Faecal Microbiota in Infants and Young Children with Functional Gastrointestinal Disorders: A Systematic Review
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy and Search Terms
2.2. Inclusion and Exclusion Criteria
2.2.1. Participants
2.2.2. Intervention
2.2.3. Control(s)
2.2.4. Outcome Measures
2.2.5. Design
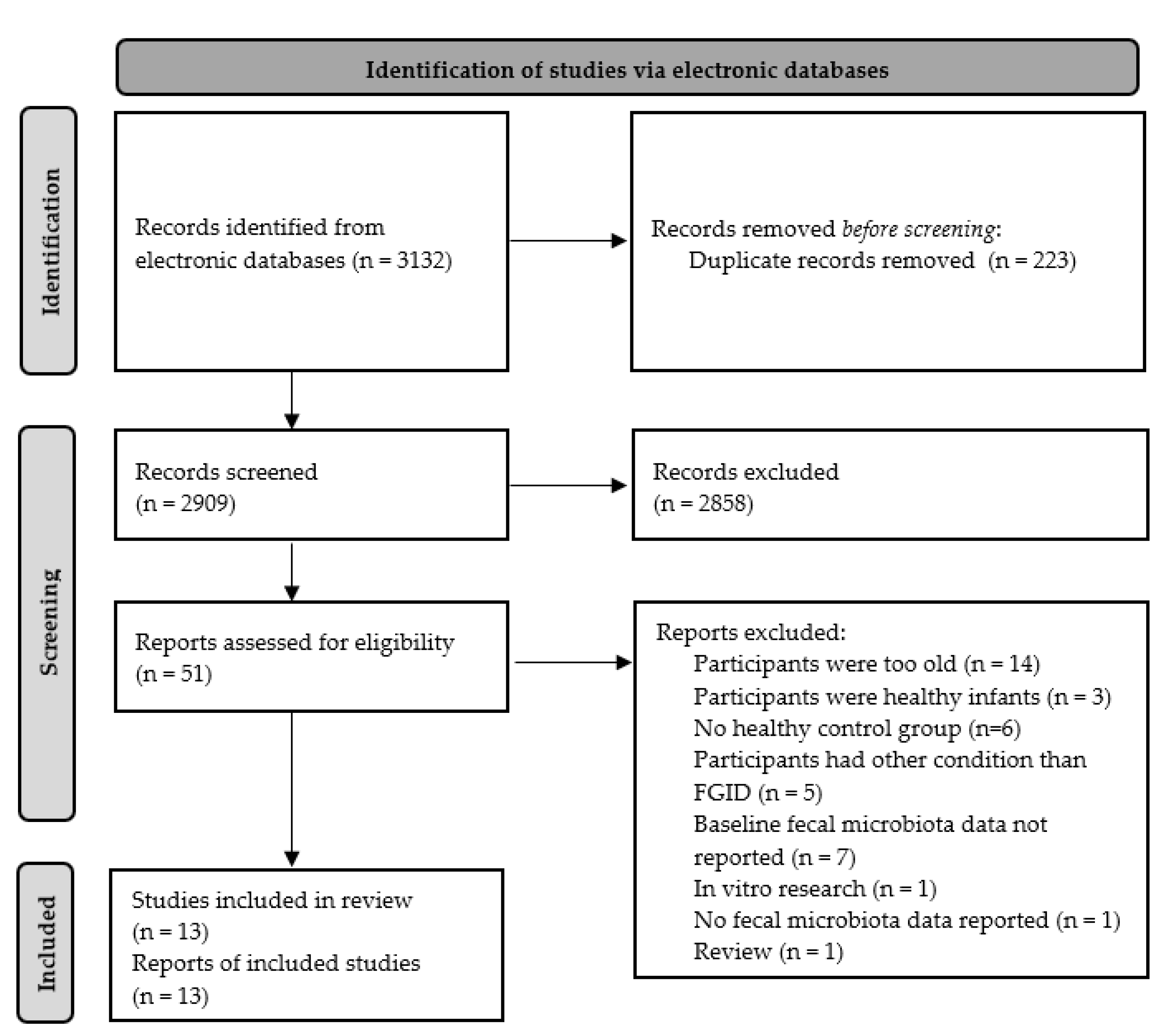
2.3. Study Selection Process
2.4. Quality Assessment
2.5. Data Extraction
3. Results
3.1. Selected Studies
3.2. Confounding Factors That Could Influence Microbiota Composition
3.3. Infantile Colic
3.3.1. Diagnostic Criteria Used
3.3.2. Sampling and Storage of Stool Samples
3.3.3. Method of Analysis of Faecal Microbiota
3.3.4. Faecal Microbiota Composition
qPCR and 16S Sequencing Results
| Phylum | Class | Order | Family | Genus | Species | Studies | Compared to Controls | References | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | ↑ | = | ↓ | Unknown * | |||||||
| Actinobacteria | 1 | 1 | [34] | ||||||||
| Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacteria | 4 | 1 | 2 | 1 | [25,26,27,34] | |||
| B. Breve | |||||||||||
| B. longum | 1 | 1 | [32] | ||||||||
| Coriobacteriales | Coriobacteriaceae | Collinsella | 1 | 1 | [27] | ||||||
| Bacteroidetes | 3 | 1 | 2 | [26,27,34] | |||||||
| Firmicutes | 2 | 1 | 1 | [22,34] | |||||||
| Bacilli | Lactobacillalles | Lactobacillaceae | Lactobacilli | 4 | 1 | 2 | 1 | [22,25,26,27] | |||
| L. iners | 1 | 1 | [34] | ||||||||
| Enterococcaceae | Enterococcus | 1 | 1 | [27] | |||||||
| Streptococcaceae | Streptococcus | 1 | 1 | [27] | |||||||
| S. thermophilus | 1 | 1 | [34] | ||||||||
| Eubacteriaceae | Eubacterium | E. hallii | 1 | 1 | [23] | ||||||
| Erysipelatoclostridium | 1 | 1 | [27] | ||||||||
| Negativicutes | Vellionellales | Veillonellaceae | Veillonella | 2 | 1 | 1 | [23,27] | ||||
| Proteobacteria | 3 | 2 | 1 | [26,30,34] | |||||||
| Gammaproteobateria | Enterobacteriales | Enterobacteriaceae | 1 | 1 | [26] | ||||||
| Escherichia | 1 | 1 | [27] | ||||||||
| E. coli | 2 | 1 | 1 | [25,32] | |||||||
| Klebsiella | |||||||||||
| K. pneumoniae | 1 | 1 | [32] | ||||||||
| K. oxytoca | |||||||||||
| Shigella | 2 | 2 | [27,32] | ||||||||
| Enterobacter | |||||||||||
| E. clocae | 1 | 1 | [32] | ||||||||
| Pseudomonadales | Moraxellaceae | Acinetobacter | 1 | [34] | |||||||
| Verrucomicrobia | 1 | 1 | [34] | ||||||||
Results Using Other Methods of Analysis
| Phylum | Class | Order | Family | Genus | Species | Studies | Compared to Controls | References | ||
|---|---|---|---|---|---|---|---|---|---|---|
| n | ↑ | = | ↓ | |||||||
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacteria | 1 | 1 | [31] | |||
| Bacteroidetes | n/a | |||||||||
| Firmicutes | Bacilli | Lactobacillalles | Lactobacillaceae | Lactobacilli | 2 | 1 | 1 | [28,31] | ||
| Enterococcaceae | 2 | 2 | [28,31] | |||||||
| Enterococcus | E. faecalis | 1 | 1 | [29] | ||||||
| E. aerogenes | 1 | 1 | [29] | |||||||
