Structural Characteristic and In-Vitro Anticancer Activities of Dandelion Leaf Polysaccharides from Pressurized Hot Water Extraction
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction and Purification
2.3. Chemical Composition Analysis
2.4. Molecular Weight Analysis
2.5. Monosaccharide Composition Analysis
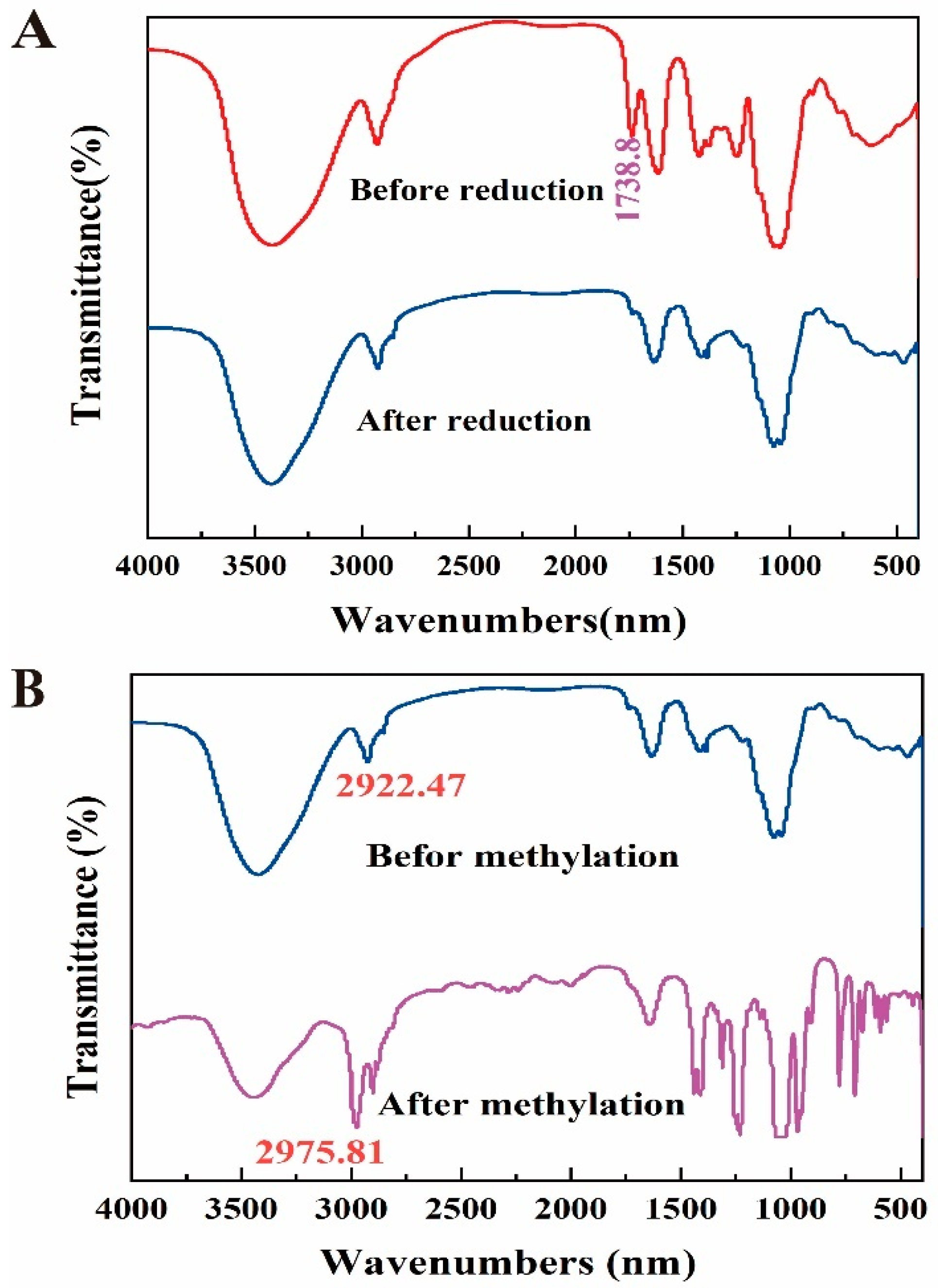
2.6. Fourier Transform Infrared Spectroscopy (FT-IR) Analysis
2.7. Reduction of Uronic Acids
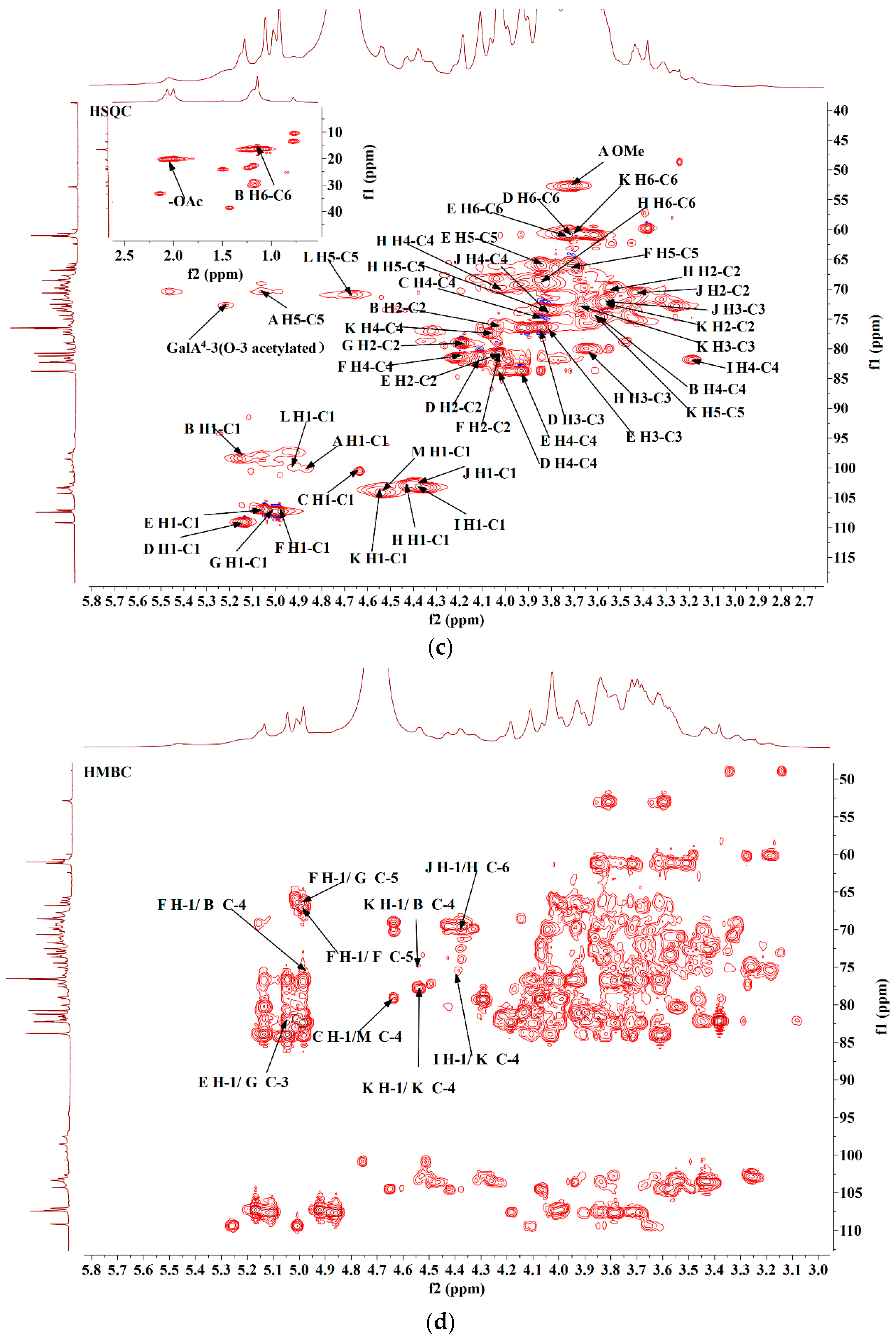
2.8. Methylation Analysis
2.9. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.10. Anticancer Activity In Vitro
2.10.1. Cell Culture
2.10.2. Cell Viability and Proliferation
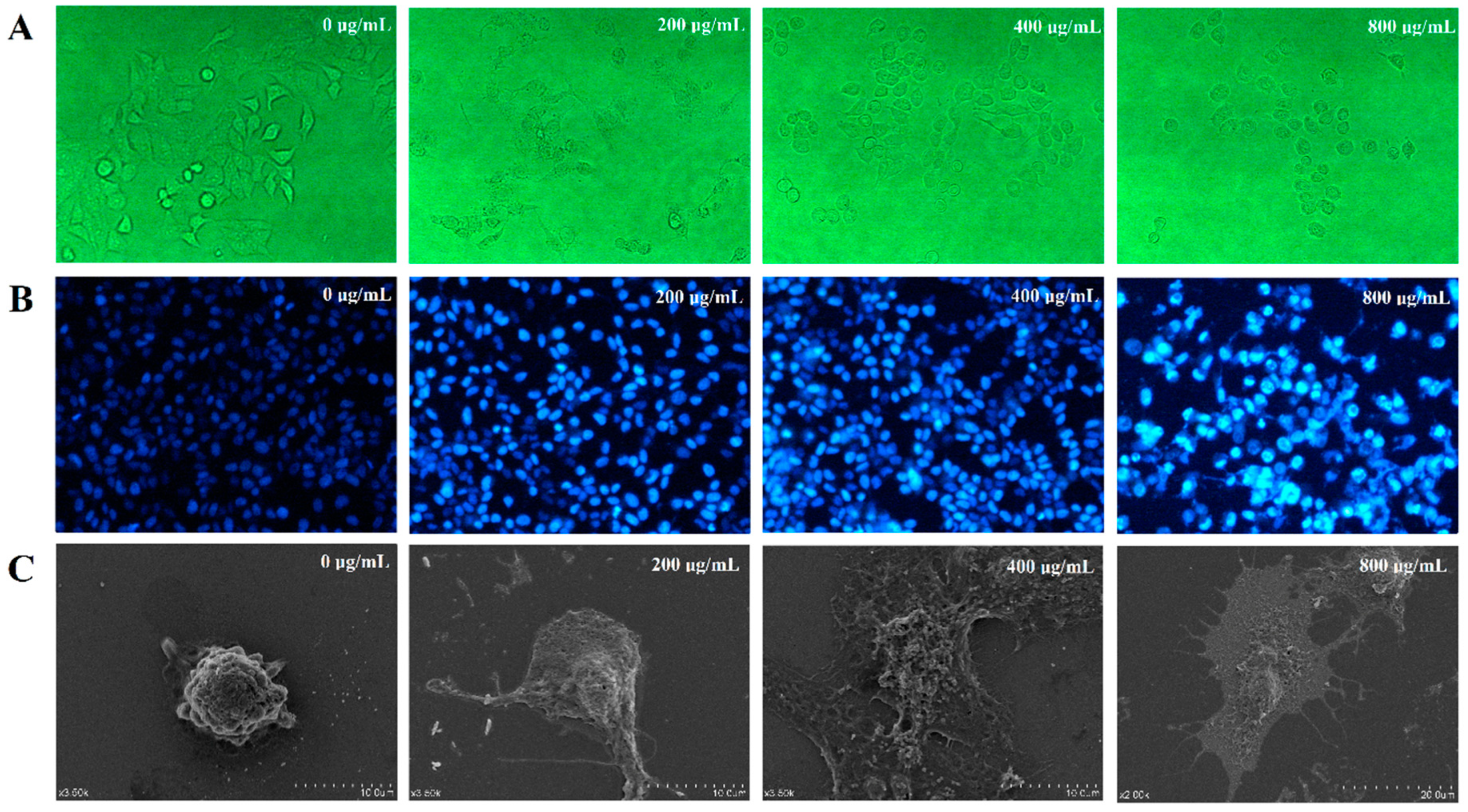
2.10.3. Observation of Cell Morphology by Scanning Electron Microscope (SEM)
2.10.4. Apoptosis Assay
2.10.5. Cell Cycle Assay
2.10.6. Annexin V-FITC Assay
2.11. Statistical Analysis
3. Results
3.1. Chemical Compositions
3.2. Monosaccharide Composition
3.3. FT-IR Analysis
3.4. Methylation Analysis
3.5. NMR Analysis
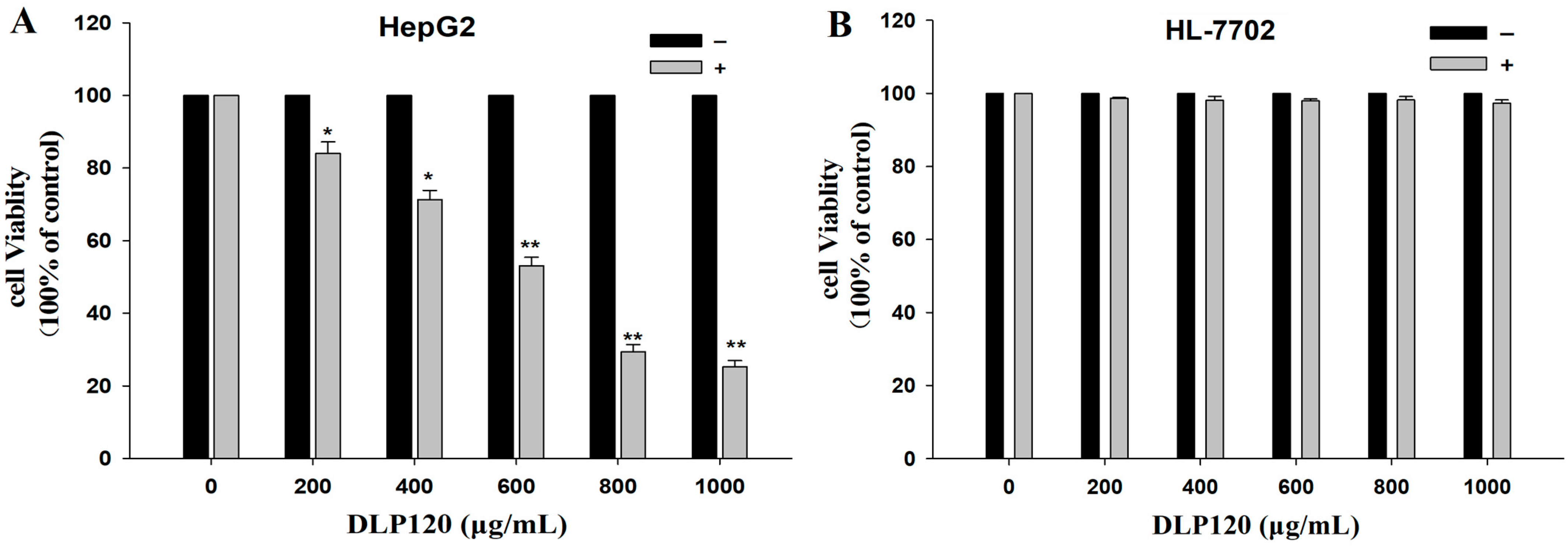
3.6. Anticancer Activity In Vitro
3.6.1. Effect of DLP120 on HepG2 Cell Proliferation
3.6.2. Effect of DLP120 on HepG2 Cell Morphology
3.6.3. Effect of DLP120 on HepG2 Cell Cycle Distribution
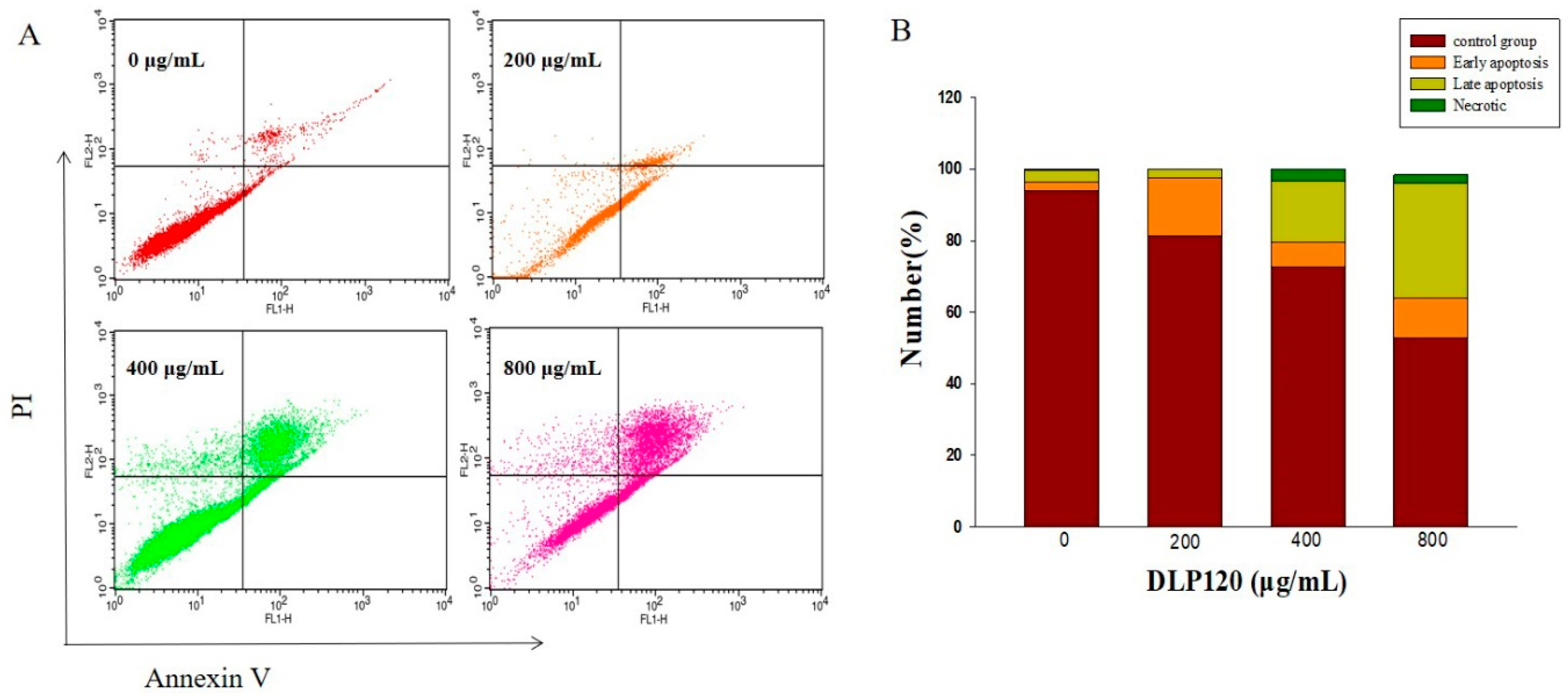
3.6.4. Effect of DLP120 on HepG2 Cell Apoptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Giovannucci, E.; Chan, A.T. Role of Vitamin and Mineral Supplementation and Aspirin Use in Cancer Survivors. J. Clin. Oncol. 2010, 28, 4081–4085. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-N.; Deng, G.; Mao, J.J. Practical Application of “About Herbs” Website Herbs and Dietary Supplement Use in Oncology Settings. Cancer J. 2019, 25, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, M.H.; Malik, A.; Haider, G.; Akhtar, M.S.; Gilani, A.H. Herbs for Health: An alternative approach to identify newer therapeutic option for the treatment of diabetes. Biochem. Pharmacol. 2017, 139, 105–141. [Google Scholar] [CrossRef]
- Martinez, M.; Poirrier, P.; Chamy, R.; Prüfer, D.; Schulze-Gronover, C.; Jorquera, L.; Ruiz, G. Taraxacum officinale and related species—An ethnopharmacological review and its potential as a commercial medicinal plant. J. Ethnopharmacol. 2015, 169, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Clare, B.A.; Conroy, R.S.; Spelman, K. The Diuretic Effect in Human Subjects of an Extract of Taraxacum officinale Folium over a Single Day. J. Altern. Complement. Med. 2009, 15, 929–934. [Google Scholar] [CrossRef]
