Vitamin D and Its Target Genes
Abstract
:1. Introduction
2. VDR: A Transcription Factor Activated by Vitamin D
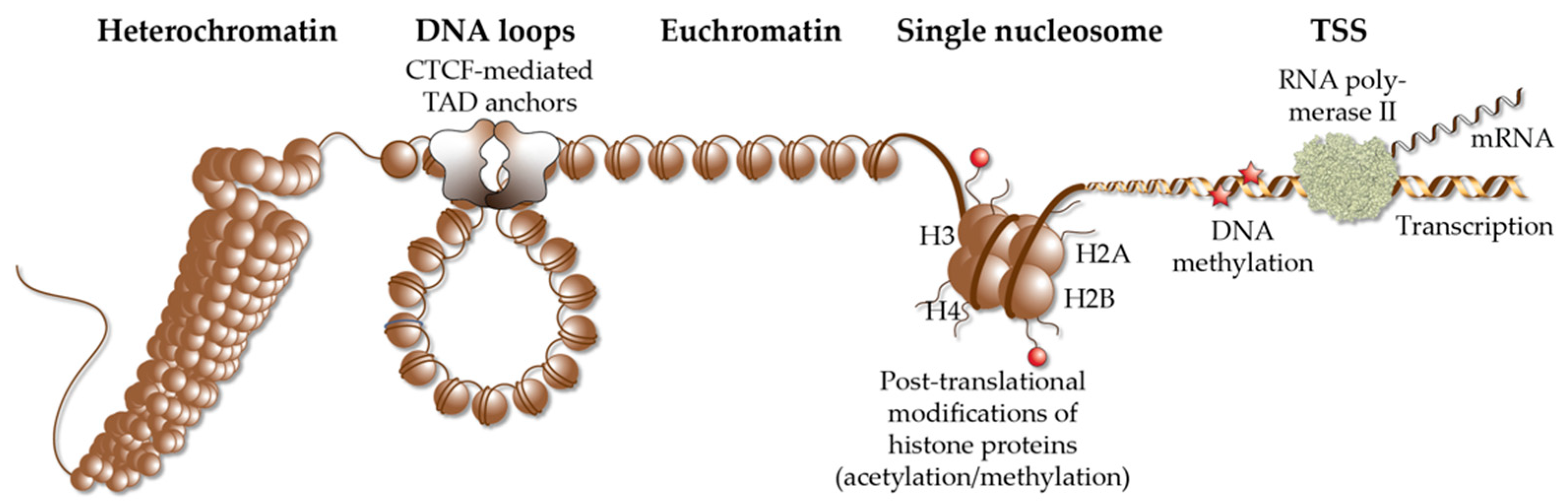
3. Vitamin D Target Gene Regulation in the Context of Chromatin
- VDR initiates the demethylation of its binding sites via interaction with TET2 [79];
- The binding of CTCF, to more than 1000 of its genomic sites, is modulated by 1,25(OH)2D3 [82];
- The organization of some 400 TADs is dependent on 1,25(OH)2D3 [82], i.e., vitamin D affects the 3-dimensional chromatin structure.
4. Genome-Wide Location of VDR
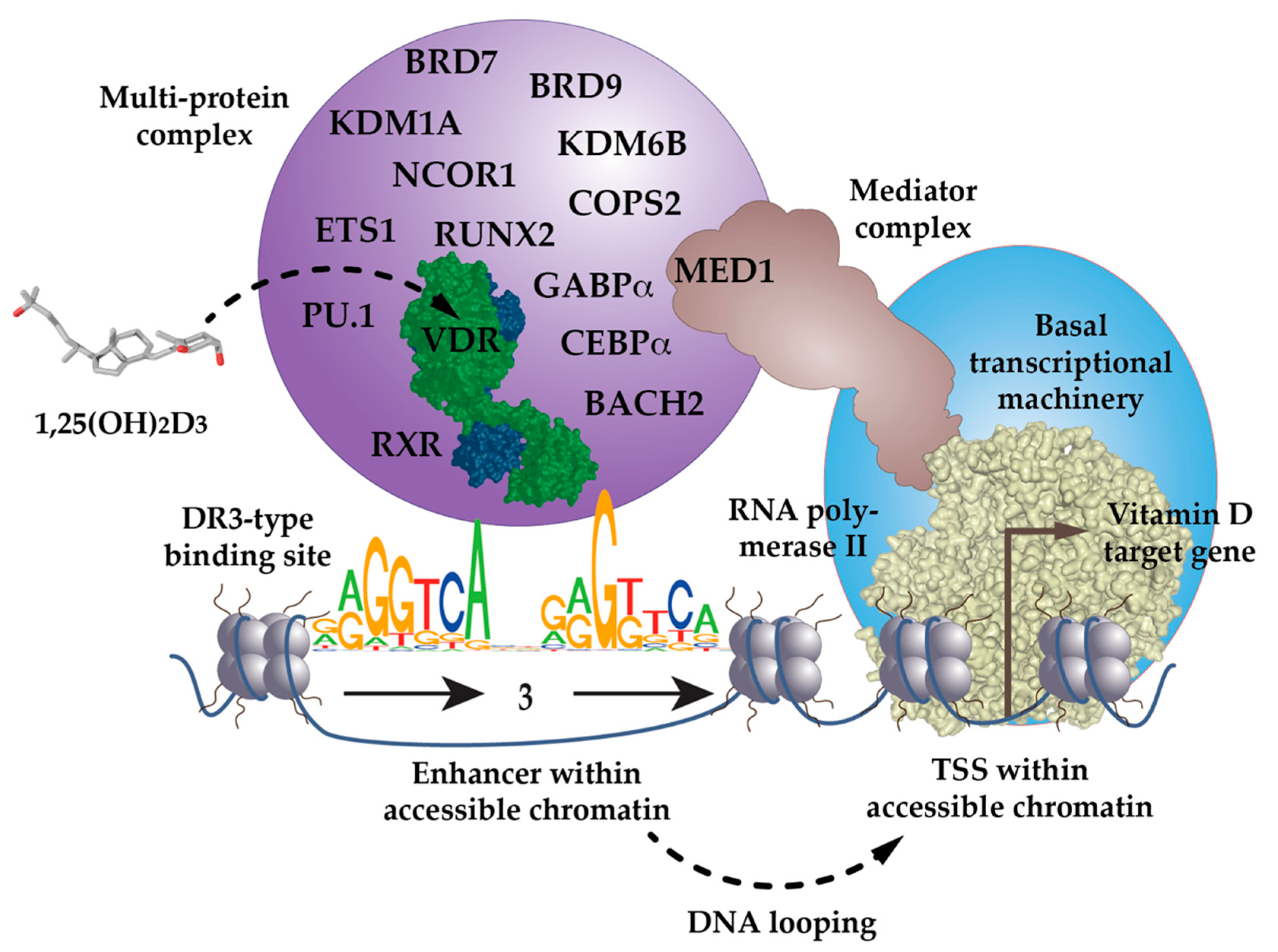
5. Model of Vitamin D Signaling
- The pioneer factors within the complex may take the first contact to enhancer regions. With the help of chromatin-remodeling proteins, they optimize the access of VDR to suitable binding motifs within the enhancer region, including the demethylation of genomic DNA.
- Chromatin modifiers within the complex then leave marks, such as H3K27ac, to the local chromatin region.
- Although VDR may not be the first protein of the complex making contact with the enhancer region, its specific activation by 1,25(OH)2D3 drives the activity of the other members of the complex. This may explain the epigenetic effects of 1,25(OH)2D3, such as chromatin opening, histone marks and the recruitment of pioneer factors.
- When the complex is established on the enhancer region, DNA looping events to TSS regions within the same TAD region, which are complexed with a basal transcriptional machinery, become stabilized. Via 1,25(OH)2D3-triggered effects on CTCF-dependent TAD anchor formation, this also affects the structure of the whole TAD.
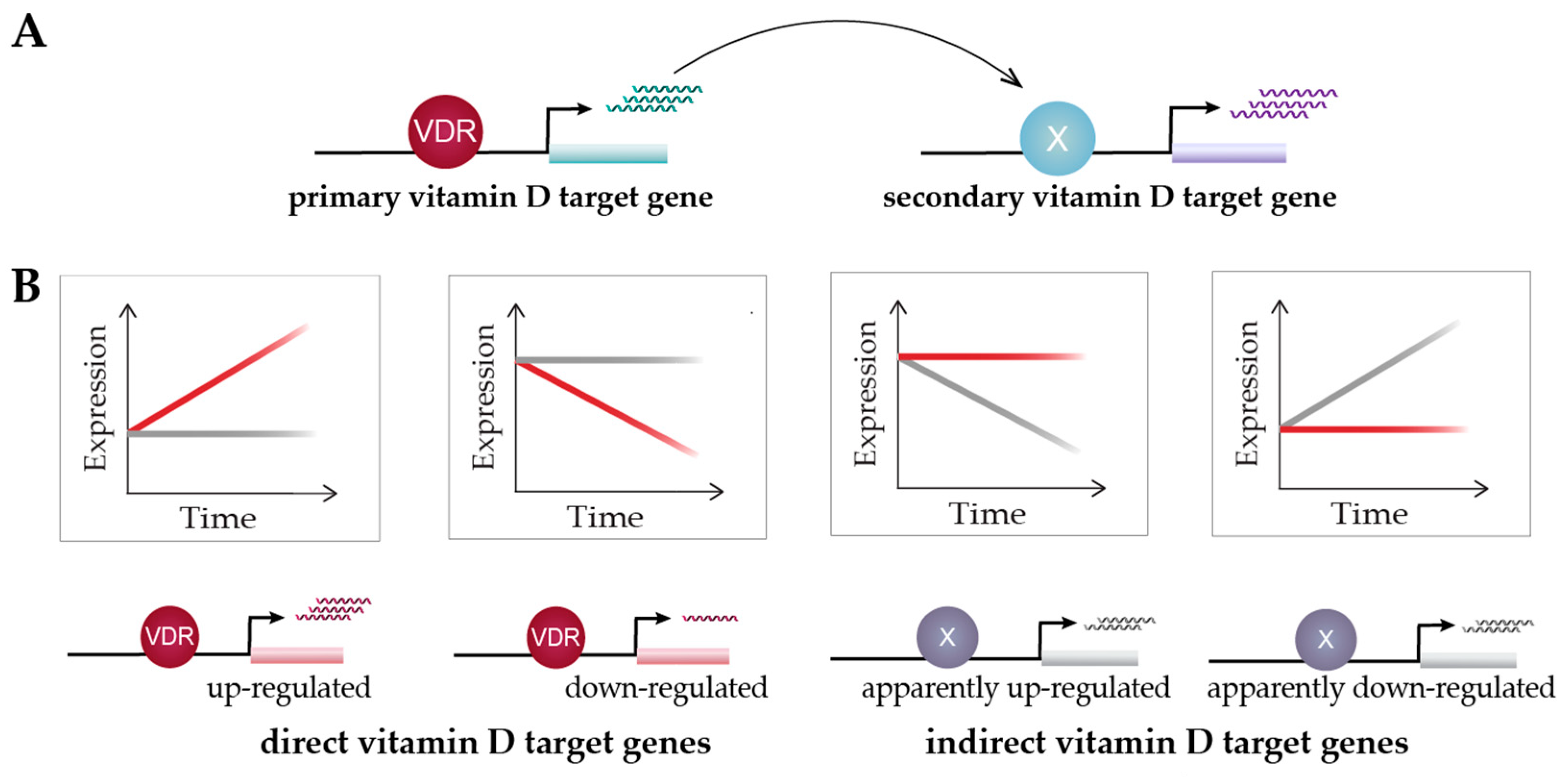
6. Vitamin D Target Genes
7. Functional Profile of Vitamin D Target Genes
8. Conclusions and Future View
Funding
Acknowledgments
Conflicts of Interest
References
- McMollum, E.V.; Simmonds, N.; Becker, J.E.; Shipley, P.G. Studies on experimental rickets: An experimental demonstration of the existence of a vitamin which promotes calcium deposition. J. Biol. Chem. 1922, 52, 293–298. [Google Scholar]
- Holick, M.F. Resurrection of vitamin D deficiency and rickets. J. Clin. Investig. 2006, 116, 2062–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renkema, K.Y.; Alexander, R.T.; Bindels, R.J.; Hoenderop, J.G. Calcium and phosphate homeostasis: Concerted interplay of new regulators. Ann. Med. 2008, 40, 82–91. [Google Scholar] [PubMed]
- Tsai, H.C.; Norman, A.W. Studies on calciferol metabolism. 8. Evidence for a cytoplasmic receptor for 1,25-dihydroxyvitamin D3 in the intestinal mucosa. J. Biol. Chem. 1973, 248, 5967–5975. [Google Scholar] [PubMed]
- Brumbaugh, P.F.; Hughes, M.R.; Haussler, M.R. Cytoplasmic and nuclear binding components for 1α,25-dihydroxyvitamin D3 in chick parathyroid glands. Proc. Natl. Acad. Sci. USA 1975, 72, 4871–4875. [Google Scholar]
- McDonnell, D.P.; Mangelsdorf, D.J.; Pike, J.W.; Haussler, M.R.; O’Malley, B.W. Molecular cloning of complementary DNA encoding the avian receptor for vitamin D. Science 1987, 235, 1214–1217. [Google Scholar]
