Hempseed (Cannabis sativa) Peptide H3 (IGFLIIWV) Exerts Cholesterol-Lowering Effects in Human Hepatic Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. HepG2 Cell Culture Conditions and Treatment
2.3. HMGCoAR Activity Assay
2.4. MTT Assay
2.5. In-Cell Western (ICW) Assay
2.6. Fluorescent LDL Uptake
2.7. Western Blot Analysis
2.8. Quantification through ELISA of PCSK9 Secreted by HepG2 Cells
2.9. Computational Methods
2.10. Statistical Analysis
3. Results
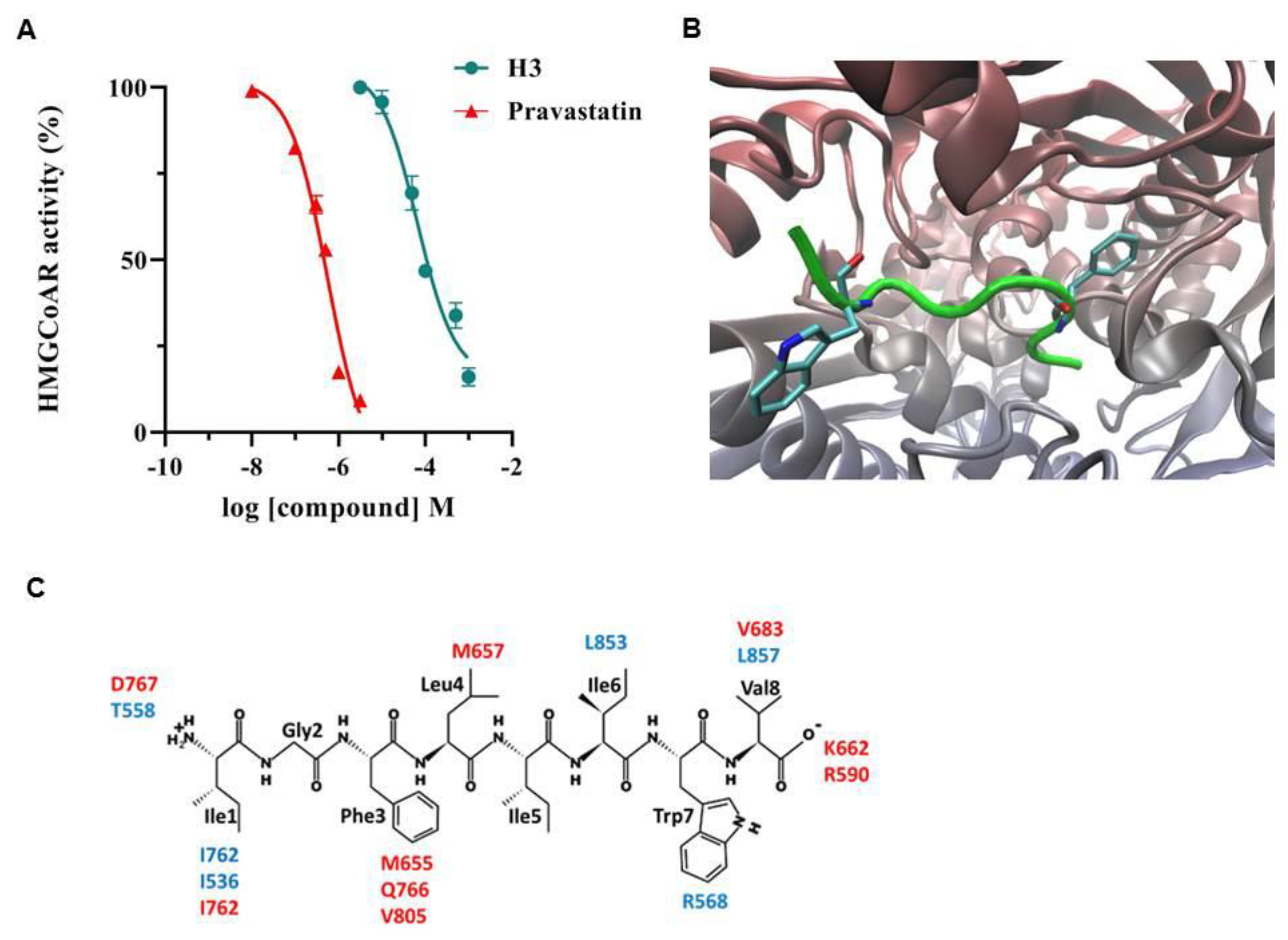
3.1. Peptide H3 Drops HMGCoAR Activity In Vitro
3.2. Peptide H3 Effects on the Cell Vitality of HepG2
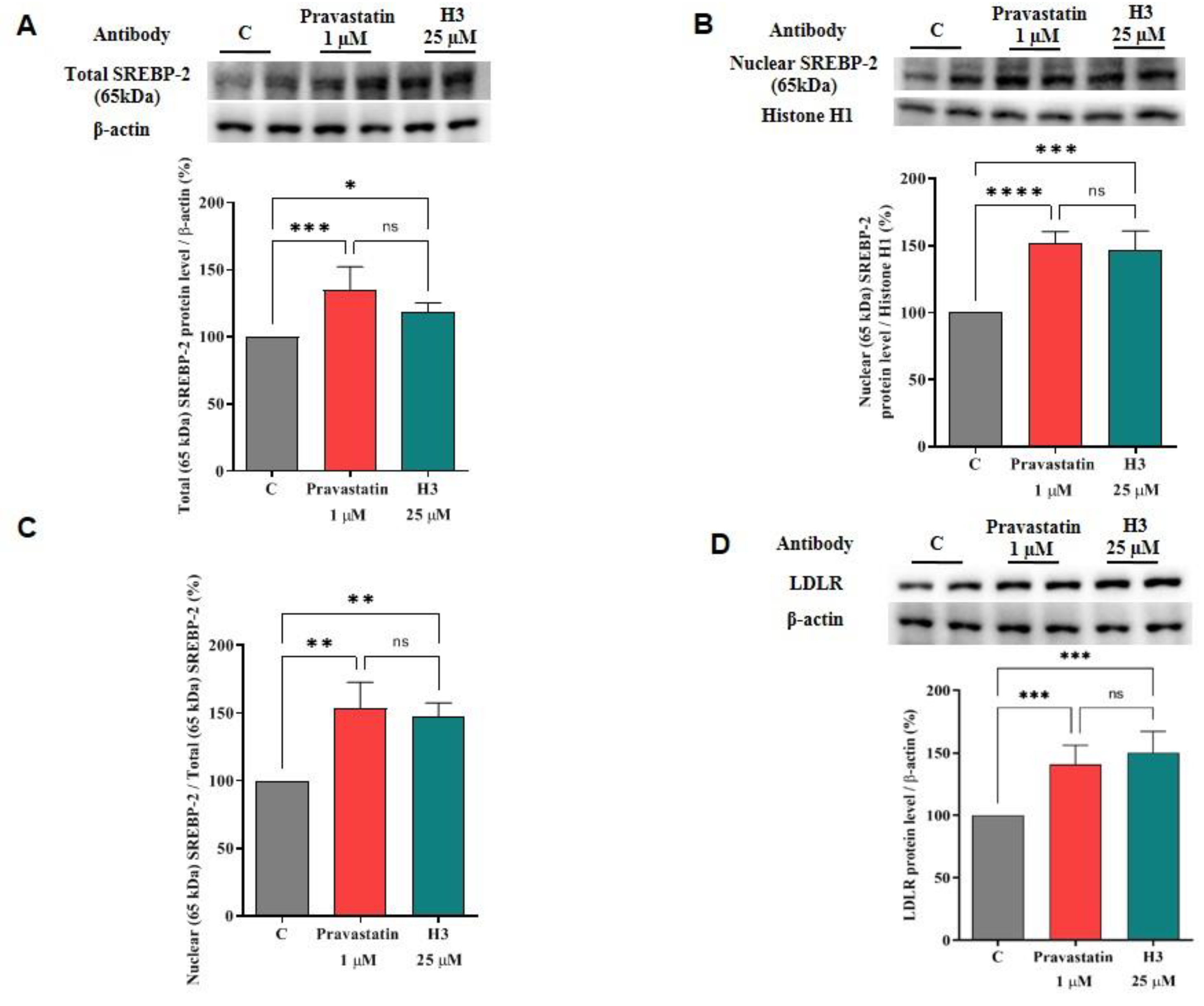
3.3. Peptide H3 Modulates the LDLR Pathway
3.4. Peptide H3 Modulates Intracellular HMGCoAR Protein Levels
3.5. Peptide H3 Increases the Expression of LDLR Localized on Cellular Membranes and Modulates LDL-Uptake in the HepG2 Cell Environment
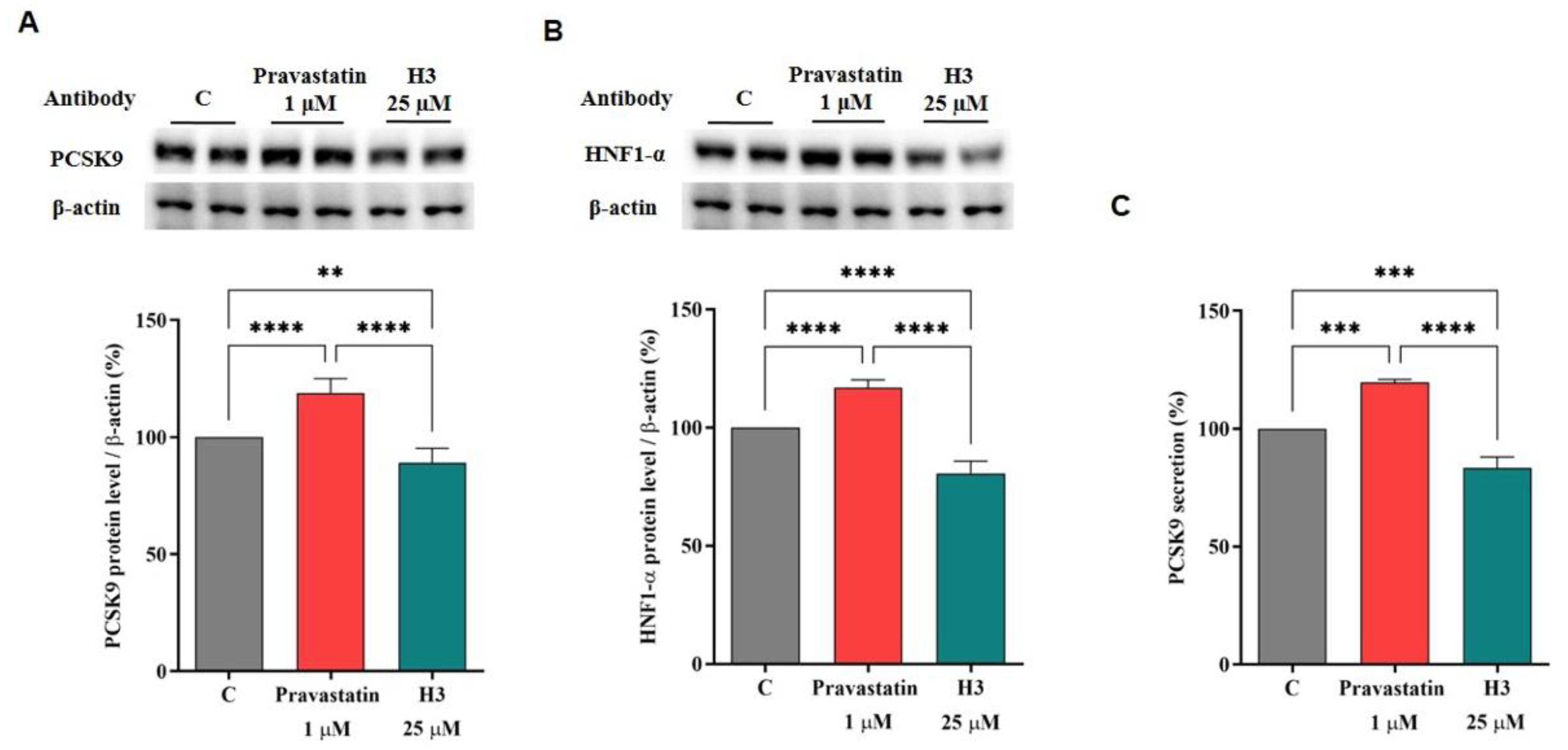
3.6. The Effect of Peptide H3 on the Modulation of PCSK9 Protein and Release Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Go, G.W.; Mani, A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J. Biol. Med. 2012, 85, 19–28. [Google Scholar] [PubMed]
- Mattiuzzi, C.; Sanchis-Gomar, F.; Lippi, G. Worldwide burden of LDL cholesterol: Implications in cardiovascular disease. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMPK—Sensing Energy while Talking to Other Signaling Pathways. Cell Metab. 2014, 20, 939–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, M.S.; Goldstein, J.L.J.C. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Shimano, H. Sterol regulatory element-binding proteins (SREBPs): Transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001, 40, 439–452. [Google Scholar] [CrossRef]