- Schuetz, K.; Carle, R.; Schieber, A. Taraxacum—A review on its phytochemical and pharmacological profile. J. Ethnopharmacol. 2006, 107, 313–323. [Google Scholar] [CrossRef]
- El-Nagar, D.M.; Al-Dahmash, B.A.; Alkahtani, S.; Kalu, A.A.; Rady, A. Dandelion (Taraxacum officinale) seeds extract attenuates hypercholesterolemia in swiss albino mice. J. King Saud Univ.-Sci. 2022, 34, 102198. [Google Scholar] [CrossRef]
- Hu, C.; Kitts, D.D. Dandelion (Taraxacum officinale) flower extract suppresses both reactive oxygen species and nitric oxide and prevents lipid oxidation in vitro. Phytomedicine 2005, 12, 588–597. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.-F.; Li, W.; Xu, G.-Y.; Wang, K.-R.; Li, L.; Luo, H.; Zou, L.; Wu, J.-S. Updates and advances on pharmacological properties of Taraxacum mongolicum Hand.-Mazz and its potential applications. Food Chem. 2022, 373, 131380. [Google Scholar] [CrossRef]
- Hu, C. Taraxacum: Phytochemistry and health benefits. Chin. Herb. Med. 2018, 10, 353–361. [Google Scholar] [CrossRef]
- Davaatseren, M.; Hur, H.J.; Yang, H.J.; Hwang, J.-T.; Park, J.H.; Kim, H.-J.; Kim, M.J.; Kwon, D.Y.; Sung, M.J. Taraxacum official (dandelion) leaf extract alleviates high-fat diet-induced nonalcoholic fatty liver. Food Chem. Toxicol. 2013, 58, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Jana, S.; Khawas, S.; Kicuntod, J.; Marschall, M.; Ray, B.; Ray, S. Synthesis, molecular features and biological activities of modified plant polysaccharides. Carbohydr. Polym. 2022, 289, 119299. [Google Scholar] [CrossRef] [PubMed]
- Lis, B.; Rolnik, A.; Jedrejek, D.; Soluch, A.; Stochmal, A.; Olas, B. Dandelion (Taraxacum officinale L.) root components exhibit anti-oxidative and antiplatelet action in an in vitro study. J. Funct. Foods 2019, 59, 16–24. [Google Scholar] [CrossRef]
- Wang, L.; Li, T.; Liu, F.; Liu, D.; Xu, Y.; Yang, Y.; Zhao, Y.; Wei, H. Ultrasonic-assisted enzymatic extraction and characterization of polysaccharides from dandelion (Taraxacum officinale) leaves. Int. J. Biol. Macromol. 2019, 126, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Feng, K.-L.; Yang, J.-C.; He, Y.-S.; Guo, H.; Wang, S.-P.; Gan, R.-Y.; Wu, D.-T. Polysaccharides from dandelion (Taraxacum mongolicum) leaves: Insights into innovative drying techniques on their structural characteristics and biological activities. Int. J. Biol. Macromol. 2021, 167, 995–1005. [Google Scholar] [CrossRef]
