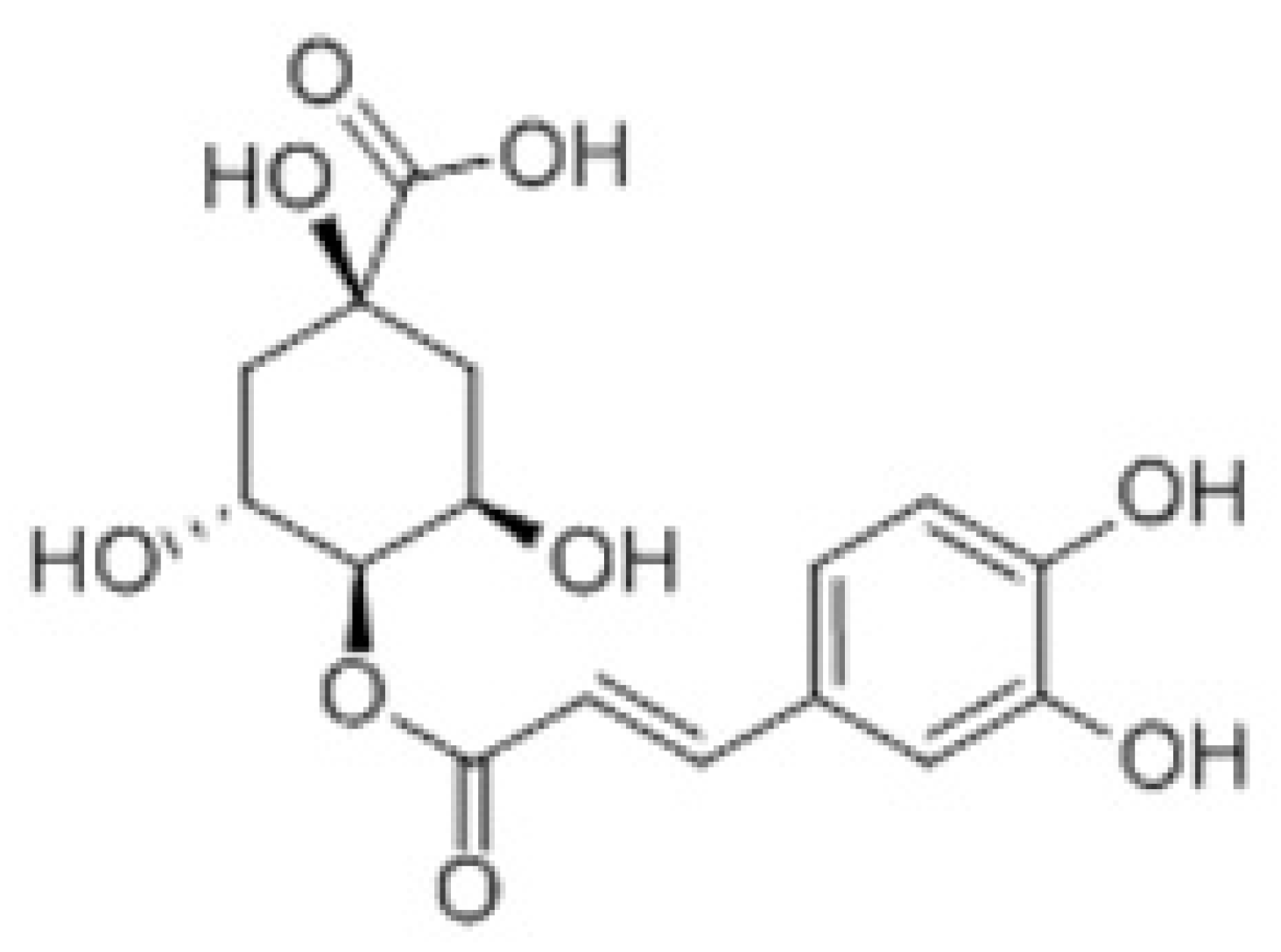
Anti-Inflammatory Activity and Mechanism of Cryptochlorogenic Acid from Ageratina adenophora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Sample, Chemicals, and Reagents
2.2. Extraction of CCGA
Preparation of Aqueous Extract from Ageratina adenophora
2.3. Separation of Cryptochlorogenic Acid by Macroporous Resin
2.3.1. Macroporous Resin Pretreatment
2.3.2. Sample Loading and Elution System
2.4. Purification of Cryptochlorogenic Acid by Silica Gel Column Chromatography
2.5. Identification of Monomer Purity by Ultra High Performance Liquid Chromatography
2.5.1. Chromatographic Conditions
2.5.2. Making the Standard Curve
2.5.3. Determination of Cryptochlorogenic Acid in Ageratina adenophora
