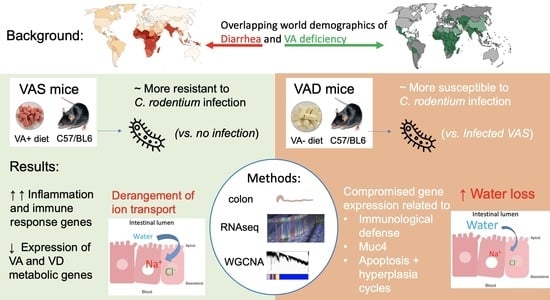
Transcriptional Profiling of the Small Intestine and the Colon Reveals Modulation of Gut Infection with Citrobacter rodentium According to the Vitamin A Status
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Infection of C. rodentium
2.3. Tissue Collection and RNA Extraction
2.4. RNAseq Library Preparation, Sequencing, and Mapping
2.5. Differential Expression
2.6. WGCNA
2.7. Functional Enrichment Analysis
3. Results
3.1. Model Validation: Infection Status and VA Status
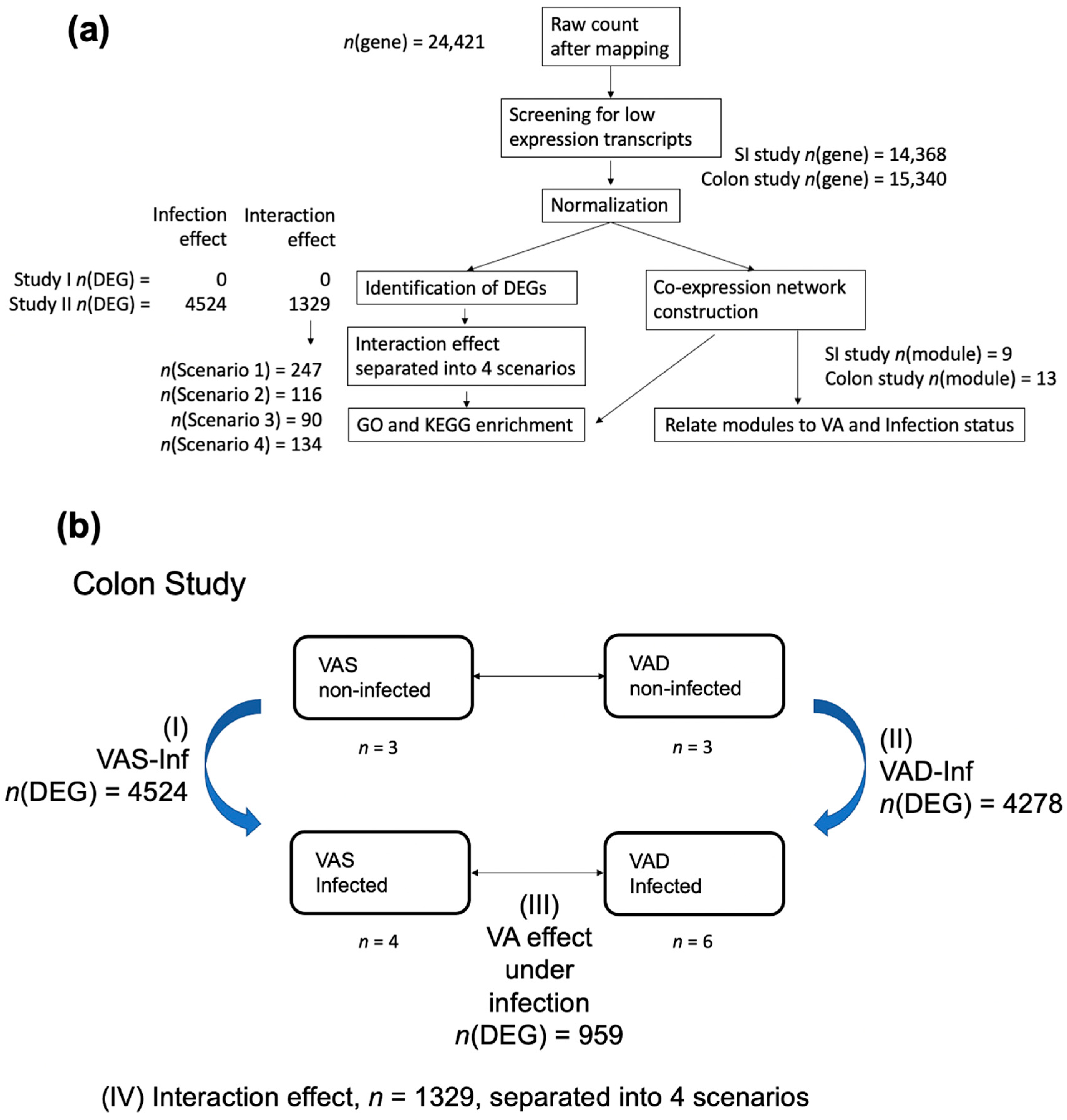
3.2. Mapping and Overview of the Differential Expression Results in the SI and the Colon
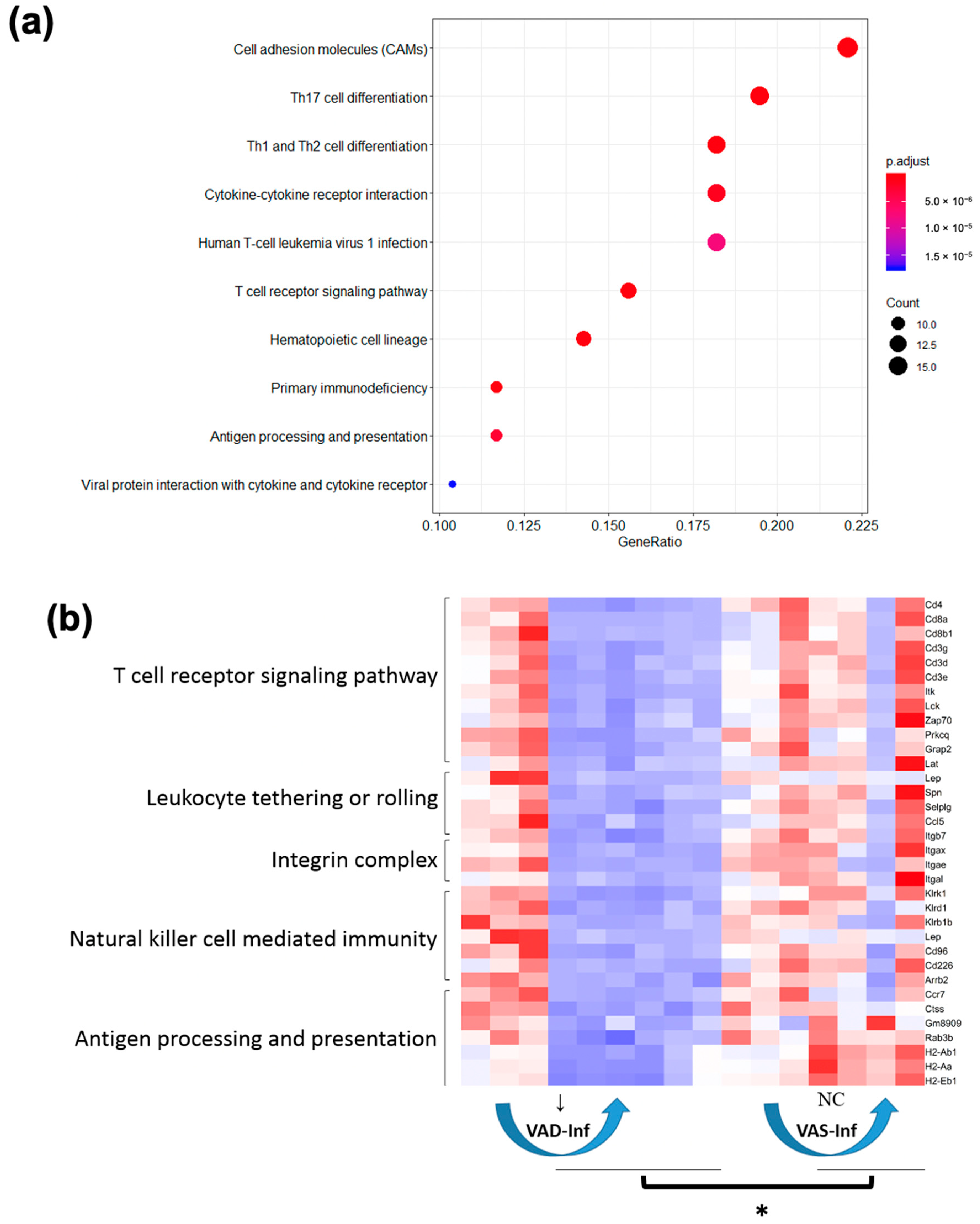
3.3. Functional Categories of Colon DEGs in the VAS Infected vs. Non-Infected States (VAS-Inf Effect)
3.4. The Effect of Infection under VA Deficiency (VAD-Inf) and the Effect of the VA Status under Infection
3.5. GO and KEGG Enrichment of DEGs: Four Interaction Scenarios Comparing Up- and Downregulated Gene Patterns in the Colon of VAS-Infected and VAD-Infected Mice
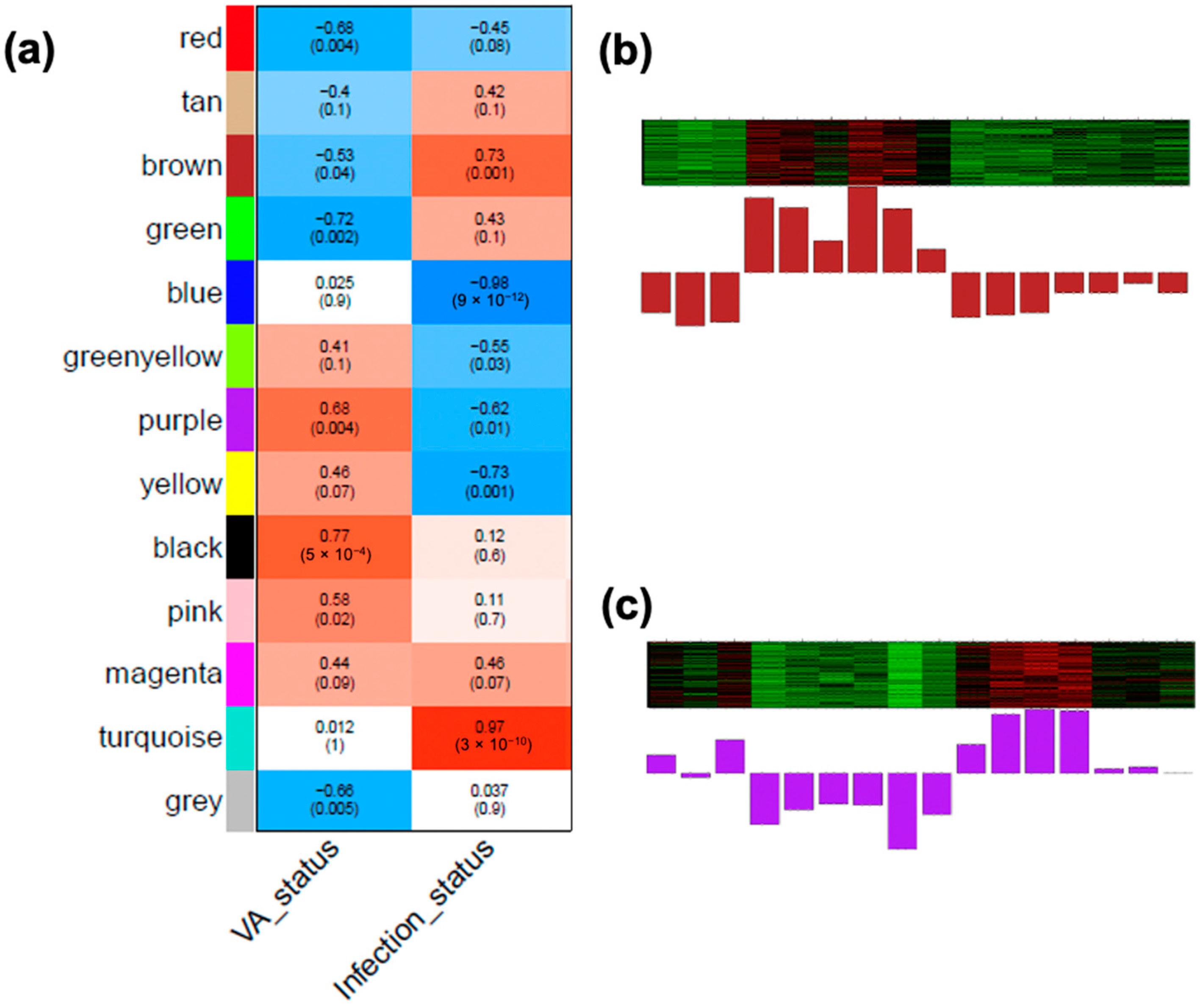
3.6. Identifying WGCNA Modules That Were Significantly Correlated with Infection Effects and the Interaction of the VA Status and Infection in the Colon
4. Discussion
4.1. Approach and Validation
4.2. DEGs Positively Associated with Host Resistance under the VAS Status
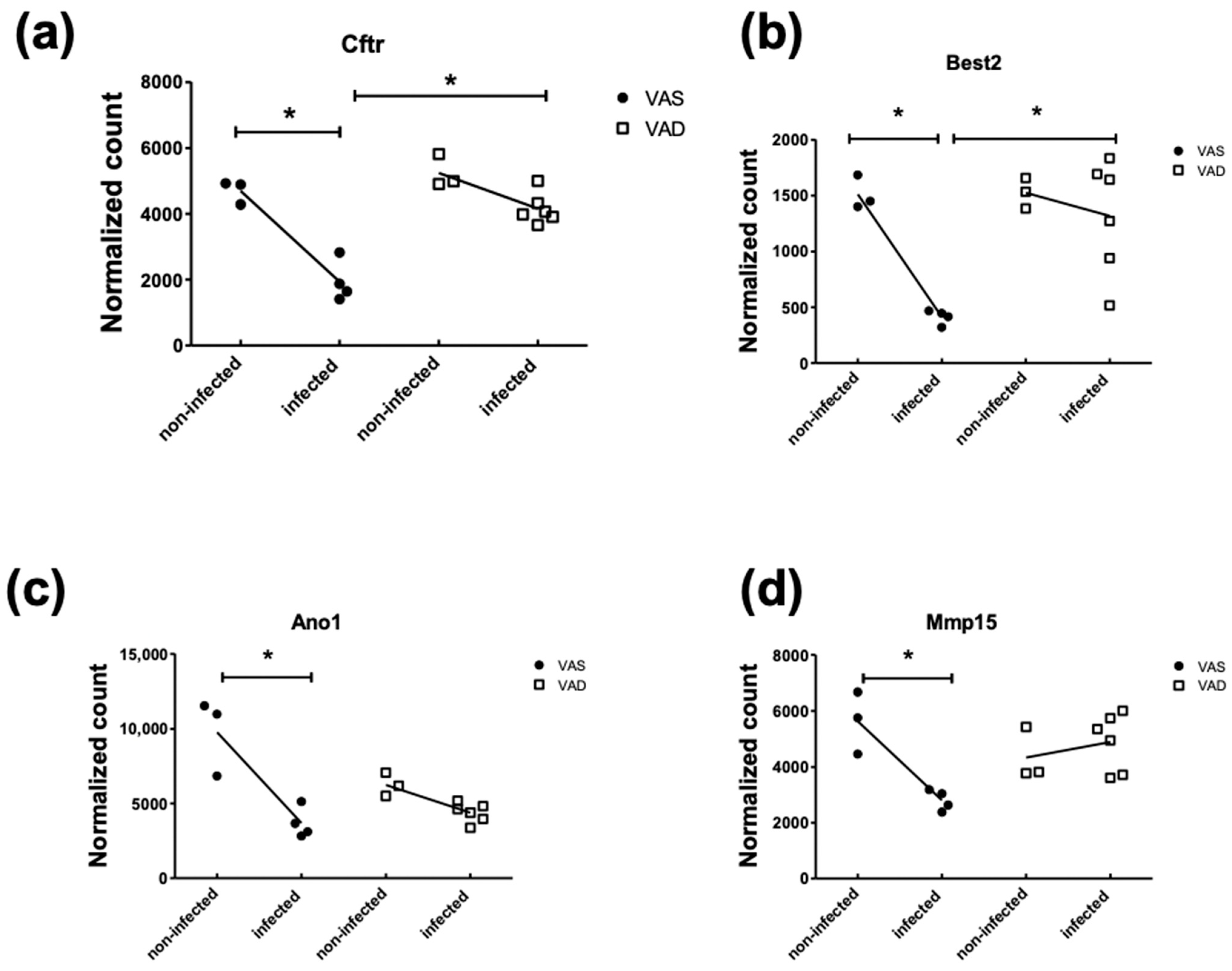
4.3. Three Categories of Downregulated DEGs Related to Nutritient Utilization and Ion Transport Relevant to Diarrheal Disease
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2161. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Petri, W.A.; Miller, M.; Binder, H.J.; Levine, M.M.; Dillingham, R.; Guerrant, R.L. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Investig. 2008, 118, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.K.; Hodges, K.; Hecht, G. Enteric infection meets intestinal function: How bacterial pathogens cause diarrhoea. Nat. Rev. Microbiol. 2008, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Kosek, M.; Bern, C.; Guerrant, R.L. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. World Health Organ. 2003, 81, 197–204. [Google Scholar] [PubMed]
- Collins, J.W.; Keeney, K.M.; Crepin, V.F.; Rathinam, V.A.; Fitzgerald, K.A.; Finlay, B.B.; Frankel, G. Citrobacter rodentium: Infection, inflammation and the microbiota. Nat. Rev. Microbiol. 2014, 12, 612–623. [Google Scholar] [CrossRef]
- Silberger, D.J.; Zindl, C.L.; Weaver, C.T. Citrobacter rodentium: A model enteropathogen for understanding the interplay of innate and adaptive components of type 3 immunity. Mucosal Immunol. 2017, 10, 1108. [Google Scholar] [CrossRef]
- Wiles, S.; Clare, S.; Harker, J.; Huett, A.; Young, D.; Dougan, G.; Frankel, G. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell. Microbiol. 2004, 6, 963–972. [Google Scholar] [CrossRef]
- Wiles, S.; Pickard, K.M.; Peng, K.; MacDonald, T.T.; Frankel, G. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect. Immun. 2006, 74, 5391–5396. [Google Scholar] [CrossRef]
- Mundy, R.; MacDonald, T.T.; Dougan, G.; Frankel, G.; Wiles, S. Citrobacter rodentium of mice and man. Cell. Microbiol. 2005, 7, 1697–1706. [Google Scholar] [CrossRef]
- Sano, T.; Huang, W.D.; Hall, J.A.; Yang, Y.; Chen, A.; Gavzy, S.J.; Lee, J.Y.; Ziel, J.W.; Miraldi, E.R.; Domingos, A.I.; et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 2015, 163, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Ando, M.; Kamada, N.; Nagano, Y.; Narushima, S.; Suda, W.; Imaoka, A.; Setoyama, H.; Nagamori, T.; et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 2015, 163, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dai, C.; Brown, K.; Rajendiran, E.; Makarenko, S.; Baker, J.; Ma, C.; Halder, S.; Montero, M.; Ionescu, V.-A. Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G39–G49. [Google Scholar] [CrossRef] [PubMed]