- Wu, D.-T.; He, Y.; Yuan, Q.; Wang, S.; Gan, R.-Y.; Hu, Y.-C.; Zou, L. Effects of molecular weight and degree of branching on microbial fermentation characteristics of okra pectic-polysaccharide and its selective impact on gut microbial composition. Food Hydrocoll. 2022, 132, 107897. [Google Scholar] [CrossRef]
- Abuduwaili, A.; Rozi, P.; Mutailifu, P.; Gao, Y.; Nuerxiati, R.; Aisa, H.A.; Yili, A. Effects of different extraction techniques on physicochemical properties and biological activities of polysaccharides from Fritillaria pallidiflora Schrenk. Process Biochem. 2019, 83, 189–197. [Google Scholar] [CrossRef]
- Plaza, M.; Marina, M.L. Pressurized hot water extraction of bioactives. TrAC Trends Anal. Chem. 2019, 116, 236–247. [Google Scholar] [CrossRef]
- Getachew, A.T.; Cho, Y.J.; Chun, B.S. Effect of pretreatments on isolation of bioactive polysaccharides from spent coffee grounds using subcritical water. Int. J. Biol. Macromol. 2018, 109, 711–719. [Google Scholar] [CrossRef]
- Sakdasri, W.; Arnutpongchai, P.; Phonsavat, S.; Bumrungthaichaichan, E.; Sawangkeaw, R. Pressurized hot water extraction of crude polysaccharides, β-glucan, and phenolic compounds from dried gray oyster mushroom. LWT 2022, 168, 113895. [Google Scholar] [CrossRef]
- Lei, J.; Li, W.; Fu, M.-X.; Wang, A.-Q.; Wu, D.-T.; Guo, H.; Hu, Y.-C.; Gan, R.-Y.; Zou, L.; Liu, Y. Pressurized hot water extraction, structural properties, biological effects, and in vitro microbial fermentation characteristics of sweet tea polysaccharide. Int. J. Biol. Macromol. 2022, 222, 3215–3228. [Google Scholar] [CrossRef]
- Savag, M.G.; Lackman, D.B.; Smolens, J. The isolation of the components of streptococcal nucleoproteins in serologically active form. J. Biol. Chem. 1938, 124, 425–436. [Google Scholar] [CrossRef]
- Zhai, X.; Zhu, C.; Li, Y.; Zhang, Y.; Duan, Z.; Yang, X. Optimization for pectinase-assisted extraction of polysaccharides from pomegranate peel with chemical composition and antioxidant activity. Int. J. Biol. Macromol. 2018, 109, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Blumenkrantz, N.; Asboe-Hansen, G. New method for quantitative determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.-T.; Zhang, X.-W.; Liu, H.-P.; Wen, Y.-H.; Li, H.-R.; Gao, J. Structural characterization of an acid polysaccharide from Pinellia ternata and its induction effect on apoptosis of Hep G2 cells. Int. J. Biol. Macromol. 2020, 153, 451–460. [Google Scholar] [CrossRef]
