detecting Disease. JACC Cardiovasc. Imaging 2022, 15, 1619–1621. [Google Scholar] [CrossRef]
- Ralph, B.D.A.; Ramachandran, S.V.; Michael, J.P.; Philip, A.W.; Mark, C.; Joseph, M.M.; William, B.K. General Cardiovascular Risk Profile for Use in Primary Care. Circulation 2008, 117, 743–753. [Google Scholar] [CrossRef]
- Nishimura, K.; Okamura, T.; Watanabe, M.; Nakai, M.; Takegami, M.; Higashiyama, A.; Kokubo, Y.; Okayama, A.; Miyamoto, Y. Predicting Coronary Heart Disease Using Risk Factor Categories for a Japanese Urban Population, and Comparison with the Framingham Risk Score: The Suita Study. J. Atheroscler. Thromb. 2014, 21, 784–798. [Google Scholar] [CrossRef] [PubMed]
- Vrablik, M.; Dlouha, D.; Todorovova, V.; Stefler, D.; Hubacek, J.A. Genetics of Cardiovascular Disease: How Far Are We from Personalized CVD Risk Prediction and Management? Int. J. Mol. Sci. 2021, 22, 4182. [Google Scholar] [CrossRef] [PubMed]
- Hongmei, Y.; Yongping, J.; Jiyuan, L. Interleukin-6 polymorphisms and risk of coronary artery diseases in a Chinese population: A case-control study. Pak. J. Med. Sci. 2016, 32, 880–8855. [Google Scholar] [CrossRef]
- Schunkert, H.; König, I.R.; Kathiresan, S.; Reilly, M.P.; Assimes, T.L.; Holm, H.; Preuss, M.; Stewart, A.F.; Barbalic, M.; Gieger, C.; et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011, 43, 333–338. [Google Scholar] [CrossRef]
- Kathiresan, S.; Srivastava, D. Genetics of human cardiovascular disease. Cell 2012, 148, 1242–1257. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Hoffman-Andrews, L. The known unknown: The challenges of genetic variants of uncertain significance in clinical practice. J. Law Biosci. 2018, 4, 648–657. [Google Scholar] [CrossRef]
- Jackson, M.; Marks, L.; May, G.H.W.; Wilson, J.B. The genetic basis of disease. Essays Biochem. 2018, 62, 643–723. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chen, J.; Shirkey, G.; John, R.; Wu, S.R.; Park, H.; Shao, C. Applications of structural equation modeling (SEM) in ecological studies: An updated review. Ecol. Process. 2016, 5, 19. [Google Scholar] [CrossRef]
- Zsido, A.N.; Arato, N.; Lang, A.; Labadi, B.; Stecina, D.; Bandi, S.A. The connection and background mechanisms of social fears and problematic social networking site use: A structural equation modeling analysis. Psychiatry Res. 2020, 292, 113323. [Google Scholar] [CrossRef] [PubMed]
- Moen, G.-H.; Nivard, M.; Bhatta, L.; Warrington, N.M.; Willer, C.; Åsvold, B.O.; Brumpton, B.; Evans, D.M. Using Genomic Structural Equation Modeling to Partition the Genetic Covariance Between Birthweight and Cardiometabolic Risk Factors into Maternal and Offspring Components in the Norwegian HUNT Study. Behav. Genet. 