- Baker, A.R.; McDonnell, D.P.; Hughes, M.; Crisp, T.M.; Mangelsdorf, D.J.; Haussler, M.R.; Pike, J.W.; Shine, J.; O’Malley, B. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 3294–3298. [Google Scholar]
- Evans, R.; Mangelsdorf, D. Nuclear Receptors, RXR, and the Big Bang. Cell 2014, 157, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Evans, R.M. The nuclear receptor superfamily: A rosetta stone for physiology. Mol. Endocrinol. 2005, 19, 1429–1438. [Google Scholar] [CrossRef]
- Carlberg, C.; Polly, P. Gene regulation by vitamin D3. Crit. Rev. Eukaryot. Gene Expr. 1998, 8, 19–42. [Google Scholar]
- Haussler, M.R.; Haussler, C.A.; Jurutka, P.W.; Thompson, P.D.; Hsieh, J.C.; Remus, L.S.; Selznick, S.H.; Whitfield, G.K. The vitamin D hormone and its nuclear receptor: Molecular actions and disease states. J. Endocrinol. 1997, 154, S57–S73. [Google Scholar] [PubMed]
- Pike, J.W. Vitamin D3 receptors: Structure and function in transcription. Annu. Rev. Nutr. 1991, 11, 189–216. [Google Scholar] [PubMed]
- Veldurthy, V.; Wei, R.; Oz, L.; Dhawan, P.; Jeon, Y.H.; Christakos, S. Vitamin D, calcium homeostasis and aging. Bone Res. 2016, 4, 16041. [Google Scholar] [CrossRef] [Green Version]
- Muller, V.; de Boer, R.J.; Bonhoeffer, S.; Szathmary, E. An evolutionary perspective on the systems of adaptive immunity. Biol. Rev. Camb. Philos. Soc. 2018, 93, 505–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanherwegen, A.S.; Gysemans, C.; Mathieu, C. Vitamin D endocrinology on the cross-road between immunity and metabolism. Mol. Cell. Endocrinol. 2017, 453, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Cortes, M.; Chen, M.J.; Stachura, D.L.; Liu, S.Y.; Kwan, W.; Wright, F.; Vo, L.T.; Theodore, L.N.; Esain, V.; Frost, I.M.; et al. Developmental vitamin D availability impacts hematopoietic stem cell production. Cell Rep. 2016, 17, 458–468. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C. Vitamin D genomics: From in vitro to in vivo. Front. Endocrinol. 2018, 9, 250. [Google Scholar] [CrossRef]
- Khanal, R.C.; Nemere, I. The ERp57/GRp58/1,25D3-MARRS receptor: Multiple functional roles in diverse cell systems. Curr. Med. Chem. 2007, 14, 1087–1093. [Google Scholar] [CrossRef]
- Zmijewski, M.A.; Carlberg, C. Vitamin D receptor(s): In the nucleus but also at membranes? Exp. Dermatol. 2020, 29, 876–884. [Google Scholar] [CrossRef]
- van Etten, E.; Stoffels, K.; Gysemans, C.; Mathieu, C.; Overbergh, L. Regulation of vitamin D homeostasis: Implications for the immune system. Nutr. Rev. 2008, 66, S125–S134. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [PubMed]
- Carlberg, C.; Molnár, F. Transcription factors. In Mechanisms of Gene Regulation, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 57–73. [Google Scholar]
- Huang, P.; Chandra, V.; Rastinejad, F. Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu. Rev. Physiol. 2010, 72, 247–272. [Google Scholar] [PubMed] [Green Version]
- Carlberg, C.; Molnár, F. Switching genes on and off: The example of nuclear receptors. In Mechanisms of Gene Regulation, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 95–108. [Google Scholar]