- Miao, Y.-Z.; Lin, Q.; Cao, Y.; He, G.-H.; Qiao, D.-R.; Cao, Y. Extraction of water-soluble polysaccharides (WSPS) from Chinese truffle and its application in frozen yogurt. Carbohydr. Polym. 2011, 86, 566–573. [Google Scholar] [CrossRef]
- Su, C.-H.; Lai, M.-N.; Ng, L.-T. Effects of different extraction temperatures on the physicochemical properties of bioactive polysaccharides from Grifola frondosa. Food Chem. 2017, 220, 400–405. [Google Scholar] [CrossRef]
- Wang, L.B.; Gao, J.Y.; Li, L.Y.; Huang, J.; Yang, Y.; Xu, Y.Q.; Wang, Y.B.; Liu, Y. Characterization and Biological Activities of Polysaccharides from Dandelion (Taraxacum offificinale) Leaves. Starch-Starke 2021, 73, 2000051. [Google Scholar] [CrossRef]
- Zhang, S.J.; Song, Z.T.; Shi, L.J.; Zhou, L.A.; Zhang, J.; Cui, J.L.; Li, Y.H.; Jin, D.Q.; Ohizumi, Y.; Xu, J.; et al. A dandelion polysaccharide and its selenium nanoparticles: Structure features and evaluation of anti-tumor activity in zebrafish models. Carbohydr. Polym. 2021, 270, 118365. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, M.-Y.; Nie, S.-P.; Li, C.; Wang, Y.-X. Purification, composition analysis and antioxidant activity of a polysaccharide from the fruiting bodies of Ganoderma atrum. Food Chem. 2008, 107, 231–241. [Google Scholar] [CrossRef]
- Gong, G.; Zhao, J.; Wang, C.; Wei, M.; Dang, T.; Deng, Y.; Sun, J.; Song, S.; Huang, L.; Wang, Z. Structural characterization and antioxidant activities of the degradation products from Porphyra haitanensis polysaccharides. Process Biochem. 2018, 74, 185–193. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, L.-M. Chemical structural and chain conformational characterization of some bioactive polysaccharides isolated from natural sources. Carbohydr. Polym. 2009, 76, 349–361. [Google Scholar] [CrossRef]
- Sims, I.M.; Carnachan, S.M.; Bell, T.J.; Hinkley, S.F.R. Methylation analysis of polysaccharides: Technical advice. Carbohydr. Polym. 2018, 188, 1–7. [Google Scholar] [CrossRef]
- Kang, J.; Cui, S.W.; Phillips, G.O.; Chen, J.; Guo, Q.; Wang, Q. New studies on gum ghatti (Anogeissus latifolia) Part III: Structure characterization of a globular polysaccharide fraction by 1D, 2D NMR spectroscopy and methylation analysis. Food Hydrocoll. 2011, 25, 1999–2007. [Google Scholar] [CrossRef]
- Kim, J.S.; Reuhs, B.L.; Michon, F.; Kaiser, R.E.; Arumugham, R.G. Addition of glycerol for improved methylation linkage analysis of polysaccharides. Carbohydr. Res. 2006, 341, 1061–1064. [Google Scholar] [CrossRef]