- Xiao, C.; Dash, S.; Morgantini, C.; Hegele, R.A.; Lewis, G.F. Pharmacological Targeting of the Atherogenic Dyslipidemia Complex: The Next Frontier in CVD Prevention Beyond Lowering LDL Cholesterol. Diabetes 2016, 65, 1767–1778. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.-J.; Juillerat, M.A.; Lee, C.-H. Cholesterol Lowering Mechanism of Soybean Protein Hydrolysate. J. Agric. Food Chem. 2007, 55, 10599–10604. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, Y.L. Processing, Nutrition, and Functionality of Hempseed Protein: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 936–952. [Google Scholar] [CrossRef] [Green Version]
- Leonard, W.; Zhang, P.; Ying, D.; Fang, Z. Hempseed in food industry: Nutritional value, health benefits, and industrial applications. Compr. Rev. Food Sci. Food Saf. 2020, 19, 282–308. [Google Scholar] [CrossRef] [Green Version]
- Aiello, G.; Lammi, C.; Boschin, G.; Zanoni, C.; Arnoldi, A. Exploration of Potentially Bioactive Peptides Generated from the Enzymatic Hydrolysis of Hempseed Proteins. J. Agric. Food Chem. 2017, 65, 10174–10184. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, C.; Aiello, G.; Arnoldi, A.; Lammi, C. Hempseed Peptides Exert Hypocholesterolemic Effects with a Statin-Like Mechanism. J. Agric. Food Chem. 2017, 65, 8829–8838. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Meng, J.; Wang, Z.; Yuan, T.; Qian, H.; Chen, W.; Tong, J.; Xie, Y.; Zhang, Y.; Zhao, J.; et al. Proanthocyanidins Attenuation of H2O2Induced Oxidative Damage in Tendon-Derived Stem Cells via Upregulating Nrf-2 Signaling Pathway. BioMed Res. Int. 2017, 2017, 7529104. [Google Scholar] [CrossRef] [Green Version]
- Bollati, C.; Cruz-Chamorro, I.; Aiello, G.; Li, J.; Bartolomei, M.; Santos-Sánchez, G.; Ranaldi, G.; Ferruzza, S.; Sambuy, Y.; Arnoldi, A.; et al. Investigation of the intestinal trans-epithelial transport and antioxidant activity of two hempseed peptides WVSPLAGRT (H2) and IGFLIIWV (H3). Food Res. Int. 2022, 152, 110720. [Google Scholar] [CrossRef]