- Molnár, F.; Peräkylä, M.; Carlberg, C. Vitamin D receptor agonists specifically modulate the volume of the ligand-binding pocket. J. Biol. Chem. 2006, 281, 10516–10526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef]
- Molnár, F. Structural considerations of vitamin D signaling. Front. Physiol. 2014, 5, 191. [Google Scholar] [CrossRef] [Green Version]
- Tagami, T.; Lutz, W.H.; Kumar, R.; Jameson, J.L. The interaction of the vitamin D receptor with nuclear receptor corepressors and coactivators. Biochem. Biophys. Res. Commun. 1998, 253, 358–363. [Google Scholar]
- Polly, P.; Herdick, M.; Moehren, U.; Baniahmad, A.; Heinzel, T.; Carlberg, C. VDR-Alien: A novel, DNA-selective vitamin D3 receptor-corepressor partnership. FASEB J. 2000, 14, 1455–1463. [Google Scholar]
- Herdick, M.; Carlberg, C. Agonist-triggered modulation of the activated and silent state of the vitamin D3 receptor by interaction with co-repressors and co-activators. J. Mol. Biol. 2000, 304, 793–801. [Google Scholar]
- Rachez, C.; Lemon, B.D.; Suldan, Z.; Bromleigh, V.; Gamble, M.; Näär, A.M.; Erdjument-Bromage, H.; Tempst, P.; Freedman, L.P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 1999, 398, 824–828. [Google Scholar]
- Belorusova, A.Y.; Bourguet, M.; Hessmann, S.; Chalhoub, S.; Kieffer, B.; Cianferani, S.; Rochel, N. Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer. Nucleic Acids Res. 2020, 48, 11199–11213. [Google Scholar] [CrossRef]
- Yuan, C.-X.; Ito, M.; Fondell, J.D.; Fu, Z.-Y.; Roeder, R.G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 1998, 95, 7939–7944. [Google Scholar] [PubMed] [Green Version]
- Pereira, F.; Barbachano, A.; Silva, J.; Bonilla, F.; Campbell, M.J.; Munoz, A.; Larriba, M.J. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum. Mol. Genet. 2011, 20, 4655–4665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battaglia, S.; Karasik, E.; Gillard, B.; Williams, J.; Winchester, T.; Moser, M.T.; Smiraglia, D.J.; Foster, B.A. LSD1 dual function in mediating epigenetic corruption of the vitamin D signaling in prostate cancer. Clin. Epigenet. 2017, 9, 82. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Yoshihara, E.; He, N.; Hah, N.; Fan, W.; Pinto, A.F.M.; Huddy, T.; Wang, Y.; Ross, B.; Estepa, G.; et al. Vitamin D switches BAF complexes to protect beta cells. Cell 2018, 173, 1135–1149.e15. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Pertile, R.; Eyles, D.W. The vitamin D receptor (VDR) binds to the nuclear matrix via its hinge domain: A potential mechanism for the reduction in VDR mediated transcription in mitotic cells. Mol. Cell. Endocrinol. 2018, 472, 18–25. [Google Scholar] [CrossRef]
- Carlberg, C.; Bendik, I.; Wyss, A.; Meier, E.; Sturzenbecker, L.J.; Grippo, J.F.; Hunziker, W. Two nuclear signalling pathways for vitamin D. Nature 1993, 361, 657–660. [Google Scholar] [CrossRef]
- Sone, T.; Ozono, K.; Pike, J.W. A 55-kilodalton accessory factor facilitates vitamin D receptor DNA binding. Mol. Endocrinol. 1991, 5, 1578–1586. [Google Scholar]