- Vallance, B.A.; Deng, W.; Jacobson, K.; Finlay, B.B. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect. Immun. 2003, 71, 3443–3453. [Google Scholar] [CrossRef] [PubMed]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar] [CrossRef]
- Sonnenburg, J.L.; Xu, J.; Leip, D.D.; Chen, C.-H.; Westover, B.P.; Weatherford, J.; Buhler, J.D.; Gordon, J.I. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005, 307, 1955–1959. [Google Scholar] [CrossRef]
- Bergstrom, K.S.; Kissoon-Singh, V.; Gibson, D.L.; Ma, C.; Montero, M.; Sham, H.P.; Ryz, N.; Huang, T.; Velcich, A.; Finlay, B.B. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010, 6, e1000902. [Google Scholar] [CrossRef]
- Gustafsson, J.K.; Navabi, N.; Rodriguez-Piñeiro, A.M.; Alomran, A.H.; Premaratne, P.; Fernandez, H.R.; Banerjee, D.; Sjövall, H.; Hansson, G.C.; Lindén, S.K. Dynamic changes in mucus thickness and ion secretion during Citrobacter rodentium infection and clearance. PLoS ONE 2013, 8, e84430. [Google Scholar] [CrossRef]
- Morampudi, V.; Dalwadi, U.; Bhinder, G.; Sham, H.; Gill, S.; Chan, J.; Bergstrom, K.; Huang, T.; Ma, C.; Jacobson, K. The goblet cell-derived mediator RELM-β drives spontaneous colitis in Muc2-deficient mice by promoting commensal microbial dysbiosis. Mucosal Immunol. 2016, 9, 1218. [Google Scholar] [CrossRef]
- Bergstrom, K.S.; Morampudi, V.; Chan, J.M.; Bhinder, G.; Lau, J.; Yang, H.; Ma, C.; Huang, T.; Ryz, N.; Sham, H.P. Goblet cell derived RELM-β recruits CD4+ T cells during infectious colitis to promote protective intestinal epithelial cell proliferation. PLoS Pathog. 2015, 11, e1005108. [Google Scholar] [CrossRef]
- Erickson, N.A.; Nyström, E.E.L.; Mundhenk, L.; Arike, L.; Glauben, R.; Heimesaat, M.M.; Fischer, A.; Bereswill, S.; Birchenough, G.M.H.; Gruber, A.D.; et al. The Goblet Cell Protein Clca1 (Alias mClca3 or Gob-5) Is Not Required for Intestinal Mucus Synthesis, Structure and Barrier Function in Naive or DSS-Challenged Mice. PLoS ONE 2015, 10, e0131991. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.A.N.; Clare, S.; Goulding, D.; Arasteh, J.M.; Stares, M.D.; Browne, H.P.; Keane, J.A.; Page, A.J.; Kumasaka, N.; Kane, L. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 2014, 16, 504–516. [Google Scholar] [CrossRef]
- McDaniel, K.L.; Restori, K.H.; Dodds, J.W.; Kennett, M.J.; Ross, A.C.; Cantorna, M.T. Vitamin A-deficient hosts become nonsymptomatic reservoirs of Escherichia coli-like enteric infections. Infect. Immun. 2015, 83, 2984–2991. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Waddell, A.; Lin, Y.-D.; Cantorna, M.T. Dysbiosis caused by vitamin D receptor deficiency confers colonization resistance to Citrobacter rodentium through modulation of innate lymphoid cells. Mucosal Immunol. 2015, 8, 618. [Google Scholar] [CrossRef]
- Snyder, L.M.; McDaniel, K.L.; Tian, Y.; Wei, C.-H.; Kennett, M.J.; Patterson, A.D.; Ross, A.C.; Cantorna, M.T. Retinoic acid mediated clearance of Citrobacter rodentium in vitamin A deficient mice requires CD11b+ and T Cells. Front. Immunol. 2019, 9, 3090. [Google Scholar] [CrossRef]
- Mangan, P.R.; Harrington, L.E.; O’Quinn, D.B.; Helms, W.S.; Bullard, D.C.; Elson, C.O.; Hatton, R.D.; Wahl, S.M.; Schoeb, T.R.; Weaver, C.T. Transforming growth factor-β induces development of the T H 17 lineage. Nature 2006, 441, 231. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, G.F.; Monticelli, L.A.; Elloso, M.M.; Fouser, L.A.; Artis, D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 2011, 34, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Borenshtein, D.; Schlieper, K.A.; Rickman, B.H.; Chapman, J.M.; Schweinfest, C.W.; Fox, J.G.; Schauer, D.B. Decreased expression of colonic Slc26a3 and carbonic anhydrase iv as a cause of fatal infectious diarrhea in mice. Infect. Immun. 2009, 77, 3639–3650. [Google Scholar] [CrossRef]
- Borenshtein, D.; Fry, R.C.; Groff, E.B.; Nambiar, P.R.; Carey, V.J.; Fox, J.G.; Schauer, D.B. Diarrhea as a cause of mortality in a mouse model of infectious colitis. Genome Biol. 2008, 9, R122. [Google Scholar] [CrossRef]