- Sarkar, D.; Saha, P.; Gamre, S.; Bhattacharjee, S.; Hariharan, C.; Ganguly, S.; Sen, R.; Mandal, G.; Chattopadhyay, S.; Majumdar, S.; et al. Anti-inflammatory effect of allylpyrocatechol in LPS-induced macrophages is mediated by suppression of iNOS and COX-2 via the NF-κB pathway. Int. Immunopharmacol. 2008, 8, 1264–1271. [Google Scholar] [CrossRef]
- Cruz-Chamorro, I.; Santos-Sánchez, G.; Bollati, C.; Bartolomei, M.; Li, J.; Arnoldi, A.; Lammi, C. Hempseed (Cannabis sativa) Peptides WVSPLAGRT and IGFLIIWV Exert Anti-inflammatory Activity in the LPS-Stimulated Human Hepatic Cell Line. J. Agric. Food Chem. 2022, 70, 577–583. [Google Scholar] [CrossRef]
- Zanoni, C.; Aiello, G.; Arnoldi, A.; Lammi, C. Investigations on the hypocholesterolaemic activity of LILPKHSDAD and LTFPGSAED, two peptides from lupin β-conglutin: Focus on LDLR and PCSK9 pathways. J. Funct. Foods 2017, 32, 1–8. [Google Scholar] [CrossRef]
- Lammi, C.; Sgrignani, J.; Roda, G.; Arnoldi, A.; Grazioso, G. Inhibition of PCSK9D374Y/LDLR Protein–Protein Interaction by Computationally Designed T9 Lupin Peptide. ACS Med. Chem. Lett. 2019, 10, 425–430. [Google Scholar] [CrossRef]
- Pedretti, A.; Mazzolari, A.; Gervasoni, S.; Fumagalli, L.; Vistoli, G. The VEGA suite of programs: An versatile platform for cheminformatics and drug design projects. Bioinformatics 2021, 37, 1174–1175. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical Scoring Functions for Advanced Protein−Ligand Docking with PLANTS. J. Chem. Inf. Modeling 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Vistoli, G.; Mazzolari, A.; Testa, B.; Pedretti, A. Binding Space Concept: A New Approach to Enhance the Reliability of Docking Scores and Its Application to Predicting Butyrylcholinesterase Hydrolytic Activity. J. Chem. Inf. Modeling 2017, 57, 1691–1702. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.S.; Deisenhofer, J. Structural Mechanism for Statin Inhibition of HMG-CoA Reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosley, S.T.; Kalinowski, S.S.; Schafer, B.L.; Tanaka, R.D. Tissue-selective acute effects of inhibitors of 3-hydroxy-3-methylglutaryl coenzyme A reductase on cholesterol biosynthesis in lens. J. Lipid Res. 1989, 30, 1411–1420. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: Cambridge, MA, USA, 1965; pp. 97–166. [Google Scholar]