| Clostridia | Clostridiales | Clotridiaceae | Clostridium | 1 | 1 | [28] | ||||
| Proteobacteria | Gammaproteobateria | Enterobacteriales | 2 | 1 | 1 | [28,29] | ||||
| Escherichia | ||||||||||
| E. coli | 1 | 1 | [29] | |||||||
| Klebsiella | ||||||||||
| K. pneumoniae | 1 | 1 | [29] | |||||||
| K. oxytoca | 1 | 1 | [29] | |||||||
| Enterobacter | ||||||||||
| E. clocae | 1 | 1 | [29] | |||||||
| Verrucomicrobia | n/a | |||||||||
3.3.5. Microbiota Diversity
3.3.6. Metabolites
3.3.7. Markers of Inflammation
3.4. Constipation
3.4.1. Sampling, Storage, and Method of Analysis
3.4.2. Faecal Microbiota Composition
3.5. Other FGID
4. Discussion
4.1. Infantile Colic
4.1.1. Faecal Microbiota and Metabolites
4.1.2. Markers of Inflammation
4.2. Functional Constipation
4.3. Limitations of Included Studies
4.3.1. Study Population Selection
4.3.2. Stool Sampling, Storage, and Processing
4.3.3. Analysis of Stool Samples
4.3.4. Reported Outcome Measures
4.4. Limitations of This Review
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeevenhooven, J.; Koppen, I.J.N.; Benninga, M.A. The New Rome IV Criteria for Functional Gastrointestinal Disorders in Infants and Toddlers. Pediatr. Gastroenterol. Hepatol. Nutr. 2017, 20, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenplas, Y.; Hauser, B.; Salvatore, S. Functional Gastrointestinal Disorders in Infancy: Impact on the Health of the Infant and Family. Pediatr. Gastroenterol. Hepatol. Nutr. 2019, 22, 207. [Google Scholar] [CrossRef] [PubMed]
- Wessel, M.A.; Cobb, J.C.; Jackson, E.B.; Harris, G.S.; Detwiler, A.C. Paroxysmal Fussing in Infancy, Sometimes Called Colic. Pediatrics 1954, 14, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Van Tilburg, M.A.L.; Hyman, P.E.; Walker, L.; Rouster, A.; Palsson, O.S.; Kim, S.M.; Whitehead, W.E. Prevalence of Functional Gastrointestinal Disorders in Infants and Toddlers. J. Pediatr. 2015, 166, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Rautava, P.; Lehtonen, L.; Helenius, H.; Sillanpää, M. Infantile Colic: Child and Family Three Years Later. Pediatrics 1995, 96, 43–47. [Google Scholar] [CrossRef]
- Indrio, F.; Di Mauro, A.; Riezzo, G.; Cavallo, L.; Francavilla, R. Infantile Colic, Regurgitation, and Constipation: An Early Traumatic Insult in the Development of Functional Gastrointestinal Disorders in Children? Eur. J. Pediatr. 2015, 174, 841–842. [Google Scholar] [CrossRef]
- Ouald Chaib, A.; Levy, I.E.; Ouald Chaib, M.; Vandenplas, Y. The Influence of the Gastrointestinal Microbiome on Infant Colic. Expert Rev. Gastroenterol. Hepatol. 2020, 14, 919–932. [Google Scholar] [CrossRef]
- Bellaiche, M.; Oozeer, R.; Gerardi-Temporel, G.; Faure, C.; Vandenplas, Y. Multiple Functional Gastrointestinal Disorders Are Frequent in Formula-Fed Infants and Decrease Their Quality of Life. Acta Paediatr. Int. J. Paediatr. 2018, 107, 1276–1282. [Google Scholar] [CrossRef] [Green Version]