- Barrett, K.E.; Keely, S.J. Chloride secretion by the intestinal epithelium: Molecular basis and regulatory aspects. Annu. Rev. Physiol. 2000, 62, 535–572. [Google Scholar] [CrossRef]
- Ardizzone, S.; Cassinotti, A.; Trabattoni, D.; Manzionna, G.; Rainone, V.; Bevilacqua, M.; Massari, A.; Manes, G.; Maconi, G.; Clerici, M. Immunomodulatory effects of 1, 25-dihydroxyvitamin D3 on TH1/TH2 cytokines in inflammatory bowel disease: An in vitro study. Int. J. Immunopathol. Pharmacol. 2009, 22, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Bartels, L.E.; Jørgensen, S.P.; Agnholt, J.; Kelsen, J.; Hvas, C.L.; Dahlerup, J.F. 1, 25-dihydroxyvitamin D3 and dexamethasone increase interleukin-10 production in CD4+ T cells from patients with Crohn’s disease. Int. Immunopharmacol. 2007, 7, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Laverny, G.; Penna, G.; Vetrano, S.; Correale, C.; Nebuloni, M.; Danese, S.; Adorini, L. Efficacy of a potent and safe vitamin D receptor agonist for the treatment of inflammatory bowel disease. Immunol. Lett. 2010, 131, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Stio, M.; Bonanomi, A.G.; d’Albasio, G.; Treves, C. Suppressive effect of 1, 25-dihydroxyvitamin D3 and its analogues EB 1089 and KH 1060 on T lymphocyte proliferation in active ulcerative colitis. Biochem. Pharmacol. 2001, 61, 365–371. [Google Scholar] [CrossRef]
- Martinesi, M.; Treves, C.; d’Albasio, G.; Bagnoli, S.; Bonanomi, A.G.; Stio, M. Vitamin D derivatives induce apoptosis and downregulate ICAM-1 levels in peripheral blood mononuclear cells of inflammatory bowel disease patients. Inflamm. Bowel Dis. 2008, 14, 597–604. [Google Scholar] [CrossRef]
- Gieng, S.H.; Green, M.H.; Green, J.B.; Rosales, F.J. Model-based compartmental analysis indicates a reduced mobilization of hepatic vitamin A during inflammation in rats. J. Lipid Res. 2007, 48, 904–913. [Google Scholar] [CrossRef]
- Rosales, F.J.; Ritter, S.J.; Zolfaghari, R.; Smith, J.E.; Ross, A.C. Effects of acute inflammation on plasma retinol, retinol-binding protein, and its mRNA in the liver and kidneys of vitamin A-sufficient rats. J. Lipid Res. 1996, 37, 962–971. [Google Scholar] [CrossRef]
- Restori, K.H.; McDaniel, K.L.; Wray, A.E.; Cantorna, M.T.; Ross, A.C. Streptococcus pneumoniae-induced pneumonia and Citrobacter rodentium-induced gut infection differentially alter vitamin A concentrations in the lung and liver of mice. J. Nutr. 2014, 144, 392–398. [Google Scholar] [CrossRef]
- Krutzik, S.R.; Hewison, M.; Liu, P.T.; Robles, J.A.; Stenger, S.; Adams, J.S.; Modlin, R.L. IL-15 links TLR2/1-induced macrophage differentiation to the vitamin D-dependent antimicrobial pathway. J. Immunol. 2008, 181, 7115–7120. [Google Scholar] [CrossRef]
- Liu, P.T.; Stenger, S.; Li, H.; Wenzel, L.; Tan, B.H.; Krutzik, S.R.; Ochoa, M.T.; Schauber, J.; Wu, K.; Meinken, C. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 2006, 311, 1770–1773. [Google Scholar] [CrossRef]
- Stoffels, K.; Overbergh, L.; Giulietti, A.; Verlinden, L.; Bouillon, R.; Mathieu, C. Immune regulation of 25-hydroxyvitamin-D3-1α-hydroxylase in human monocytes. J. Bone Miner. Res. 2006, 21, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Glasziou, P.P.; Mackerras, D.E. Vitamin A supplementation in infectious diseases: A meta-analysis. Br. Med. J. 1993, 306, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Sirisinha, S. The pleiotropic role of vitamin A in regulating mucosal immunity. Asian Pac. J. Allergy Immunol. 2015, 33, 71–89. [Google Scholar] [PubMed]
- Chai, Z.; Lyu, Y.; Chen, Q.; Wei, C.-H.; Snyder, L.M.; Weaver, V.; Sebastian, A.; Albert, I.; Li, Q.; Cantorna, M.T. RNAseq studies reveal distinct transcriptional response to vitamin A deficiency in small intestine versus colon, uncovering novel vitamin A-regulated genes. J. Nutr. Biochem. 2021, 98, 108814. [Google Scholar] [CrossRef] [PubMed]