- Liao, J.; Ozano, K.; Sone, T.; McDonnell, D.P.; Pike, J.W. Vitamin D receptor requires a nuclear protein and 1,25-dihydroxyvitamin D3. Proc. Natl. Acad. Sci. USA 1990, 87, 9751–9755. [Google Scholar]
- Shaffer, P.L.; Gewirth, D.T. Structural analysis of RXR-VDR interactions on DR3 DNA. J. Steroid Biochem. Mol. Biol. 2004, 89–90, 215–219. [Google Scholar]
- Umesono, K.; Murakami, K.K.; Thompson, C.C.; Evans, R.M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 1991, 65, 1255–1266. [Google Scholar]
- Ozono, K.; Liao, J.; Kerner, S.A.; Scott, R.A.; Pike, J.W. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J. Biol. Chem. 1990, 265, 21881–21888. [Google Scholar] [PubMed]
- Tuoresmäki, P.; Väisänen, S.; Neme, A.; Heikkinen, S.; Carlberg, C. Patterns of genome-wide VDR locations. PLoS ONE 2014, 9, e96105. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [PubMed] [Green Version]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta 2017, 1860, 952–961. [Google Scholar] [CrossRef] [Green Version]
- Schräder, M.; Bendik, I.; Becker-Andre, M.; Carlberg, C. Interaction between retinoic acid and vitamin D signaling pathways. J. Biol. Chem. 1993, 268, 17830–17836. [Google Scholar] [PubMed]
- Schräder, M.; Müller, K.M.; Carlberg, C. Specificity and flexibility of vitamin D signaling.: Modulation of the activation of natural vitamin D response elements by thyroid hormone. J. Biol. Chem. 1994, 269, 5501–5504. [Google Scholar]
- Schräder, M.; Müller, K.M.; Nayeri, S.; Kahlen, J.P.; Carlberg, C. VDR-T3R receptor heterodimer polarity directs ligand sensitivity of transactivation. Nature 1994, 370, 382–386. [Google Scholar]
- Carlberg, C.; Molnár, F. Vitamin D receptor signaling and its therapeutic implications: Genome-wide and structural view. Can. J. Physiol. Pharmacol. 2015, 93, 311–318. [Google Scholar] [CrossRef] [Green Version]
- Seuter, S.; Neme, A.; Carlberg, C. Epigenomic PU.1-VDR crosstalk modulates vitamin D signaling. Biochim. Biophys. Acta 2017, 1860, 405–415. [Google Scholar] [CrossRef]
- Novershtern, N.; Subramanian, A.; Lawton, L.N.; Mak, R.H.; Haining, W.N.; McConkey, M.E.; Habib, N.; Yosef, N.; Chang, C.Y.; Shay, T.; et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 2011, 144, 296–309. [Google Scholar] [CrossRef] [Green Version]
- Nurminen, V.; Neme, A.; Seuter, S.; Carlberg, C. Modulation of vitamin D signaling by the pioneer factor CEBPA. Biochim. Biophys. Acta 2019, 1862, 96–106. [Google Scholar] [CrossRef]
- Seuter, S.; Neme, A.; Carlberg, C. ETS transcription factor family member GABPA contributes to vitamin D receptor target gene regulation. J. Steroid Biochem. Mol. Biol. 2018, 177, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Warwick, T.; Schulz, M.H.; Gunther, S.; Gilsbach, R.; Neme, A.; Carlberg, C.; Brandes, R.P.; Seuter, S. A hierarchical regulatory network analysis of the vitamin D induced transcriptome reveals novel regulators and complete VDR dependency in monocytes. Sci. Rep. 2021, 11, 6518. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.B.; Benkusky, N.A.; Sen, B.; Rubin, J.; Pike, J.W. Epigenetic plasticity drives adipogenic and osteogenic differentiation of marrow-derived mesenchymal stem cells. J. Biol. Chem. 2016, 291, 17829–17847. [Google Scholar] [CrossRef] [Green Version]