- Izumi, Y.; Shiota, M.; Kusakabe, H.; Hikita, Y.; Nakao, T.; Nakamura, Y.; Muro, T.; Miura, K.; Yoshiyama, M.; Iwao, H. Pravastatin accelerates ischemia-induced angiogenesis through AMP-activated protein kinase. Hypertens. Res. 2009, 32, 675–679. [Google Scholar] [CrossRef]
- Dehnavi, S.; Kiani, A.; Sadeghi, M.; Biregani, A.F.; Banach, M.; Atkin, S.L.; Jamialahmadi, T.; Sahebkar, A. Targeting AMPK by Statins: A Potential Therapeutic Approach. Drugs 2021, 81, 923–933. [Google Scholar] [CrossRef]
- Athyros, V.G.; Boutari, C.; Stavropoulos, K.; Anagnostis, P.; Imprialos, K.P.; Doumas, M.; Karagiannis, A.J.C.V.P. Statins: An under-appreciated asset for the prevention and the treatment of NAFLD or NASH and the related cardiovascular risk. Curr. Vasc. Pharmacol. 2018, 16, 246–253. [Google Scholar] [CrossRef]
- Viollet, B.; Foretz, M.; Guigas, B.; Horman, S.; Dentin, R.; Bertrand, L.; Hue, L.; Andreelli, F. Activation of AMP-activated protein kinase in the liver: A new strategy for the management of metabolic hepatic disorders. J. Physiol. 2006, 574, 41–53. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A. A simple and high-throughput in-cell Western assay using HepG2 cell line for investigating the potential hypocholesterolemic effects of food components and nutraceutics. Food Chem. 2015, 169, 59–64. [Google Scholar] [CrossRef]
- Bays, H.E.; Taub, P.R.; Epstein, E.; Michos, E.D.; Ferraro, R.A.; Bailey, A.L.; Kelli, H.M.; Ferdinand, K.C.; Echols, M.R.; Weintraub, H.; et al. Ten things to know about ten cardiovascular disease risk factors. Am. J. Prev. Cardiol. 2021, 5, 100149. [Google Scholar] [CrossRef]