2022, 53, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Nock, N.L.; Wang, X.; Thompson, C.L.; Song, Y.; Baechle, D.; Raska, P.; Stein, C.M.; Gray-McGuire, C. Defining genetic determinants of the Metabolic Syndrome in the Framingham Heart Study using association and structural equation modeling methods. BMC Proc. 2009, 3, S50. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Shin, J.-Y.; Yee, J.; Park, T.; Park, M. Structural equation modeling for hypertension and type 2 diabetes based on multiple SNPs and multiple phenotypes. PLoS ONE 2019, 14, e0217189. [Google Scholar] [CrossRef]
- Adedia, D.; Adebanji, A.O.; Appiah, S.K. Comparative Analysis of Some Structural Equation Model Estimation Methods with Application to Coronary Heart Disease Risk. J. Probab. Stat. 2020, 2020, 4181426. [Google Scholar] [CrossRef]
- Keates, A.K.; Mocumbi, A.O.; Ntsekhe, M.; Sliwa, K.; Stewart, S. Cardiovascular disease in Africa: Epidemiological profile and challenges. Nat. Rev. Cardiol. 2017, 14, 273–293. [Google Scholar] [CrossRef]
- Chalwe, J.M.; Mukherjee, U.; Grobler, C.; Mbambara, S.H.; Oldewage-Theron, W. Association between hypertension, obesity and dietary intake in post-menopausal women from rural Zambian communities. Health SA Gesondheid 2021, 26, a1496. [Google Scholar] [CrossRef]
- Bansal, S.K.; Yadav, R. A Study of the Extended Lipid Profile including Oxidized LDL, Small Dense LDL, Lipoprotein (a) and Apolipoproteins in the Assessment of Cardiovascular Risk in Hypothyroid Patients. J. Clin. Diagn. Res. JCDR 2016, 10, BC04–BC08. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-K.; Han, K.; Kim, M.K.; Koh, E.S.; Kim, E.S.; Nam, G.E.; Kwon, H.-S. Changes in metabolic syndrome and its components and the risk of type 2 diabetes: A nationwide cohort study. Sci. Rep. 2020, 10, 2313. [Google Scholar] [CrossRef]
- Anderson, D.; Patarapichayatham, C.; Nese, J.F.T. Basic Concepts of Structural Equation Modeling; University of Oregon: Eugene, OR, USA, 2012; pp. 1–36. [Google Scholar]
- Rosseel, Y. Lavaan: An R Package for Structural Equation Modeling. J. Stat. Softw. 2012, 48, 1–36. [Google Scholar] [CrossRef]
- Bentler, P.; Bonett, D. Significance Tests and Goodness-of-Fit in Analysis of Covariance Structures. Psychol. Bull. 1980, 88, 588–606. [Google Scholar] [CrossRef]
- Hu, L.T.; Bentler, P.M. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Struct. Equ. Model. A Multidiscip. J. 1999, 6, 1–55. [Google Scholar] [CrossRef]
- Mulaik, S.; James, L.; Alstine, J.; Bennett, N.; Lind, S.; Stilwell, C. Evaluation of Goodness-of-Fit Indices for Structural Equation Models. Psychol. Bull. 1989, 105, 430–445. [Google Scholar] [CrossRef]
- Browne, M.W.; Cudeck, R. Alternative Ways of Assessing Model Fit. Sociol. Methods Res. 1992, 21, 230–258. [Google Scholar] [CrossRef]