- West, K.P., Jr. Extent of vitamin A deficiency among preschool children and women of reproductive age. J. Nutr. 2002, 132, 2857S–2866S. [Google Scholar] [CrossRef]
- Carman, J.; Smith, S.; Hayes, C. Characterization of a helper T lymphocyte defect in vitamin A-deficient mice. J. Immunol. 1989, 142, 388–393. [Google Scholar]
- Smith, S.M.; Levy, N.S.; Hayes, C.E. Impaired immunity in vitamin A-deficient mice. J. Nutr. 1987, 117, 857–865. [Google Scholar] [CrossRef]
- Smith, S.M.; Hayes, C.E. Contrasting impairments in IgM and IgG responses of vitamin A-deficient mice. Proc. Natl. Acad. Sci. USA 1987, 84, 5878–5882. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007, 1, 54. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Witcomb, L.A.; Collins, J.W.; McCarthy, A.J.; Frankel, G.; Taylor, P.W. Bioluminescent Imaging Reveals Novel Patterns of Colonization and Invasion in Systemic Escherichia coli K1 Experimental Infection in the Neonatal Rat. Infect. Immun. 2015, 83, 4528–4540. [Google Scholar] [CrossRef] [PubMed]
- Luperchio, S.A.; Schauer, D.B. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 2001, 3, 333–340. [Google Scholar] [CrossRef]
- Arike, L.; Holmén-Larsson, J.; Hansson, G.C. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 2017, 27, 318–328. [Google Scholar] [CrossRef]
- Bry, L.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A model of host-microbial interactions in an open mammalian ecosystem. Science 1996, 273, 1380–1383. [Google Scholar] [CrossRef]
- Goto, Y.; Obata, T.; Kunisawa, J.; Sato, S.; Ivanov, I.I.; Lamichhane, A.; Takeyama, N.; Kamioka, M.; Sakamoto, M.; Matsuki, T. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014, 345, 1254009. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Maurice, C.F.; Kinnebrew, M.A.; Abt, M.C.; Schenten, D.; Golovkina, T.V.; Bogatyrev, S.R.; Ismagilov, R.F.; Pamer, E.G.; Turnbaugh, P.J. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature 2014, 514, 638. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.; Ma, C.; Bergstrom, K.; Huang, J.; Man, C.; Vallance, B. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell. Microbiol. 2008, 10, 618–631. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptor and RIG-1-like receptor signaling. Ann. N. Y. Acad. Sci. 2008, 1143, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Bommarius, B.; Parkos, C.A.; Sherman, M.A.; Kalman, D. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J. Immunol. 2007, 179, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.P.; Clare, S.; Ghaem-Maghami, M.; Uren, T.K.; Rankin, J.; Huett, A.; Goldin, R.; Lewis, D.J.; MacDonald, T.T.; Strugnell, R.A. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect. Immun. 2003, 71, 5077–5086. [Google Scholar] [CrossRef]
- Vallance, B.A.; Deng, W.; Knodler, L.A.; Finlay, B.B. Mice lacking T and B lymphocytes develop transient colitis and crypt hyperplasia yet suffer impaired bacterial clearance during Citrobacter rodentium infection. Infect. Immun. 2002, 70, 2070–2081. [Google Scholar] [CrossRef]
- Higgins, L.M.; Frankel, G.; Douce, G.; Dougan, G.; MacDonald, T.T. Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect. Immun. 1999, 67, 3031–3039. [Google Scholar] [CrossRef]
- Simmons, C.P.; Goncalves, N.S.; Ghaem-Maghami, M.; Bajaj-Elliott, M.; Clare, S.; Neves, B.; Frankel, G.; Dougan, G.; MacDonald, T.T. Impaired resistance and enhanced pathology during infection with a noninvasive, attaching-effacing enteric bacterial pathogen, Citrobacter rodentium, in mice lacking IL-12 or IFN-γ. J. Immunol. 2002, 168, 1804–1812. [Google Scholar] [CrossRef]
- Ishigame, H.; Kakuta, S.; Nagai, T.; Kadoki, M.; Nambu, A.; Komiyama, Y.; Fujikado, N.; Tanahashi, Y.; Akitsu, A.; Kotaki, H. Differential roles of interleukin-17A and-17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 2009, 30, 108–119. [Google Scholar] [CrossRef]