- Chauss, D.; Freiwald, T.; McGregor, R.; Yan, B.; Wang, L.; Nova-Lamperti, E.; Kumar, D.; Zhang, Z.; Teague, H.; West, E.E.; et al. Autocrine vitamin D signaling switches off pro-inflammatory programs of TH1 cells. Nat. Immunol. 2022, 23, 62–74. [Google Scholar] [CrossRef]
- Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Kenneth Baillie, J.; de Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef]
- ENCODE-Project-Consortium; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Ali, T.; Renkawitz, R.; Bartkuhn, M. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016, 37, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghirlando, R.; Felsenfeld, G. CTCF: Making the right connections. Genes Dev. 2016, 30, 881–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenfeld, G.; Burgess-Beusse, B.; Farrell, C.; Gaszner, M.; Ghirlando, R.; Huang, S.; Jin, C.; Litt, M.; Magdinier, F.; Mutskov, V.; et al. Chromatin boundaries and chromatin domains. Cold Spring Harb. Symp. Quant. Biol. 2004, 69, 245–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlberg, C.; Molnár, F. The impact of chromatin. In Mechanisms of Gene Regulation, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Bell, O.; Tiwari, V.K.; Thoma, N.H.; Schubeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011, 12, 554–564. [Google Scholar] [CrossRef]
- Song, L.; Crawford, G.E. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5384. [Google Scholar] [CrossRef] [Green Version]
- Giresi, P.G.; Kim, J.; McDaniell, R.M.; Iyer, V.R.; Lieb, J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007, 17, 877–885. [Google Scholar]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Carlberg, C.; Molnár, F. The epigenome. In Mechanisms of Gene Regulation, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Carlberg, C.; Molnár, F. Chromatin modifiers. In Mechanisms of Gene Regulation, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Roadmap Epigenomics, C.; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C.; Molnár, F. Human Epigenetics: How Science Works; Springer: New York, NY, USA, 2019. [Google Scholar]
- Nurminen, V.; Neme, A.; Seuter, S.; Carlberg, C. The impact of the vitamin D-modulated epigenome on VDR target gene regulation. Biochim. Biophys. Acta 2018, 1861, 697–705. [Google Scholar] [CrossRef]
- Catala-Moll, F.; Ferrete-Bonastre, A.G.; Godoy-Tena, G.; Morante-Palacios, O.; Ciudad, L.; Barbera, L.; Fondelli, F.; Martinez-Caceres, E.M.; Rodriguez-Ubreva, J.; Li, T.; et al. Vitamin D receptor, STAT3, and TET2 cooperate to establish tolerogenesis. Cell Rep. 2022, 38, 110244. [Google Scholar] [CrossRef] [PubMed]
- Seuter, S.; Neme, A.; Carlberg, C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016, 44, 4090–4104. [Google Scholar] [CrossRef] [Green Version]
- Seuter, S.; Pehkonen, P.; Heikkinen, S.; Carlberg, C. Dynamics of 1α,25-dihydroxyvitamin D-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim. Biophys. Acta 2013, 1829, 1266–1275. [Google Scholar] [CrossRef]