- Kesavelu, D.; Sethi, G.; Bangale, N.; Anwar, F.; Rao, S. Common Gastrointestinal Distress among Infants: Role of Optimal Nutritional Interventions. Clin. Epidemiol. Glob. Health 2018, 6, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Zeevenhooven, J.; Browne, P.D.; L’Hoir, M.P.; de Weerth, C.; Benninga, M.A. Infant Colic: Mechanisms and Management. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 479–496. [Google Scholar] [CrossRef]
- Fouhy, F.; Watkins, C.; Hill, C.J.; O’Shea, C.A.; Nagle, B.; Dempsey, E.M.; O’Toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. Perinatal Factors Affect the Gut Microbiota up to Four Years after Birth. Nat. Commun. 2019, 10, 1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of Delivery Mode-Associated Gut Microbiota Dynamics on Health in the First Year of Life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, N.T.; Li, F.; Lee-Sarwar, K.A.; Tun, H.M.; Brown, B.P.; Pannaraj, P.S.; Bender, J.M.; Azad, M.B.; Thompson, A.L.; Weiss, S.T.; et al. Meta-Analysis of Effects of Exclusive Breastfeeding on Infant Gut Microbiota across Populations. Nat. Commun. 2018, 9, 4169. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [Green Version]
- Dogra, S.; Sakwinska, O.; Soh, S.-E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Karnani, N.; Lee, Y.S.; Yap, F. Rate of Establishing the Gut Microbiota in Infancy Has Consequences for Future Health. Gut Microbes 2015, 6, 321–325. [Google Scholar] [CrossRef] [Green Version]
- Korpela, K.; de Vos, W.M. Antibiotic Use in Childhood Alters the Gut Microbiota and Predisposes to Overweight. Microb. Cell 2016, 3, 296. [Google Scholar] [CrossRef]
- van den Bogert, B.; de Vos, W.M.; Zoetendal, E.G.; Kleerebezem, M. Microarray Analysis and Barcoded Pyrosequencing Provide Consistent Microbial Profiles Depending on the Source of Human Intestinal Samples. Appl. Environ. Microbiol. 2011, 77, 2071–2080. [Google Scholar] [CrossRef] [Green Version]
- Lepage, P.; Seksik, P.; Sutren, M.; de la Cochetiere, M.-F.; Jian, R.; Marteau, P.; Doré, J. Biodiversity of the Mucosa-Associated Microbiota Is Stable along the Distal Digestive Tract in Healthy Individuals and Patients with IBD. Inflamm. Bowel Dis. 2005, 11, 473–480. [Google Scholar] [CrossRef]
- Couch, R.D.; Navarro, K.; Sikaroodi, M.; Gillevet, P.; Forsyth, C.B.; Mutlu, E.; Engen, P.A.; Keshavarzian, A. The Approach to Sample Acquisition and Its Impact on the Derived Human Fecal Microbiome and VOC Metabolome. PLoS ONE 2013, 8, e81163. [Google Scholar] [CrossRef] [Green Version]
- Sirriyeh, R.; Lawton, R.; Gardner, P.; Armitage, G. Reviewing Studies with Diverse Designs: The Development and Evaluation of a New Tool. J. Eval. Clin. Pract. 2012, 18, 746–752. [Google Scholar] [CrossRef]
- De Moraes, J.G.; Motta, M.E.F.; Beltrão, M.F.; Salviano, T.L.; da Silva, G.A.P. Fecal Microbiota and Diet of Children with Chronic Constipation. Int. J. Pediatr. 2016, 2016, 6787269. [Google Scholar] [CrossRef] [Green Version]