- Guo, Q.B.; Cui, S.W.; Kang, J.; Ding, H.H.; Wang, Q.; Wang, C. Non-starch polysaccharides from American ginseng: Physicochemical investigation and structural characterization. Food Hydrocoll. 2015, 44, 320–327. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, X.J.; Wang, S.W.; Guo, Q.B.; Li, Z.J.; Liu, H.H.; Wang, C.L. Structural characterisation and immunomodulatory activity of polysaccharides from white asparagus skin. Carbohydr. Polym. 2020, 227, 115314. [Google Scholar] [CrossRef]
- do Prado, S.B.R.; Santos, G.R.C.; Mourão, P.A.S.; Fabi, J.P. Chelate-soluble pectin fraction from papaya pulp interacts with galectin-3 and inhibits colon cancer cell proliferation. Int. J. Biol. Macromol. 2019, 126, 170–178. [Google Scholar] [CrossRef]
- Makarova, E.N.; Shakhmatov, E.G. Characterization of pectin-xylan-glucan-arabinogalactan proteins complex from Siberian fir Abies sibirica Ledeb. Carbohydr. Polym. 2021, 260, 117825. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-T.; He, Y.; Fu, M.-X.; Gan, R.-Y.; Hu, Y.-C.; Peng, L.-X.; Zhao, G.; Zou, L. Structural characteristics and biological activities of a pectic-polysaccharide from okra affected by ultrasound assisted metal-free Fenton reaction. Food Hydrocoll. 2022, 122, 107085. [Google Scholar] [CrossRef]
- Huang, L.; Zhao, J.; Wei, Y.; Yu, G.; Li, F.; Li, Q. Structural characterization and mechanisms of macrophage immunomodulatory activity of a pectic polysaccharide from Cucurbita moschata Duch. Carbohydr. Polym. 2021, 269, 118288. [Google Scholar] [CrossRef] [PubMed]
- Makarova, E.N.; Shakhmatov, E.G. Structural characteristics of oxalate-soluble polysaccharides from Norway spruce (Picea abies) foliage. Carbohydr. Polym. 2020, 246, 116544. [Google Scholar] [CrossRef]
- Yue, F.; Xu, J.; Zhang, S.; Hu, X.; Wang, X.; Lü, X. Structural features and anticancer mechanisms of pectic polysaccharides: A review. Int. J. Biol. Macromol. 2022, 209, 825–839. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef]
- Feng, Y.-Y.; Ji, H.-Y.; Dong, X.-D.; Liu, A.-J. An alcohol-soluble polysaccharide from Atractylodes macrocephala Koidz induces apoptosis of Eca-109 cells. Carbohydr. Polym. 2019, 226, 115136. [Google Scholar] [CrossRef]
- de Camargo, M.R.; Frazon, T.F.; Inacio, K.K.; Smiderle, F.R.; Amôr, N.G.; Dionísio, T.J.; Santos, C.F.; Rodini, C.O.; Lara, V.S. Ganoderma lucidum polysaccharides inhibit in vitro tumorigenesis, cancer stem cell properties and epithelial-mesenchymal transition in oral squamous cell carcinoma. J. Ethnopharmacol. 2022, 286, 114891. [Google Scholar] [CrossRef]