- Neme, A.; Seuter, S.; Carlberg, C. Vitamin D-dependent chromatin association of CTCF in human monocytes. Biochim. Biophys. Acta 2016, 1859, 1380–1388. [Google Scholar] [CrossRef]
- Nurminen, V.; Neme, A.; Ryynanen, J.; Heikkinen, S.; Seuter, S.; Carlberg, C. The transcriptional regulator BCL6 participates in the secondary gene regulatory response to vitamin D. Biochim. Biophys. Acta 2015, 1849, 300–308. [Google Scholar] [CrossRef]
- Nurminen, V.; Seuter, S.; Carlberg, C. Primary vitamin D target genes of human monocytes. Front. Physiol. 2019, 10, 194. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C.; Campbell, M.J. Vitamin D receptor signaling mechanisms: Integrated actions of a well-defined transcription factor. Steroids 2013, 78, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Ramagopalan, S.V.; Heger, A.; Berlanga, A.J.; Maugeri, N.J.; Lincoln, M.R.; Burrell, A.; Handunnetthi, L.; Handel, A.E.; Disanto, G.; Orton, S.M.; et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010, 20, 1352–1360. [Google Scholar]
- Handel, A.E.; Sandve, G.K.; Disanto, G.; Berlanga-Taylor, A.J.; Gallone, G.; Hanwell, H.; Drablos, F.; Giovannoni, G.; Ebers, G.C.; Ramagopalan, S.V. Vitamin D receptor ChIP-seq in primary CD4+ cells: Relationship to serum 25-hydroxyvitamin D levels and autoimmune disease. BMC Med. 2013, 11, 163. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Goetsch, P.D.; Pike, J.W. VDR/RXR and TCF4/beta-catenin cistromes in colonic cells of colorectal tumor origin: Impact on c-FOS and c-MYC gene expression. Mol. Endocrinol. 2012, 26, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Fleet, J.C.; Kovalenko, P.L.; Li, Y.; Smolinski, J.; Spees, C.; Yu, J.G.; Thomas-Ahner, J.M.; Cui, M.; Neme, A.; Carlberg, C.; et al. Vitamin D signaling suppresses early prostate carcinogenesis in TgAPT121 mice. Cancer Prev. Res. 2019, 12, 343–356. [Google Scholar] [CrossRef]
- Ding, N.; Yu, R.T.; Subramaniam, N.; Sherman, M.H.; Wilson, C.; Rao, R.; Leblanc, M.; Coulter, S.; He, M.; Scott, C.; et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 2013, 153, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Lee, S.M.; Carlson, A.H.; Benkusky, N.A.; Kaufmann, M.; Jones, G.; Pike, J.W. A chromatin-based mechanism controls differential regulation of the cytochrome P450 gene Cyp24a1 in renal and non-renal tissues. J. Biol. Chem. 2019, 294, 14467–14481. [Google Scholar] [CrossRef] [Green Version]
- Heikkinen, S.; Väisänen, S.; Pehkonen, P.; Seuter, S.; Benes, V.; Carlberg, C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011, 39, 9181–9193. [Google Scholar] [CrossRef] [Green Version]
- Siersbaek, R.; Rabiee, A.; Nielsen, R.; Sidoli, S.; Traynor, S.; Loft, A.; La Cour Poulsen, L.; Rogowska-Wrzesinska, A.; Jensen, O.N.; Mandrup, S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014, 7, 1443–1455. [Google Scholar] [CrossRef] [Green Version]