- Korpela, K.; Renko, M.; Paalanne, N.; Vänni, P.; Salo, J.; Tejesvi, M.; Koivusaari, P.; Pokka, T.; Kaukola, T.; Pirttilä, A.M.; et al. Microbiome of the First Stool after Birth and Infantile Colic. Pediatr. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.T.; Lacroix, C.; Braegger, C.P.; Chassard, C. Lactate-Utilizing Community Is Associated with Gut Microbiota Dysbiosis in Colicky Infants. Sci. Rep. 2017, 7, 11176. [Google Scholar] [CrossRef] [Green Version]
- Pärtty, A.; Kalliomäki, M.; Salminen, S.; Isolauri, E. Infantile Colic Is Associated with Low-Grade Systemic Inflammation. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Savino, F.; Garro, M.; Montanari, P.; Galliano, I.; Bergallo, M. Crying Time and RORγ/FOXP3 Expression in Lactobacillus Reuteri DSM17938-Treated Infants with Colic: A Randomized Trial. J. Pediatr. 2018, 192, 171–177.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Weerth, C.; Fuentes, S.; Puylaert, P.; De Vos, W.M. Intestinal Microbiota of Infants with Colic: Development and Specific Signatures. Pediatrics 2013, 131, e550–e558. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, M.; Alba, C.; Cam Public Health Area, P.S.G.O.; Rodríguez, J.M.; Fernández, L. Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study. Nutrients 2020, 12, 634. [Google Scholar] [CrossRef] [Green Version]
- Savino, F.; Cresi, F.; Pautasso, S.; Palumeri, E.; Tullio, V.; Roana, J.; Silvestro, L.; Oggero, R. Intestinal Microflora in Breastfed Colicky and Non-Colicky Infants. Acta Paediatr. 2004, 93, 825–829. [Google Scholar] [CrossRef]
- Savino, F.; Cordisco, L.; Tarasco, V.; Calabrese, R.; Palumeri, E.; Matteuzzi, D. Molecular Identification of Coliform Bacteria from Colicky Breastfed Infants. Acta Paediatr. 2009, 98, 1582–1588. [Google Scholar] [CrossRef]
- Savino, F.; Cordisco, L.; Tarasco, V.; Locatelli, E.; Di Gioia, D.; Oggero, R.; Matteuzzi, D. Antagonistic Effect of Lactobacillus Strains against Gas-Producing Coliforms Isolated from Colicky Infants. BMC Microbiol. 2011, 11, 157. [Google Scholar] [CrossRef] [Green Version]
- Savino, F.; Quartieri, A.; De Marco, A.; Garro, M.; Amaretti, A.; Raimondi, S.; Simone, M.; Rossi, M. Comparison of Formula-Fed Infants with and without Colic Revealed Significant Differences in Total Bacteria, Enterobacteriaceae and Faecal Ammonia. Acta Paediatr. Int. J. Paediatr. 2017, 106, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, J.M.; Fatheree, N.Y.; Norori, J.; Liu, Y.; Lucke, J.F.; Tyson, J.E.; Ferris, M.J. Altered Fecal Microflora and Increased Fecal Calprotectin in Infants with Colic. J. Pediatr. 2009, 155, 823–828.e1. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S.; Ernst, F. Microbiome Diversity. In Orchestrating Microbiome Analysis; 2021; Available online: https://microbiome.github.io/OMA/ (accessed on 15 December 2021).
- Rhoads, J.M.; Collins, J.; Fatheree, N.Y.; Hashmi, S.S.; Taylor, C.M.; Luo, M.; Hoang, T.K.; Gleason, W.A.; Van Arsdall, M.R.; Navarro, F.; et al. Infant Colic Represents Gut Inflammation and Dysbiosis. J. Pediatr. 2018, 203, 55–61.e3. [Google Scholar] [CrossRef] [PubMed]