- Zhong, S.; Ji, D.-F.; Li, Y.-G.; Lin, T.-B.; Lv, Z.-Q.; Chen, H.-P. Activation of P27kip1-cyclin D1/E-CDK2 pathway by polysaccharide from Phellinus linteus leads to S-phase arrest in HT-29 cells. Chem.-Biol. Interact. 2013, 206, 222–229. [Google Scholar] [CrossRef]
- El-Emam, S.Z.; Abo El-Ella, D.M.; Fayez, S.M.; Asker, M.; Nazeam, J.A. Novel dandelion mannan-lipid nanoparticle: Exploring the molecular mechanism underlying the potent anticancer effect against non-small lung carcinoma. J. Funct. Foods 2021, 87, 104781. [Google Scholar] [CrossRef]
- Khotimchenko, M. Pectin polymers for colon-targeted antitumor drug delivery. Int. J. Biol. Macromol. 2020, 158, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Dick-Perez, M.; Wang, T.; Salazar, A.; Zabotina, O.A.; Hong, M. Multidimensional solid-state NMR studies of the structure and dynamics of pectic polysaccharides in uniformly 13C-labeled Arabidopsis primary cell walls. Magn. Reson. Chem. 2012, 50, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, E.G.; Colquhoun, I.J.; Chau, H.K.; Hotchkiss, A.T.; Waldron, K.W.; Morris, V.J.; Belshaw, N.J. Rhamnogalacturonan I containing homogalacturonan inhibits colon cancer cell proliferation by decreasing ICAM1 expression. Carbohydr. Polym. 2015, 132, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; He, F.; Chen, X.; Ding, K. Isolation and structural characterization of a pectin from Lycium ruthenicum Murr and its anti-pancreatic ductal adenocarcinoma cell activity. Carbohydr. Polym. 2019, 223, 115104. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, P.; Lu, S.-M.; Ling, Z.-Q. Chemoprevention of Low-Molecular-Weight Citrus Pectin (LCP) in Gastrointestinal Cancer Cells. Int. J. Biol. Sci. 2016, 12, 746–756. [Google Scholar] [CrossRef]
- Azzopardi, M.; Farrugia, G.; Balzan, R. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech. Ageing Dev. 2017, 161, 211–224. [Google Scholar] [CrossRef]
- Prado, S.B.R.d.; Ferreira, G.F.; Harazono, Y.; Shiga, T.M.; Raz, A.; Carpita, N.C.; Fabi, J.P. Ripening-induced chemical modifications of papaya pectin inhibit cancer cell proliferation. Sci. Rep. 2017, 7, 16564. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, Z.; Leng, J.; Liu, D.; Hao, M.; Gao, X.; Tai, G.; Zhou, Y. The inhibitory effects and mechanisms of rhamnogalacturonan I pectin from potato on HT-29 colon cancer cell proliferation and cell cycle progression. Int. J. Food Sci. Nutr. 2013, 64, 36–43. [Google Scholar] [CrossRef]