- Chen, F.; Curran, P.J.; Bollen, K.A.; Kirby, J.; Paxton, P. An Empirical Evaluation of the Use of Fixed Cutoff Points in RMSEA Test Statistic in Structural Equation Models. Sociol. Methods Res. 2008, 36, 462–494. [Google Scholar] [CrossRef]
- MacCallum, R.C.; Hong, S. Power Analysis in Covariance Structure Modeling Using GFI and AGFI. Multivar. Behav. Res. 1997, 32, 193–210. [Google Scholar] [CrossRef]
- Sharma, S.; Mukherjee, S.; Kumar, A.; Dillon, W.R. A simulation study to investigate the use of cutoff values for assessing model fit in covariance structure models. J. Bus. Res. 2005, 58, 935–943. [Google Scholar] [CrossRef]
- Bentler, P.M. Comparative fit indexes in structural models. Psychol. Bull. 1990, 107, 238. [Google Scholar] [CrossRef] [PubMed]
- Aryadoust, V.; Raquel, M. Quantitative Data Analysis for Language Assessment Volume II—Advanced Methods, 1st ed.; Routledge: London, UK, 2019; Volume 2, p. 260. [Google Scholar] [CrossRef]
- Little, T.D.; Card, N.A. Longitudinal Structural Equation Modeling; Guilford Publications: New York, NY, USA, 2013. [Google Scholar]
- Cook, K.F.; Kallen, M.A.; Amtmann, D. Having a fit: Impact of number of items and distribution of data on traditional criteria for assessing IRT’s unidimensionality assumption. Qual. Life Res. Int. J. Qual. Life Asp. Treat. Care Rehabil. 2009, 18, 447–460. [Google Scholar] [CrossRef]
- Kline, R.B. Principles and Practice of Structural Equation Modeling, 4th ed.; Guilford Publications: New York, NY, USA, 2015. [Google Scholar]
- Rasool, S.U.A.; Nabi, M.; Ashraf, S.; Amin, S. Insulin Receptor Substrate 1 Gly972Arg (rs1801278) Polymorphism Is Associated with Obesity and Insulin Resistance in Kashmiri Women with Polycystic Ovary Syndrome. Genes 2022, 13, 1463. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Deng, Y.; Xie, L.; Yu, L.; Liu, L.; Liu, H.; Dai, L. Genetic polymorphisms in MTR are associated with non-syndromic congenital heart disease from a family-based case-control study in the Chinese population. Sci. Rep. 2019, 9, 5065. [Google Scholar] [CrossRef]
- Iwanicka, J.; Iwanicki, T.; Niemiec, P.; Balcerzyk, A.; Krauze, J.; Górczyńska-Kosiorz, S.; Ochalska-Tyka, A.; Grzeszczak, W.; Żak, I. Relationship between CETP gene polymorphisms with coronary artery disease in Polish population. Mol. Biol. Rep. 2018, 45, 1929–1935. [Google Scholar] [CrossRef]
- Chikowore, T.; Sahibdeen, V.; Hendry, L.M.; Norris, S.A.; Goedecke, J.H.; Micklesfield, L.K.; Lombard, Z. C679X loss-of-function PCSK9 variant is associated with lower fasting glucose in black South African adolescents: Birth to Twenty Plus Cohort. J. Clin. Transl. Endocrinol. 2019, 16, 100186. [Google Scholar] [CrossRef] [PubMed]
- Rhee, E.-J.; Kim, H.C.; Kim, J.H.; Lee, E.Y.; Kim, B.J.; Kim, E.M.; Song, Y.; Lim, J.H.; Kim, H.J.; Choi, S.; et al. 2018 Guidelines for the management of dyslipidemia. Korean J. Intern. Med. 2019, 34, 723–771. [Google Scholar] [CrossRef]