- Liang, S.C.; Tan, X.-Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9, 265. [Google Scholar] [CrossRef] [PubMed]
- Geddes, K.; Rubino, S.J.; Magalhaes, J.G.; Streutker, C.; Le Bourhis, L.; Cho, J.H.; Robertson, S.J.; Kim, C.J.; Kaul, R.; Philpott, D.J. Identification of an innate T helper type 17 response to intestinal bacterial pathogens. Nat. Med. 2011, 17, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Valdez, P.A.; Danilenko, D.M.; Hu, Y.; Sa, S.M.; Gong, Q.; Abbas, A.R.; Modrusan, Z.; Ghilardi, N.; De Sauvage, F.J. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008, 14, 282–289. [Google Scholar] [CrossRef]
- Li, L.; Shi, Q.G.; Lin, F.; Liang, Y.G.; Sun, L.J.; Mu, J.S.; Wang, Y.G.; Su, H.B.; Xu, B.; Ji, C.C. Cytokine IL-6 is required in Citrobacter rodentium infection-induced intestinal Th17 responses and promotes IL-22 expression in inflammatory bowel disease. Mol. Med. Rep. 2014, 9, 831–836. [Google Scholar] [CrossRef]
- Molenaar, R.; Knippenberg, M.; Goverse, G.; Olivier, B.J.; de Vos, A.F.; O’Toole, T.; Mebius, R.E. Expression of retinaldehyde dehydrogenase enzymes in mucosal dendritic cells and gut-draining lymph node stromal cells is controlled by dietary vitamin A. J. Immunol. 2011, 186, 1934–1942. [Google Scholar] [CrossRef]
- Ross, A.C.; Zolfaghari, R.; Weisz, J. Vitamin A: Recent advances in the biotransformation, transport, and metabolism of retinoids. Curr. Opin. Gastroenterol. 2001, 17, 184–192. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Zolfaghari, R.; Ross, A.C. Regulation of CYP26 (cytochrome P450RAI) mRNA expression and retinoic acid metabolism by retinoids and dietary vitamin A in liver of mice and rats. FASEB J. 2000, 14, 2119–2127. [Google Scholar] [CrossRef]
- Wu, L.; Guo, X.; Hartson, S.D.; Davis, M.A.; He, H.; Medeiros, D.M.; Wang, W.; Clarke, S.L.; Lucas, E.A.; Smith, B.J. Lack of β, β-carotene-9′, 10′-oxygenase 2 leads to hepatic mitochondrial dysfunction and cellular oxidative stress in mice. Mol. Nutr. Food Res. 2017, 61, 1600576. [Google Scholar] [CrossRef]
- Lane, M.A.; Xu, J.; Wilen, E.W.; Sylvester, R.; Derguini, F.; Gudas, L.J. LIF removal increases CRABPI and CRABPII transcripts in embryonic stem cells cultured in retinol or 4-oxoretinol. Mol. Cell. Endocrinol. 2008, 280, 63–74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Napoli, J.L. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2012, 1821, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Said, H.M. Recent advances in transport of water-soluble vitamins in organs of the digestive system: A focus on the colon and the pancreas. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G601–G610. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, C.-H.; Hodges, J.K.; Green, M.H.; Ross, A.C. Priming with Retinoic Acid, an Active Metabolite of Vitamin A, Increases Vitamin A Uptake in the Small Intestine of Neonatal Rats. Nutrients 2021, 13, 4275. [Google Scholar] [CrossRef] [PubMed]
- De Barboza, G.D.; Guizzardi, S.; de Talamoni, N.T. Molecular aspects of intestinal calcium absorption. World J. Gastroenterol. WJG 2015, 21, 7142. [Google Scholar] [CrossRef]
- Tang, L.; Cheng, C.Y.; Sun, X.; Pedicone, A.J.; Mohamadzadeh, M.; Cheng, S.X. The extracellular calcium-sensing receptor in the intestine: Evidence for regulation of colonic absorption, secretion, motility, and immunity. Front. Physiol. 2016, 7, 245. [Google Scholar]
- Cantorna, M.T.; Rogers, C.J.; Arora, J. Aligning the Paradoxical Role of Vitamin D in Gastrointestinal Immunity. Trends Endocrinol. Metab. 2019, 30, 459–466. [Google Scholar] [CrossRef]
- Lin, Y.-D.; Arora, J.; Diehl, K.; Bora, S.; Cantorna, M. Vitamin D is required for ILC3 derived IL-22 and protection from Citrobacter rodentium infection. Front. Immunol. 2019, 10, 1. [Google Scholar] [CrossRef]
- Khan, M.A.; Ma, C.; Knodler, L.A.; Valdez, Y.; Rosenberger, C.M.; Deng, W.; Finlay, B.B.; Vallance, B.A. Toll-like receptor 4 contributes to colitis development but not to host defense during Citrobacter rodentium infection in mice. Infect. Immun. 2006, 74, 2522–2536. [Google Scholar] [CrossRef]