- Savino, F.; Cordisco, L.; Tarasco, V.; Palumeri, E.; Calabrese, R.; Oggero, R.; Roos, S.; Matteuzzi, D. Lactobacillus Reuteri DSM 17938 in Infantile Colic: A Randomized, Double-Blind, Placebo-Controlled Trial. Pediatrics 2010, 126, e526–e533. [Google Scholar] [CrossRef]
- Roos, S.; Dicksved, J.; Tarasco, V.; Locatelli, E.; Ricceri, F.; Grandin, U.; Savino, F. 454 Pyrosequencing Analysis on Faecal Samples from a Randomized DBPC Trial of Colicky Infants Treated with Lactobacillus Reuteri DSM 17938. PLoS ONE 2013, 8, e56710. [Google Scholar] [CrossRef] [Green Version]
- Fatheree, N.Y.; Liu, Y.; Ferris, M.; Van Arsdall, M.; McMurtry, V.; Zozaya, M.; Cai, C.; Rahbar, M.H.; Hessabi, M.; Vu, T.; et al. Hypoallergenic Formula with Lactobacillus Rhamnosus GG for Babies with Colic: A Pilot Study of Recruitment, Retention, and Fecal Biomarkers. World J. Gastrointest. Pathophysiol. 2016, 7, 160–170. [Google Scholar] [CrossRef]
- Fatheree, N.Y.; Liu, Y.; Taylor, C.M.; Hoang, T.K.; Cai, C.; Rahbar, M.H.; Hessabi, M.; Ferris, M.; McMurtry, V.; Wong, C.; et al. Lactobacillus Reuteri for Infants with Colic: A Double-Blind, Placebo-Controlled, Randomized Clinical Trial. J. Pediatr. 2017, 191, 170–178.e2. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [Green Version]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Chassard, C. New Insights in Gut Microbiota Establishment in Healthy Breast Fed Neonates. PLoS ONE 2012, 7, e44595. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.V.; Metcalf, G.A.; et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The Human Microbiome and Child Growt—First 1000 Days and Beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fassarella, M.; Blaak, E.E.; Penders, J.; Nauta, A.; Smidt, H.; Zoetendal, E.G. Gut Microbiome Stability and Resilience: Elucidating the Response to Perturbations in Order to Modulate Gut Health. Gut 2021, 70, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Zierer, J.; Jackson, M.A.; Kastenmüller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J. The Fecal Metabolome as a Functional Readout of the Gut Microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential Beneficial Effects of Butyrate in Intestinal and Extraintestinal Diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.S.; Davies, S.S. Microbial Metabolism of Dietary Components to Bioactive Metabolites: Opportunities for New Therapeutic Interventions. Genome Med. 2016, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- St James-Roberts, I.; Alvarez, M.; Hovish, K. Emergence of a Developmental Explanation for Prolonged Crying in 1-to 4-Month-Old Infants: Review of the Evidence. J. Pediatr. Gastroenterol. Nutr. 2013, 57, S30–S36. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R. The Role of Butyrate on Colonic Function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; III, J.F.R.; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut Microbes Promote Colonic Serotonin Production through an Effect of Short-chain Fatty Acids on Enterochromaffin Cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, Tryptophan Metabolism and the Brain-Gut-Microbiome Axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Kurtoglu, S.; Üzüm, K.; Hallac, I.K.; Coskim, A. 5-Hydroxy-3-indole Acetic Acid Levels in Infantile Colic: Is Serotoninergic Tonus Responsible for This Problem? Acta Paediatr. 1997, 86, 764–765. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; Snelling, T.; Umlai, U.-K.; Nicholson, J.K.; Carding, S.R.; Glen, R.C.; McArthur, S. Microbiome–Host Systems Interactions: Protective Effects of Propionate upon the Blood–Brain Barrier. Microbiome 2018, 6, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burokas, A.; Arboleya, S.; Moloney, R.D.; Peterson, V.L.; Murphy, K.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Targeting the Microbiota-Gut-Brain Axis: Prebiotics Have Anxiolytic and Antidepressant-like Effects and Reverse the Impact of Chronic Stress in Mice. Biol. Psychiatry 2017, 82, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Wang, Q.; Wu, Q.; Mao-Draayer, Y.; Kim, C.H. Bidirectional Regulatory Potentials of Short-Chain Fatty Acids and Their G-Protein-Coupled Receptors in Autoimmune Neuroinflammation. Sci. Rep. 2019, 9, 8837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasco Rodríguez-Belvís, M.; Viada Bris, J.F.; Plata Fernández, C.; García-Salido, A.; Asensio Antón, J.; Domínguez Ortega, G.; Muñoz Codoceo, R.A. Normal Fecal Calprotectin Levels in Healthy Children Are Higher than in Adults and Decrease with Age. Paediatr. Child Health 2020, 25, 286–292. [Google Scholar] [CrossRef]
- Preidis, G.A.; Versalovic, J. Targeting the Human Microbiome with Antibiotics, Probiotics, and Prebiotics: Gastroenterology Enters the Metagenomics Era. Gastroenterology 2009, 136, 2015–2031. [Google Scholar] [CrossRef] [Green Version]
- Esaiassen, E.; Hjerde, E.; Cavanagh, J.P.; Pedersen, T.; Andresen, J.H.; Rettedal, S.I.; Støen, R.; Nakstad, B.; Willassen, N.P.; Klingenberg, C. Effects of Probiotic Supplementation on the Gut Microbiota and Antibiotic Resistome Development in Preterm Infants. Front. Pediatr. 2018, 6, 347. [Google Scholar] [CrossRef] [Green Version]
- Korpela, K.; Salonen, A.; Vepsäläinen, O.; Suomalainen, M.; Kolmeder, C.; Varjosalo, M.; Miettinen, S.; Kukkonen, K.; Savilahti, E.; Kuitunen, M. Probiotic Supplementation Restores Normal Microbiota Composition and Function in Antibiotic-Treated and in Caesarean-Born Infants. Microbiome 2018, 6, 182. [Google Scholar] [CrossRef] [Green Version]