- Dave, J.A.; Levitt, N.S.; Ross, I.L.; Lacerda, M.; Maartens, G.; Blom, D. Anti-Retroviral Therapy Increases the Prevalence of Dyslipidemia in South African HIV-Infected Patients. PLoS ONE 2016, 11, e0151911. [Google Scholar] [CrossRef]
- Ntusi, N. Dyslipidaemia in South Africa. SAMJ S. Afr. Med. J. 2018, 108, 256–257. [Google Scholar] [CrossRef]
- Reiger, S.; Jardim, T.V.; Abrahams-Gessel, S.; Crowther, N.J.; Wade, A.; Gomez-Olive, F.X.; Salomon, J.; Tollman, S.; Gaziano, T.A. Awareness, treatment, and control of dyslipidemia in rural South Africa: The HAALSI (Health and Aging in Africa: A Longitudinal Study of an INDEPTH Community in South Africa) study. PLoS ONE 2017, 12, e0187347. [Google Scholar] [CrossRef]
- Swarup, S.; Goyal, A.; Grigorova, Y.; Zeltser, R. Metabolic Syndrome; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Kruger, M.J.; Nell, T.A. The prevalence of the metabolic syndrome in a farm worker community in the Boland district, South Africa. BMC Public Health 2017, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Owolabi, E.O.; Goon, D.T.; Adeniyi, O.V.; Adedokun, A.O.; Seekoe, E. Prevalence and Correlates of Metabolic Syndrome Among Adults Attending Healthcare Facilities in Eastern Cape, South Africa. Open Public Health J. 2017, 10, 148–159. [Google Scholar] [CrossRef]
- Peer, N.; Lombard, C.; Steyn, K.; Levitt, N. High prevalence of metabolic syndrome in the Black population of Cape Town: The Cardiovascular Risk in Black South Africans (CRIBSA) study. Eur. J. Prev. Cardiol. 2015, 22, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Merriman, T.R.; Wilcox, P.L. Cardio-metabolic disease genetic risk factors among Māori and Pacific Island people in Aotearoa New Zealand: Current state of knowledge and future directions. Ann. Hum. Biol. 2018, 45, 202–214. [Google Scholar] [CrossRef]
- Baghbani-Arani, F.; Kavian Telori, M.; Asadi, J.; Samadian, E.; Shirkavand, A. Association of Two Single-Nucleotide Polymorphisms (rs1805087 and rs1801131) with Coronary Artery Disease in Golestan Population. Ann. Mil. Health Sci. Res. 2017, 15, e11473. [Google Scholar] [CrossRef]
- Paththinige, C.S.; Sirisena, N.D.; Dissanayake, V.H.W. Genetic determinants of inherited susceptibility to hypercholesterolemia—A comprehensive literature review. Lipids Health Dis. 2016, 16, 103. [Google Scholar] [CrossRef]
- Abudureyimu, S.; Abulaiti, P.; Xing, Z.; Li, H.; Liu, S.; Li, W.; Gao, Y. The Effect of Four Different Single Nucleotide Polymorphisms on Coronary Heart Disease in a Han Chinese Population in Xinjiang Region. J. Cardiol. Cardiovasc. Med. 2021, 5, 1–7. [Google Scholar] [CrossRef]