- St John, H.C.; Bishop, K.A.; Meyer, M.B.; Benkusky, N.A.; Leng, N.; Kendziorski, C.; Bonewald, L.F.; Pike, J.W. The osteoblast to osteocyte transition: Epigenetic changes and response to the vitamin D3 hormone. Mol. Endocrinol. 2014, 28, 1150–1165. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Lee, C.H.; Pike, J.W. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J. Biol. Chem. 2014, 289, 19539–19554. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Riley, E.M.; Meyer, M.B.; Benkusky, N.A.; Plum, L.A.; DeLuca, H.F.; Pike, J.W. 1,25-Dihydroxyvitamin D3 controls a cohort of vitamin D receptor target genes in the proximal intestine that Is enriched for calcium-regulating components. J. Biol. Chem. 2015, 290, 18199–18215. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C. Molecular endocrinology of vitamin D on the epigenome level. Mol. Cell. Endocrinol. 2017, 453, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeljic, K.; Supic, G.; Magic, Z. New insights into vitamin D anticancer properties: Focus on miRNA modulation. Mol. Genet. Genom. 2017, 292, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, S.; Guo, B. Vitamin D Suppresses Ovarian Cancer Growth and Invasion by Targeting Long Non-Coding RNA CCAT2. Int. J. Mol. Sci. 2020, 21, 2334. [Google Scholar] [CrossRef] [Green Version]
- Zuo, S.; Wu, L.; Wang, Y.; Yuan, X. Long Non-coding RNA MEG3 Activated by Vitamin D Suppresses Glycolysis in Colorectal Cancer via Promoting c-Myc Degradation. Front. Oncol. 2020, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Lian, J.B.; Glimcher, M.J.; Roufosse, A.H.; Hauschka, P.V.; Gallop, P.M.; Cohen-Solal, L.; Reit, B. Alterations of the gamma-carboxyglutamic acid and osteocalcin concentrations in vitamin D-deficient chick bone. J. Biol. Chem. 1982, 257, 4999–5003. [Google Scholar]
- Gombart, A.F.; Borregaard, N.; Koeffler, H.P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005, 19, 1067–1077. [Google Scholar]
- Lin, R.; Nagai, Y.; Sladek, R.; Bastien, Y.; Ho, J.; Petrecca, K.; Sotiropoulou, G.; Diamandis, E.P.; Hudson, T.J.; White, J.H. Expression profiling in squamous carcinoma cells reveals pleiotropic effects of vitamin D3 analog EB1089 signaling on cell proliferation, differentiation, and immune system regulation. Mol. Endocrinol. 2002, 16, 1243–1256. [Google Scholar] [CrossRef]
- Campbell, M.J. Vitamin D and the RNA transcriptome: More than mRNA regulation. Front. Physiol. 2014, 5, 181. [Google Scholar] [CrossRef] [Green Version]
- Craig, T.A.; Zhang, Y.; McNulty, M.S.; Middha, S.; Ketha, H.; Singh, R.J.; Magis, A.T.; Funk, C.; Price, N.D.; Ekker, S.C.; et al. Research resource: Whole transcriptome RNA sequencing detects multiple 1α,25-dihydroxyvitamin D3-sensitive metabolic pathways in developing zebrafish. Mol. Endocrinol. 2012, 26, 1630–1642. [Google Scholar] [CrossRef] [Green Version]
- Verway, M.; Bouttier, M.; Wang, T.T.; Carrier, M.; Calderon, M.; An, B.S.; Devemy, E.; McIntosh, F.; Divangahi, M.; Behr, M.A.; et al. Vitamin D induces interleukin-1beta expression: Paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 2013, 9, e1003407. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, V.; Barbier, C.; Ismailova, A.; Wang, Y.; Dmowski, K.; Salehi-Tabar, R.; Memari, B.; Groulx-Boivin, E.; White, J.H. Vitamin D-regulated gene expression profiles: Species-specificity and cell-specific effects on metabolism and immunity. Endocrinology 2021, 162, bqaa218. [Google Scholar] [CrossRef] [PubMed]