- Borenshtein, D.; Nambiar, P.R.; Groff, E.B.; Fox, J.G.; Schauer, D.B. Development of fatal colitis in FVB mice infected with Citrobacter rodentium. Infect. Immun. 2007, 75, 3271–3281. [Google Scholar] [CrossRef]
- Hecht, G. Innate mechanisms of epithelial host defense: Spotlight on intestine. Am. J. Physiol. Cell Physiol. 1999, 277, C351–C358. [Google Scholar] [CrossRef] [PubMed]
- Höglund, P.; Haila, S.; Socha, J.; Tomaszewski, L.; Saarialho-Kere, U.; Karjalainen-Lindsberg, M.-L.; Airola, K.; Holmberg, C.; de la Chapelle, A.; Kere, J. Mutations of the Down–regulated in adenoma (DRA) gene cause congenital chloride diarrhoea. Nat. Genet. 1996, 14, 316. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chatterjee, I.; Gujral, T.; Alakkam, A.; Coffing, H.; Anbazhagan, A.N.; Borthakur, A.; Saksena, S.; Gill, R.K.; Alrefai, W.A. Activation of Nuclear Factor− κB by Tumor Necrosis Factor in Intestinal Epithelial Cells and Mouse Intestinal Epithelia Reduces Expression of the Chloride Transporter SLC26A3. Gastroenterology 2017, 153, 1338–1350.e3. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.; Faria, D.; Skryabin, B.V.; Wanitchakool, P.; Rock, J.R.; Kunzelmann, K. Anoctamins support calcium-dependent chloride secretion by facilitating calcium signaling in adult mouse intestine. Pflügers Arch. Eur. J. Physiol. 2015, 467, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Zachos, N.C.; Tse, M.; Donowitz, M. Molecular physiology of intestinal N+/H+ exchange. Annu. Rev. Physiol. 2005, 67, 411–443. [Google Scholar] [CrossRef]
- Kunzelmann, K.; Mall, M. Electrolyte transport in the mammalian colon: Mechanisms and implications for disease. Physiol. Rev. 2002, 82, 245–289. [Google Scholar] [CrossRef]
- Pang, T.; Su, X.; Wakabayashi, S.; Shigekawa, M. Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J. Biol. Chem. 2001, 276, 17367–17372. [Google Scholar] [CrossRef]
- Stewart, A.; Boyd, C.; Vaughan-Jones, R. A novel role for carbonic anhydrase: Cytoplasmic pH gradient dissipation in mouse small intestinal enterocytes. J. Physiol. 1999, 516, 209–217. [Google Scholar] [CrossRef]
- Charney, A.N.; Dagher, P.C. Acid-base effects on colonic electrolyte transport revisited. Gastroenterology 1996, 111, 1358–1368. [Google Scholar] [CrossRef]
- Boedtkjer, E.; Praetorius, J.; Fuchtbauer, E.-M.; Aalkjaer, C. Antibody-independent localization of the electroneutral Na+-HCO3−cotransporter NBCn1 (slc4a7) in mice. Am. J. Physiol. Cell Physiol. 2008, 294, C591–C603. [Google Scholar] [CrossRef]
- Chen, M.; Praetorius, J.; Zheng, W.; Xiao, F.; Riederer, B.; Singh, A.K.; Stieger, N.; Wang, J.; Shull, G.E.; Aalkjaer, C. The electroneutral Na+:HCO3−cotransporter NBCn1 is a major pHi regulator in murine duodenum. J. Physiol. 2012, 590, 3317–3333. [Google Scholar] [CrossRef] [PubMed]
- Seidler, U.E.; Singh, A.K.; Xia, W.; Riederer, B.; Juric, M.; Li, J.; Zheng, W.; Xiao, F.; Bachmann, O.; Praetorius, J. The electroneutral Na+HCO3− cotransporter NBCn1 plays an essential role in duodenal acid/base balance and colonic mucus layer build-up in anaestetised mice. Faseb J. 2013, 27, 730. [Google Scholar] [CrossRef]


| Biological Process | Ref. | Evidence, VAS-Inf | Evidence Scenario 1 | Evidence Scenario 4 |
|---|---|---|---|---|
| Epithelial hyperplasia | [58] | “mitotic nuclear division”, “DNA replication”, and “cholesterol biosynthetic process” | “mitotic spindle organization” | |
| Apoptosis | [13,14,58] | “activation of cysteine-type endopeptidase activity involved in apoptotic process” | caspases 1, 3, 4, and 7, “cysteine-type endopeptidase activity involved in apoptotic process” | |
| Goblet cell depletion | [6,18] | Muc 1, 2, and 4, chloride channel accessory family that are products of goblet cells (Clca3a1, Clca3a2, Clca3b, Clca4a, and Clca4b), and enzymes that may affect the glycosylation of mucins (Glt28d2 [59] and Fut2 [60,61,62] | Muc4, Agr2 | |