- Thomas, V.; Clark, J.; Doré, J. Fecal Microbiota Analysis: An Overview of Sample Collection Methods and Sequencing Strategies. Future Microbiol. 2015, 10, 1485–1504. [Google Scholar] [CrossRef]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool Consistency Is Strongly Associated with Gut Microbiota Richness and Composition, Enterotypes and Bacterial Growth Rates. Gut 2016, 65, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falony, G.; Vieira-Silva, S.; Raes, J. Richness and Ecosystem Development across Faecal Snapshots of the Gut Microbiota. Nat. Microbiol. 2018, 3, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Li, B.U.K.; Misiewicz, L. Cyclic Vomiting Syndrome: A Brain–Gut Disorder. Gastroenterol. Clin. 2003, 32, 997–1019. [Google Scholar] [CrossRef]
- Aagaard, K.; Petrosino, J.; Keitel, W.; Watson, M.; Katancik, J.; Garcia, N.; Patel, S.; Cutting, M.; Madden, T.; Hamilton, H. The Human Microbiome Project Strategy for Comprehensive Sampling of the Human Microbiome and Why It Matters. FASEB J. 2013, 27, 1012–1022. [Google Scholar] [CrossRef]
- Kawar, N.; Park, S.G.; Schwartz, J.L.; Callahan, N.; Obrez, A.; Yang, B.; Chen, Z.; Adami, G.R. Salivary Microbiome with Gastroesophageal Reflux Disease and Treatment. Sci. Rep. 2021, 11, 188. [Google Scholar] [CrossRef]
- Okereke, I.; Hamilton, C.; Wenholz, A.; Jala, V.; Giang, T.; Reynolds, S.; Miller, A.; Pyles, R. Associations of the Microbiome and Esophageal Disease. J. Thorac. Dis. 2019, 11, S1588. [Google Scholar] [CrossRef]
| Search Terms | AND/OR |
|---|---|
| “microbio *” OR “dysbiosis” OR “Bifido *” OR “Lactobacill *” OR “Proteobacter *” OR “Escherichia” OR “Klebseilla” OR “Bacteroidetes” OR “Klebsiella” OR “Serratia” OR “Vibrio” OR “Yersinia” OR “Pseudomonas” OR “Enterobacter *” OR “bacteria” | |
| “functional gastrointestinal disorder *” OR “FGID *” OR “colic” OR “reflux” OR “regurgitat *” OR “GER *” OR “constipation” OR “diarrhoea *” OR “diarrhoea *” OR “vomiting” OR “dyschezia” OR “rumination” OR “inflammatory bowel dis *” OR “IBS” | AND |
| “infant *” OR “neonat *” OR “toddler *” OR “child *” OR “paediatric” OR “paediatric” OR “newborn” OR “baby” OR “babies” | AND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofman, D.; Kudla, U.; Miqdady, M.; Nguyen, T.V.H.; Morán-Ramos, S.; Vandenplas, Y. Faecal Microbiota in Infants and Young Children with Functional Gastrointestinal Disorders: A Systematic Review. Nutrients 2022, 14, 974. https://doi.org/10.3390/nu14050974
Hofman D, Kudla U, Miqdady M, Nguyen TVH, Morán-Ramos S, Vandenplas Y. Faecal Microbiota in Infants and Young Children with Functional Gastrointestinal Disorders: A Systematic Review. Nutrients. 2022; 14(5):974. https://doi.org/10.3390/nu14050974
Chicago/Turabian StyleHofman, Denise, Urszula Kudla, Mohamad Miqdady, Thi Viet Ha Nguyen, Sofía Morán-Ramos, and Yvan Vandenplas. 2022. "Faecal Microbiota in Infants and Young Children with Functional Gastrointestinal Disorders: A Systematic Review" Nutrients 14, no. 5: 974. https://doi.org/10.3390/nu14050974
APA StyleHofman, D., Kudla, U., Miqdady, M., Nguyen, T. V. H., Morán-Ramos, S., & Vandenplas, Y. (2022). Faecal Microbiota in Infants and Young Children with Functional Gastrointestinal Disorders: A Systematic Review. Nutrients, 14(5), 974. https://doi.org/10.3390/nu14050974