- Hanel, A.; Carlberg, C. Time-resolved gene expression analysis monitors the regulation of inflammatory mediators and attenuation of adaptive immune response by vitamin D. Int. J. Mol. Sci. 2022, 23, 911. [Google Scholar] [CrossRef] [PubMed]
- Zeitelhofer, M.; Adzemovic, M.Z.; Gomez-Cabrero, D.; Bergman, P.; Hochmeister, S.; N’Diaye, M.; Paulson, A.; Ruhrmann, S.; Almgren, M.; Tegner, J.N.; et al. Functional genomics analysis of vitamin D effects on CD4+ T cells in vivo in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2017, 114, E1678–E1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLuca, H.F. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004, 80, 1689S–1696S. [Google Scholar]
- Norman, A.W. From vitamin D to hormone D: Fundamentals of the vitamin D endocrine system essential for good health. Am. J. Clin. Nutr. 2008, 88, 491S–499S. [Google Scholar]
- Prietl, B.; Treiber, G.; Pieber, T.R.; Amrein, K. Vitamin D and immune function. Nutrients 2013, 5, 2502–2521. [Google Scholar] [CrossRef]
- Limketkai, B.N.; Mullin, G.E.; Limsui, D.; Parian, A.M. Role of vitamin D in inflammatory bowel disease. Nutr. Clin. Pract. 2017, 32, 337–345. [Google Scholar] [CrossRef]
- Munger, K.L.; Levin, L.I.; Hollis, B.W.; Howard, N.S.; Ascherio, A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA 2006, 296, 2832–2838. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C. Genome-wide (over)view on the actions of vitamin D. Front. Physiol. 2014, 5, 167. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C.; Seuter, S.; Nurmi, T.; Tuomainen, T.P.; Virtanen, J.K.; Neme, A. In vivo response of the human epigenome to vitamin D: A proof-of-principle study. J. Steroid Biochem. Mol. Biol. 2018, 180, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Neme, A.; Seuter, S.; Malinen, M.; Nurmi, T.; Tuomainen, T.P.; Virtanen, J.K.; Carlberg, C. In vivo transcriptome changes of human white blood cells in response to vitamin D. J. Steroid Biochem. Mol. Biol. 2019, 188, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Shirvani, A.; Kalajian, T.A.; Song, A.; Holick, M.F. Disassociation of vtamin D’s calcemic activity and non-calcemic genomic activity and idividual responsiveness: A randomized controlled double-blind clinical tial. Sci. Rep. 2019, 9, 17685. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlberg, C. Vitamin D and Its Target Genes. Nutrients 2022, 14, 1354. https://doi.org/10.3390/nu14071354
Carlberg C. Vitamin D and Its Target Genes. Nutrients. 2022; 14(7):1354. https://doi.org/10.3390/nu14071354
Chicago/Turabian StyleCarlberg, Carsten. 2022. "Vitamin D and Its Target Genes" Nutrients 14, no. 7: 1354. https://doi.org/10.3390/nu14071354
APA StyleCarlberg, C. (2022). Vitamin D and Its Target Genes. Nutrients, 14(7), 1354. https://doi.org/10.3390/nu14071354