- Perez-Martinez, P.; Perez-Jimenez, F.; Ordovas, J.M.; Bellido, C.; Moreno, J.A.; Gomez, P.; Marin, C.; Fernandez de la Puebla, R.A.; Paniagua, J.A.; Lopez-Miranda, J. The APOB −516C/T polymorphism has no effect on lipid and apolipoprotein response following changes in dietary fat intake in a healthy population. Nutr. Metab. Cardiovasc. Dis. 2007, 17, 224–229. [Google Scholar] [CrossRef]
- Daly, A.K.; Cholerton, S.; Gregory, W.; Idle, J.R. Metabolic Polymorphisms. Pharmacol. Ther. 1993, 57, 129–160. [Google Scholar] [CrossRef]
- de Sousa Parreira, J.; Kallaur, A.P.; Lehmann, M.F.; Oliveira, S.R.; Frizon, D.A.; Delongui, F.; de Araujo, M.C.M.; Rossato, C.; de Almeida, J.T.; Pelegrino, L.M.; et al. Tumor necrosis factor beta NcoI polymorphism (rs909253) is associated with inflammatory and metabolic markers in acute ischemic stroke. Metab. Brain Dis. 2015, 30, 159–167. [Google Scholar] [CrossRef]
- Kedderis, G.L.; McQueen, C.A. 1.07—Biotransformation of Toxicants. In Comprehensive Toxicology, 2nd ed.; Elsevier: Oxford, UK, 2010; pp. 137–151. [Google Scholar] [CrossRef]
- Lancheros, L.E.P.; Ramírez, C.P.; Martín, A.S.; Navas, J.M.G.; Martínez, F.M.; Tortosa, M.d.C.R.; Morales, A.J. Impact of Genetic Polymorphisms on the Metabolic Pathway of Vitamin D and Survival in Non-Small Cell Lung Cancer. Nutrients 2021, 13, 3783. [Google Scholar] [CrossRef]
- Ríos-González, B.E.; Ibarra-Cortés, B.; Ramírez-López, G.; Sánchez-Corona, J.; Magaña-Torres, M.T. Association of Polymorphisms of Genes Involved in Lipid Metabolism with Blood Pressure and Lipid Values in Mexican Hypertensive Individuals. Dis. Markers 2014, 2014, 150358. [Google Scholar] [CrossRef] [PubMed]
- Vardarlı, A.T.; Harman, E.; Çetintaş, V.B.; Kayıkçıoğlu, M.; Vardarlı, E.; Zengi, A.; Küçükaslan, A.Ş.; Eroğlu, Z. Polymorphisms of lipid metabolism enzyme-coding genes in patients with diabetic dyslipidemia. Anatol. J. Cardiol. 2017, 17, 313–321. [Google Scholar] [CrossRef]
- Chuluun-Erdene, A.; Sengeragchaa, O.; Altangerel, T.-A.; Sanjmyatav, P.; Dagdan, B.; Battulga, S.; Enkhbat, L.; Byambasuren, N.; Malchinkhuu, M.; Janlav, M. Association of Candidate Gene Polymorphism with Metabolic Syndrome among Mongolian Subjects: A Case-Control Study. Med. Sci. 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Foucan, L.; Vélayoudom-Céphise, F.-L.; Larifla, L.; Armand, C.; Deloumeaux, J.; Fagour, C.; Plumasseau, J.; Portlis, M.-L.; Liu, L.; Bonnet, F.; et al. Polymorphisms in GC and NADSYN1 Genes are associated with vitamin D status and metabolic profile in Non-diabetic adults. BMC Endocr. Disord. 2013, 13, 36. [Google Scholar] [CrossRef]
- Vasan, R. Biomarkers of Cardiovascular Disease: Molecular Basis and Practical Considerations. Circulation 2006, 113, 2335–2362. [Google Scholar] [CrossRef]