| Innate immune response | [63,64,65] | “neutrophil chemotaxis,” “monocyte chemotaxis,” and “regulation of leukocyte migration” | Tlr2, Myd88, Cxcl9, and Ccl8 | “natural killer cell mediated immunity,” “leukocyte tethering or rolling,” and “antigen processing and presentation” |
| T cell response | [66,67,68,69] | “adaptive immune response,” “lymphocyte mediated immunity,” “response to interferon-gamma,” and “T cell mediated cytotoxicity,” | “T cell mediated cytotoxicity,” “leukocyte mediated cytotoxicity,” “T cell mediated immunity,” and “leukocyte mediated immunity” | “T cell receptor signaling pathway,” and “integrin complex” |
| IL-17 response | [11,12,24,26,70,71,72] | nitric oxide synthase 2 (Nos2), dual oxidase 2 (Duox2), and dual oxidase maturation factor 2 (Duoxa2), Cebpd, Saa 2, 3, and 4; “reactive oxygen species metabolic process” and “IL-17 signaling pathway” | “Th17 cell differentiation” |
| Upregulated during Infection | |||||
|---|---|---|---|---|---|
| Base Mean | Fold Change | padj | ENTREZID | Full Name | |
| Akr1c18 | 54.22 | 7.46 | 2.79 × 10−6 | 105,349 | aldo–keto reductase family 1 member C18 |
| Aldh1a3 | 1061.53 | 2.32 | 4.70 × 10−4 | 56,847 | aldehyde dehydrogenase family 1 subfamily A3 |
| Dhrs9 | 1262.32 | 17.44 | 2.60 × 10−29 | 241,452 | dehydrogenase/reductase (SDR family) member 9 |
| Rdh1 | 24.83 | 2.59 | 4.37 × 10−4 | 107,605 | retinol dehydrogenase 1 (all trans) |
| Downregulated during infection | |||||
| Adh1 | 29,054.77 | −2.19 | 8.36 × 10−6 | 11,522 | alcohol dehydrogenase 1 (class I) |
| Adh7 | 14.73 | −3.50 | 4.95 × 10−3 | 11,529 | alcohol dehydrogenase 7 (class IV) mu or sigma polypeptide |
| Aldh1a1 | 505.92 | −2.12 | 3.20 × 10−4 | 11,668 | aldehyde dehydrogenase family 1 subfamily A1 |
| Aldh1a2 | 114.85 | −2.72 | 5.83 × 10−3 | 19,378 | aldehyde dehydrogenase family 1 subfamily A2 |
| Bco2 | 512.13 | −2.27 | 4.92 × 10−4 | 170,752 | beta-carotene oxygenase 2 |
| Crabp1 | 13.86 | −4.61 | 5.53 × 10−5 | 12,903 | cellular retinoic acid-binding protein I |
| Cyp26b1 | 41.71 | −5.57 | 2.94 × 10−6 | 232,174 | cytochrome P450 family 26 subfamily b polypeptide 1 |
| Lrat | 42.31 | −8.02 | 1.14 × 10−8 | 79,235 | lecithin retinol acyltransferase (phosphatidylcholine-retinol O-acyltransferase) |
| Rarb | 65.16 | −3.31 | 1.63 × 10−7 | 218,772 | retinoic acid receptor beta |
| Rbp2 | 237.32 | −3.23 | 9.94 × 10−8 | 19,660 | retinol-binding protein 2 cellular |
| Rdh5 | 120.00 | −2.24 | 8.23 × 10−9 | 19,682 | retinol dehydrogenase 5 |
| Rxrg | 18.21 | −10.12 | 9.98 × 10−9 | 20,183 | retinoid X receptor gamma |
| Stra6l | 517.21 | −3.29 | 1.76 × 10−14 | 74,152 | STRA6-like |
| Upregulated during Infection | |||||
|---|---|---|---|---|---|
| Base Mean | Fold Change | padj | ENTREZID | Full Name | |
| Trpv6 | 485.72 | 5.55 | 4.31 × 10−6 | 64,177 | transient receptor potential cation channel subfamily V member 6 |
| Downregulated during Infection | |||||
| Atp2b2 | 35.55 | −3.80 | 1.37 × 10−5 | 11,941 | ATPase Ca++-transporting plasma membrane 2 |
| Atp2b3 | 68.79 | −3.68 | 6.71 × 10−7 | 320,707 | ATPase Ca++-transporting plasma membrane 3 |
| Atp2b4 | 3441.90 | −4.27 | 1.68 × 10−12 | 381,290 | ATPase Ca++-transporting plasma membrane 4 |
| Cacna1d | 227.14 | −3.34 | 2.02 × 10−9 | 12,289 | calcium channel voltage-dependent L-type alpha 1D subunit |
| Cyp27a1 | 889.90 | −5.36 | 4.23 × 10−15 | 104,086 | cytochrome P450 family 27 subfamily a polypeptide 1 |
| S100g | 1483.02 | −3.49 | 4.79 × 10−2 | 12,309 | S100 calcium-binding protein G |
| Slc8a1 | 774.91 | −3.61 | 1.76 × 10−14 | 20,541 | solute carrier family 8 (sodium/calcium exchanger) member 1 |
| Vdr | 10,952.98 | −2.13 | 1.73 × 10−7 | 22,337 | vitamin D (1,25-dihydroxyvitamin D3) receptor |
| Upregulated during Infection | |||||
|---|---|---|---|---|---|
| Base Mean | Fold Change | padj | ENTREZID | Full Name | |