- Zhang, T.; Miller, M.C.; Zheng, Y.; Zhang, Z.; Xue, H.; Zhao, D.; Su, J.; Mayo, K.H.; Zhou, Y.; Tai, G. Macromolecular assemblies of complex polysaccharides with galectin-3 and their synergistic effects on function. Biochem. J. 2017, 474, 3849–3868. [Google Scholar] [CrossRef]
- Fortuna-Costa, A.; Gomes, A.M.; Kozlowski, E.O.; Stelling, M.P.; Pavão, M.S.G. Extracellular Galectin-3 in Tumor Progression and Metastasis. Front. Oncol. 2014, 4, 138. [Google Scholar] [CrossRef]
- Pfeifer, L.; Baumann, A.; Petersen, L.M.; Höger, B.; Beitz, E.; Classen, B. Degraded Arabinogalactans and Their Binding Properties to Cancer-Associated Human Galectins. Int. J. Mol. Sci. 2021, 22, 4058. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zheng, Y.; Zhao, D.; Yan, J.; Sun, C.; Zhou, Y.; Tai, G. Multiple approaches to assess pectin binding to galectin-3. Int. J. Biol. Macromol. 2016, 91, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, J.; Huang, R.; Tan, Y.; Zhang, F.; Zhou, Y.; Sun, L. Analysis of pectin from Panax ginseng flower buds and their binding activities to galectin-3. Int. J. Biol. Macromol. 2019, 128, 459–467. [Google Scholar] [CrossRef] [PubMed]




| Types of Linkage | RT (min) | PMAA | Molar Ratio (%) a |
|---|---|---|---|
| T-Rhap | 18.182 | 1,5-O-Ac2-2,3,4-O-Me3-rhamnitol | 0.86 |
| 2,4-Rhap | 23.160 | 1,2,4,5-O-Ac4-3-O-Me-rhamnitol | 6.88 |
| Total | 7.74 | ||
| T-Araf | 17.565 | 1,4-O-Ac2-2,3,5-O-Me3-arabinitol | 7.46 |
| 5-Araf | 21.107 | 1,4,5-O-Ac3-2,3-O-Me2-arabinitol | 10.35 |
| 3,5-Araf | 26.664 | 1,3,4,5-O-Ac4-2-O-Me-arabinitol | 7.51 |
| Total | 25.32 | ||
| T-Galp | 21.455 | 1,5-O-Ac2-2,3,4,6-O-Me4-galactitol | 11.14 |
| 4-Galp/GalpA | 24.283 | 1,4,5-O-Ac3-2,3,6-O-Me3-galactitol | 40.29 |
| 6-Galp | 25.212 | 1,5,6-O-Ac3-2,3,4-O-Me3-galactitol | 4.95 |
| Total | 56.38 | ||
| T-Glcp | 22.017 | 1,5-O-Ac2-2,3,4,6-O-Me4-glucitol | 4.19 |
| Total | 4.19 | ||
| 4-Manp | 24.050 | 1,4,5-O-Ac3-2,3,6-O-Me3-mannitol | 6.35 |
| Total | 6.35 |
| Sugar Residues | Chemical Shift (ppm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6a | 6b | CH3- of -OMe | |||
| A | 4-α-GalpA-6-OMe | H | 4.86 | 3.64 | 3.9 | 4.31 | 5.06 | 3.7 | ||
| C | 100.18 | 68.07 | 69.02 | 77.22 | 70.35 | 171.03 | 52.29 | |||
| B | 2,4-α-L-Rhap | H | 5.17 | 4.04 | 3.84 | 3.6 | 3.76 | 1.14 | ||
| C | 98.43 | 76.11 | 68.65 | 75.11 | 67.28 | 16.92 | ||||