- Soares, R.A.M.; Mendonça, S.; De Castro, L.Í.A.; Menezes, A.C.C.C.C.; Arêas, J.A.G. Major Peptides from Amaranth (Amaranthus cruentus) Protein Inhibit HMG-CoA Reductase Activity. Int. J. Mol. Sci. 2015, 16, 4150–4160. [Google Scholar] [CrossRef] [Green Version]
- Lammi, C.; Zanoni, C.; Arnoldi, A. IAVPGEVA, IAVPTGVA, and LPYP, three peptides from soy glycinin, modulate cholesterol metabolism in HepG2 cells through the activation of the LDLR-SREBP2 pathway. J. Funct. Foods 2015, 14, 469–478. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A.; Vistoli, G. Two Peptides from Soy β-Conglycinin Induce a Hypocholesterolemic Effect in HepG2 Cells by a Statin-Like Mechanism: Comparative in Vitro and in Silico Modeling Studies. J. Agric. Food Chem. 2015, 63, 7945–7951. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-W.; Lagace, T.A.; Garuti, R.; Zhao, Z.; McDonald, M.; Horton, J.D.; Cohen, J.C.; Hobbs, H.H. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 2007, 282, 18602–18612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, E.J.; Park, J.H.; Sung, M.J.; Chung, M.-Y.; Hwang, J.-T. Citrus junos Tanaka peel ameliorates hepatic lipid accumulation in HepG2 cells and in mice fed a high-cholesterol diet. BMC Complementary Altern. Med. 2016, 16, 499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, S.S.; Pal, S. Margarine phytosterols decrease the secretion of atherogenic lipoproteins from HepG2 liver and Caco2 intestinal cells. Atherosclerosis 2005, 182, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Lammi, C.; Zanoni, C.; Ferruzza, S.; Ranaldi, G.; Sambuy, Y.; Arnoldi, A. Hypocholesterolaemic Activity of Lupin Peptides: Investigation on the Crosstalk between Human Enterocytes and Hepatocytes Using a Co-Culture System Including Caco-2 and HepG2 Cells. Nutrients 2016, 8, 437. [Google Scholar] [CrossRef] [Green Version]
- Lammi, C.; Zanoni, C.; Calabresi, L.; Arnoldi, A. Lupin protein exerts cholesterol-lowering effects targeting PCSK9: From clinical evidences to elucidation of the in vitro molecular mechanism using HepG2 cells. J. Funct. Foods 2016, 23, 230–240. [Google Scholar] [CrossRef]