| Adora2b | 14.73 | 3.61 | 2.51 × 10−3 | 11,541 | adenosine A2b receptor |
| Aqp4 | 24,562.26 | 5.41 | 6.42 × 10−19 | 11,829 | aquaporin 4 |
| Atp12a | 21,642.30 | 2.38 | 1.36 × 10−2 | 192,113 | ATPase H+/K+-transporting non-gastric alpha polypeptide |
| Atp1b2 | 24,979.73 | 21.05 | 1.90 × 10−15 | 11,932 | ATPase Na+/K+-transporting beta 2 polypeptide |
| Atp2a2 | 14,010.80 | 2.36 | 1.84 × 10−12 | 11,938 | ATPase Ca++-transporting cardiac muscle slow twitch 2 |
| Car13 | 832.60 | 2.57 | 3.69 × 10−13 | 71,934 | carbonic anhydrase 13 |
| Car8 | 392.90 | 3.28 | 5.17 × 10−12 | 12,319 | carbonic anhydrase 8 |
| Clcn1 | 19.82 | 5.18 | 6.28 × 10−6 | 12,723 | chloride channel voltage-sensitive 1 |
| Clic1 | 11,306.30 | 2.58 | 6.81 × 10−47 | 114,584 | chloride intracellular channel 1 |
| Fosb | 3418.09 | 8.33 | 2.72 × 10−7 | 14,282 | FBJ osteosarcoma oncogene B |
| Slc9a3 | 695.50 | 2.75 | 8.18 × 10−3 | 105,243 | solute carrier family 9 (sodium/hydrogen exchanger) member 3 |
| Downregulated during infection | |||||
| Ano1 | 5571.44 | −2.65 | 1.79 × 10−10 | 101,772 | anoctamin 1 calcium-activated chloride channel |
| Ano2 | 23.72 | −8.24 | 2.21 × 10−8 | 243,634 | anoctamin 2 |
| Aqp1 | 1252.79 | −4.39 | 3.60 × 10−11 | 11,826 | aquaporin 1 |
| Aqp7 | 21.12 | −10.28 | 5.10 × 10−6 | 11,832 | aquaporin 7 |
| Best2 | 1165.72 | −3.66 | 2.27 × 10−9 | 212,989 | bestrophin 2 |
| Camk2a | 143.61 | −2.43 | 5.14 × 10−6 | 12,322 | calcium/calmodulin-dependent protein kinase II alpha |
| Car11 | 65.93 | −5.89 | 3.07 × 10−16 | 12,348 | carbonic anhydrase 11 |
| Car14 | 17.35 | −13.72 | 1.41 × 10−7 | 23,831 | carbonic anhydrase 14 |
| Car15 | 99.56 | −2.40 | 1.16 × 10−5 | 80,733 | carbonic anhydrase 15 |
| Car2 | 15,652.54 | −2.06 | 1.24 × 10−5 | 12,349 | carbonic anhydrase 2 |
| Car3 | 2010.26 | −4.40 | 4.29 × 10−3 | 12,350 | carbonic anhydrase 3 |
| Car4 | 18,070.39 | −2.68 | 1.21 × 10−2 | 12,351 | carbonic anhydrase 4 |
| Cftr | 3901.82 | −2.43 | 2.21 × 10−11 | 12,638 | cystic fibrosis transmembrane conductance regulator |
| Chp2 | 1689.23 | −2.26 | 9.99 × 10−7 | 70,261 | calcineurin-like EF hand protein 2 |
| Slc15a1 | 2813.63 | −5.26 | 7.65 × 10−19 | 56,643 | solute carrier family 15 (oligopeptide transporter) member 1 |
| Slc26a3 | 13,041.41 | −2.04 | 2.25 × 10−2 | 13,487 | solute carrier family 26 member 3 |
| Slc9a2 | 3212.65 | −2.22 | 1.25 × 10−5 | 226,999 | solute carrier family 9 (sodium/hydrogen exchanger) member 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, Z.; Lyu, Y.; Chen, Q.; Wei, C.-H.; Snyder, L.M.; Weaver, V.; Sebastian, A.; Albert, I.; Li, Q.; Cantorna, M.T.; et al. Transcriptional Profiling of the Small Intestine and the Colon Reveals Modulation of Gut Infection with Citrobacter rodentium According to the Vitamin A Status. Nutrients 2022, 14, 1563. https://doi.org/10.3390/nu14081563
Chai Z, Lyu Y, Chen Q, Wei C-H, Snyder LM, Weaver V, Sebastian A, Albert I, Li Q, Cantorna MT, et al. Transcriptional Profiling of the Small Intestine and the Colon Reveals Modulation of Gut Infection with Citrobacter rodentium According to the Vitamin A Status. Nutrients. 2022; 14(8):1563. https://doi.org/10.3390/nu14081563
Chicago/Turabian StyleChai, Zhi, Yafei Lyu, Qiuyan Chen, Cheng-Hsin Wei, Lindsay M. Snyder, Veronika Weaver, Aswathy Sebastian, István Albert, Qunhua Li, Margherita T. Cantorna, and et al. 2022. "Transcriptional Profiling of the Small Intestine and the Colon Reveals Modulation of Gut Infection with Citrobacter rodentium According to the Vitamin A Status" Nutrients 14, no. 8: 1563. https://doi.org/10.3390/nu14081563
APA StyleChai, Z., Lyu, Y., Chen, Q., Wei, C.-H., Snyder, L. M., Weaver, V., Sebastian, A., Albert, I., Li, Q., Cantorna, M. T., & Ross, C. (2022). Transcriptional Profiling of the Small Intestine and the Colon Reveals Modulation of Gut Infection with Citrobacter rodentium According to the Vitamin A Status. Nutrients, 14(8), 1563. https://doi.org/10.3390/nu14081563