- Brennan, L.; de Roos, B. Nutrigenomics: Lessons learned and future perspectives. Am. J. Clin. Nutr. 2021, 113, 503–516. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Fan, X.; Thompson, B.; Wang, L. Effects of sample size, estimation methods, and model specification on structural equation modeling fit indexes. Struct. Equ. Model. A Multidiscip. J. 1999, 6, 56–83. [Google Scholar] [CrossRef]
- Fritz, M.S.; Mackinnon, D.P. Required sample size to detect the mediated effect. Psychol. Sci. 2007, 18, 233–239. [Google Scholar] [CrossRef]
- Iacobucci, D. Structural equations modeling: Fit Indices, sample size, and advanced topics. J. Consum. Psychol. 2010, 20, 90–98. [Google Scholar] [CrossRef]
- Muthén, L.K.; Muthén, B.O. How to Use a Monte Carlo Study to Decide on Sample Size and Determine Power. Struct. Equ. Model. A Multidiscip. J. 2002, 9, 599–620. [Google Scholar] [CrossRef]
- Hoyle, R.H. Structural Equation Modeling: Concepts, Issues, and Applications; SAGE Publications: Thousand Oaks, CA, USA, 1995. [Google Scholar]
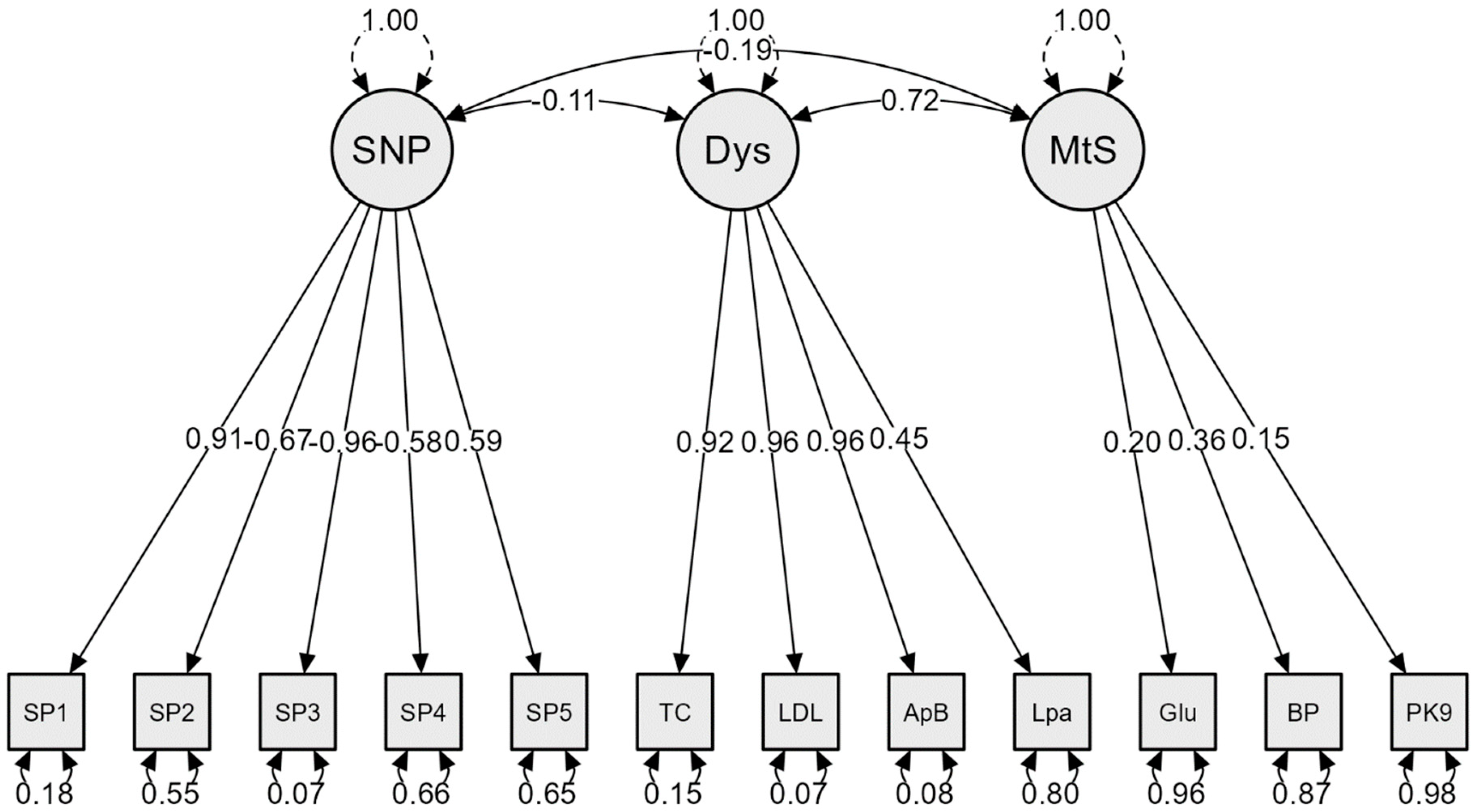
| Latent Variable | Indicator | Factor Loading | p-Value (Significance) |
|---|---|---|---|
| SNPs | SP1 | 0.91 | <0.001 |
| SP2 | −0.67 | <0.001 | |
| SP3 | −0.96 | <0.001 | |
| SP4 | −0.58 | <0.001 | |
| SP5 | 0.59 | <0.001 | |
| Dyslipidemia | TC | 0.92 | <0.001 |
| LDL-C | 0.96 | <0.001 | |
| ApoB | 0.96 | <0.001 | |
| Lipo (a) | 0.45 | 0.001 | |
| Metabolic syndrome | Glucose | 0.20 | 0.673 |
| BP | 0.36 | 0.645 | |
| PCSK9 | 0.15 | 0.576 |
| Pathway | Association | p-Value (Significance) |
|---|---|---|
| SNPs ↔ Dys | −0.114 | 0.440 |
| SNPs ↔ MetS | −0.194 | 0.612 |
| Dys ↔ MetS | 0.719 | 0.619 |
| Model | x2 | df | RMSEA | RMSEA 90% CI | SRMR | CFI | TLI |
|---|---|---|---|---|---|---|---|
| SEM | 118.483 | 51 | 0.164 | 0.126–0.203 | 0.087 | 0.825 | 0.773 |
| (p ≤ 0.001) | Marginal | Acceptable | Good | Moderate/OK | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalwe, J.M.; Grobler, C.; Oldewage-Theron, W. Development of a Structural Equation Model to Examine the Relationships between Genetic Polymorphisms and Cardiovascular Risk Factors. Nutrients 2023, 15, 2470. https://doi.org/10.3390/nu15112470
Chalwe JM, Grobler C, Oldewage-Theron W. Development of a Structural Equation Model to Examine the Relationships between Genetic Polymorphisms and Cardiovascular Risk Factors. Nutrients. 2023; 15(11):2470. https://doi.org/10.3390/nu15112470
Chicago/Turabian StyleChalwe, Joseph Musonda, Christa Grobler, and Wilna Oldewage-Theron. 2023. "Development of a Structural Equation Model to Examine the Relationships between Genetic Polymorphisms and Cardiovascular Risk Factors" Nutrients 15, no. 11: 2470. https://doi.org/10.3390/nu15112470
APA StyleChalwe, J. M., Grobler, C., & Oldewage-Theron, W. (2023). Development of a Structural Equation Model to Examine the Relationships between Genetic Polymorphisms and Cardiovascular Risk Factors. Nutrients, 15(11), 2470. https://doi.org/10.3390/nu15112470