- Donato, M.T.; Tolosa, L.; Gómez-Lechón, M.J. Culture and Functional Characterization of Human Hepatoma HepG2 Cells. In Protocols in In Vitro Hepatocyte Research; Vinken, M., Rogiers, V., Eds.; Springer: New York, NY, USA, 2015; pp. 77–93. [Google Scholar]
- Pallottini, V.; Martini, C.; Pascolini, A.; Cavallini, G.; Gori, Z.; Bergamini, E.; Incerpi, S.; Trentalance, A. 3-Hydroxy-3-methylglutaryl coenzyme A reductase deregulation and age-related hypercholesterolemia: A new role for ROS. Mech. Ageing Dev. 2005, 126, 845–851. [Google Scholar] [CrossRef]
- Ching, Y.P.; Davies, S.P.; Hardie, D.G. Analysis of the Specificity of the AMP-Activated Protein Kinase by Site-Directed Mutagenesis of Bacterially Expressed 3-hydroxy 3-methylglutaryl-CoA Reductase, Using a Single Primer Variant of the Unique-site-elimination Method. Eur. J. Biochem. 1996, 237, 800–808. [Google Scholar] [CrossRef]
- Oliaro-Bosso, S.; Calcio Gaudino, E.; Mantegna, S.; Giraudo, E.; Meda, C.; Viola, F.; Cravotto, G. Regulation of HMGCoA Reductase Activity by Policosanol and Octacosadienol, a New Synthetic Analogue of Octacosanol. Lipids 2009, 44, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Lee, T.-S.; Zhu, M.; Gu, C.; Wang, Y.; Zhu, Y.; Shyy, J.Y.-J. Statins Activate AMP-Activated Protein Kinase In Vitro and In Vivo. Circulation 2006, 114, 2655–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, E.; Ouadda, A.B.D.; Spahis, S.; Sane, A.T.; Garofalo, C.; Grenier, É.; Emonnot, L.; Yara, S.; Couture, P.; Beaulieu, J.-F.; et al. PCSK9 plays a significant role in cholesterol homeostasis and lipid transport in intestinal epithelial cells. Atherosclerosis 2013, 227, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Dubuc, G.; Chamberland, A.; Wassef, H.; Davignon, J.; Seidah, N.G.; Bernier, L.; Prat, A. Statins Upregulate PCSK9, the Gene Encoding the Proprotein Convertase Neural Apoptosis-Regulated Convertase-1 Implicated in Familial Hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1454–1459. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, K.N.; Fisher, E.A.; Breslow, J.L. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA 2005, 102, 2069–2074. [Google Scholar] [CrossRef] [Green Version]
- Dong, B.; Li, H.; Singh, A.B.; Cao, A.; Liu, J. Inhibition of PCSK9 transcription by berberine involves down-regulation of hepatic HNF1α protein expression through the ubiquitin-proteasome degradation pathway. J. Biol. Chem. 2015, 290, 4047–4058. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, R.; Garg, J.; Shah, N.; Sumner, A. PCSK9 inhibitors: A new era of lipid lowering therapy. World J. Cardiol. 2017, 9, 76–91. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Bollati, C.; Bartolomei, M.; Mazzolari, A.; Arnoldi, A.; Vistoli, G.; Lammi, C. Hempseed (Cannabis sativa) Peptide H3 (IGFLIIWV) Exerts Cholesterol-Lowering Effects in Human Hepatic Cell Line. Nutrients 2022, 14, 1804. https://doi.org/10.3390/nu14091804
Li J, Bollati C, Bartolomei M, Mazzolari A, Arnoldi A, Vistoli G, Lammi C. Hempseed (Cannabis sativa) Peptide H3 (IGFLIIWV) Exerts Cholesterol-Lowering Effects in Human Hepatic Cell Line. Nutrients. 2022; 14(9):1804. https://doi.org/10.3390/nu14091804
Chicago/Turabian StyleLi, Jianqiang, Carlotta Bollati, Martina Bartolomei, Angelica Mazzolari, Anna Arnoldi, Giulio Vistoli, and Carmen Lammi. 2022. "Hempseed (Cannabis sativa) Peptide H3 (IGFLIIWV) Exerts Cholesterol-Lowering Effects in Human Hepatic Cell Line" Nutrients 14, no. 9: 1804. https://doi.org/10.3390/nu14091804
APA StyleLi, J., Bollati, C., Bartolomei, M., Mazzolari, A., Arnoldi, A., Vistoli, G., & Lammi, C. (2022). Hempseed (Cannabis sativa) Peptide H3 (IGFLIIWV) Exerts Cholesterol-Lowering Effects in Human Hepatic Cell Line. Nutrients, 14(9), 1804. https://doi.org/10.3390/nu14091804