| C | T-α-L-Rhap | H | 4.63 | 3.84 | 3.66 | 3.33 | 3.93 | 1.14 | ||
| C | 100.61 | 71.27 | 69.22 | 71.75 | 69.21 | 16.92 | ||||
| D | T-α-L-Araf | H | 5.13 | 4.1 | 3.91 | 4.02 | 3.72 | 3.62 | ||
| C | 109.18 | 82.01 | 76.69 | 83.86 | 60.96 | |||||
| E | T-α-L-Araf | H | 5.04 | 4.02 | 3.84 | 3.94 | 3.72 | 3.62 | ||
| C | 107.09 | 80.99 | 76.95 | 83.84 | 60.96 | |||||
| F | 5-α-L-Araf | H | 4.99 | 4.03 | 3.84 | 4.11 | 3.7 | 3.78 | ||
| C | 107.43 | 80.87 | 76.69 | 81.82 | 66.2 | |||||
| G | 3,5-α-L-Araf | H | 5.01 | 4.19 | 3.99 | 3.93 | 3.84 | |||
| C | 107.32 | 79.12 | 82.22 | 83.97 | 66.04 | |||||
| H | 3,6-β-D-Galp | H | 4.43 | 3.54 | 3.64 | 4.01 | 3.81 | 3.82 | 3.92 | |
| C | 103.14 | 69.67 | 80.05 | 69.93 | 73.33 | 69.26 | ||||
| I | T-β-D-Galp | H | 4.38 | 3.25 | 3.44 | 3.18 | 3.6 | -- | ||
| C | 103.21 | 72.77 | 74.93 | 81.7 | 74.36 | 62.38 | ||||
| J | 6-β-D-Galp | H | 4.37 | 3.44 | 3.55 | 3.82 | 3.94 | 3.92 | ||
| C | 103.25 | 70.6 | 72.66 | 73.59 | 68.84 | 69.24 | ||||
| K | 4-β-D-Galp | H | 4.54 | 3.57 | 3.66 | 4.06 | 3.6 | 3.67 | ||
| C | 104.33 | 72.09 | 73.33 | 77.77 | 74.93 | 60.89 | ||||
| L | 4-α-GalpA | H | 4.92 | 3.63 | 3.84 | 4.26 | 4.67 | |||
| C | 99.71 | 67.81 | 69.21 | 79.37 | 71.14 | 172.99 | ||||
| M | 4-β-GlcpA | H | 4.56 | 3.43 | 3.65 | 3.47 | 3.80 | |||
| C | 104.00 | 74.46 | 75.73 | 79.02 | 76.35 | 171.70 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.; Ding, S.; Yan, Z.; Liu, H.; Tu, J.; Chen, Y.; Zhang, X. Structural Characteristic and In-Vitro Anticancer Activities of Dandelion Leaf Polysaccharides from Pressurized Hot Water Extraction. Nutrients 2023, 15, 80. https://doi.org/10.3390/nu15010080
Chen P, Ding S, Yan Z, Liu H, Tu J, Chen Y, Zhang X. Structural Characteristic and In-Vitro Anticancer Activities of Dandelion Leaf Polysaccharides from Pressurized Hot Water Extraction. Nutrients. 2023; 15(1):80. https://doi.org/10.3390/nu15010080
Chicago/Turabian StyleChen, Pei, Suyun Ding, Zhiqian Yan, Huiping Liu, Jianqiu Tu, Yi Chen, and Xiaowei Zhang. 2023. "Structural Characteristic and In-Vitro Anticancer Activities of Dandelion Leaf Polysaccharides from Pressurized Hot Water Extraction" Nutrients 15, no. 1: 80. https://doi.org/10.3390/nu15010080
APA StyleChen, P., Ding, S., Yan, Z., Liu, H., Tu, J., Chen, Y., & Zhang, X. (2023). Structural Characteristic and In-Vitro Anticancer Activities of Dandelion Leaf Polysaccharides from Pressurized Hot Water Extraction. Nutrients, 15(1), 80. https://doi.org/10.3390/nu15010080