2.6. Cell Viability Using CCK8 Method
2.7. Measurement of Nitric Oxide in RAW264.7 Cells
2.8. Enzyme-Linked Immunosorbent Assay (ELISA) for Detecting TNF-α, IL-1β, IL-6, IL-8, INOS, COX-2, and IL-10 Production
2.9. RNA Extraction and Real Time RT-PCR Analysis for Detecting TNF-α, IL-1β, IL-6, IL-8, IL-10, COX-2, and INOS Production
2.10. Transient Transfection and Luciferase Activity Assay
2.11. Western Blot Analysis
2.12. Statistical Analysis
3. Results
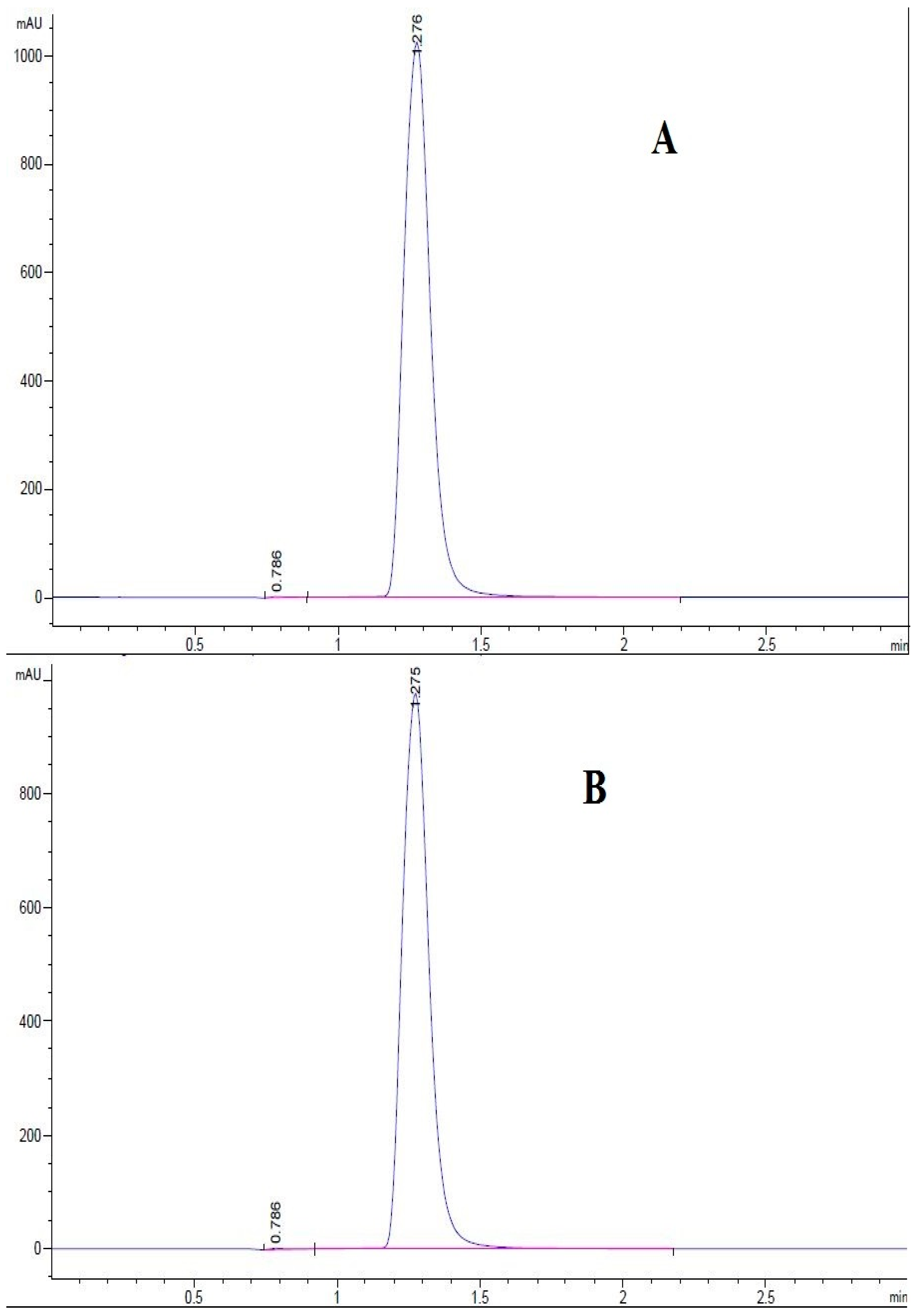
3.1. Detection of Cryptochlorogenic Acid by Thin Layer Chromatography
3.2. Determination of the Purity of Cryptochlorogenic Acid by Ultra High Performance Liquid Chromatography
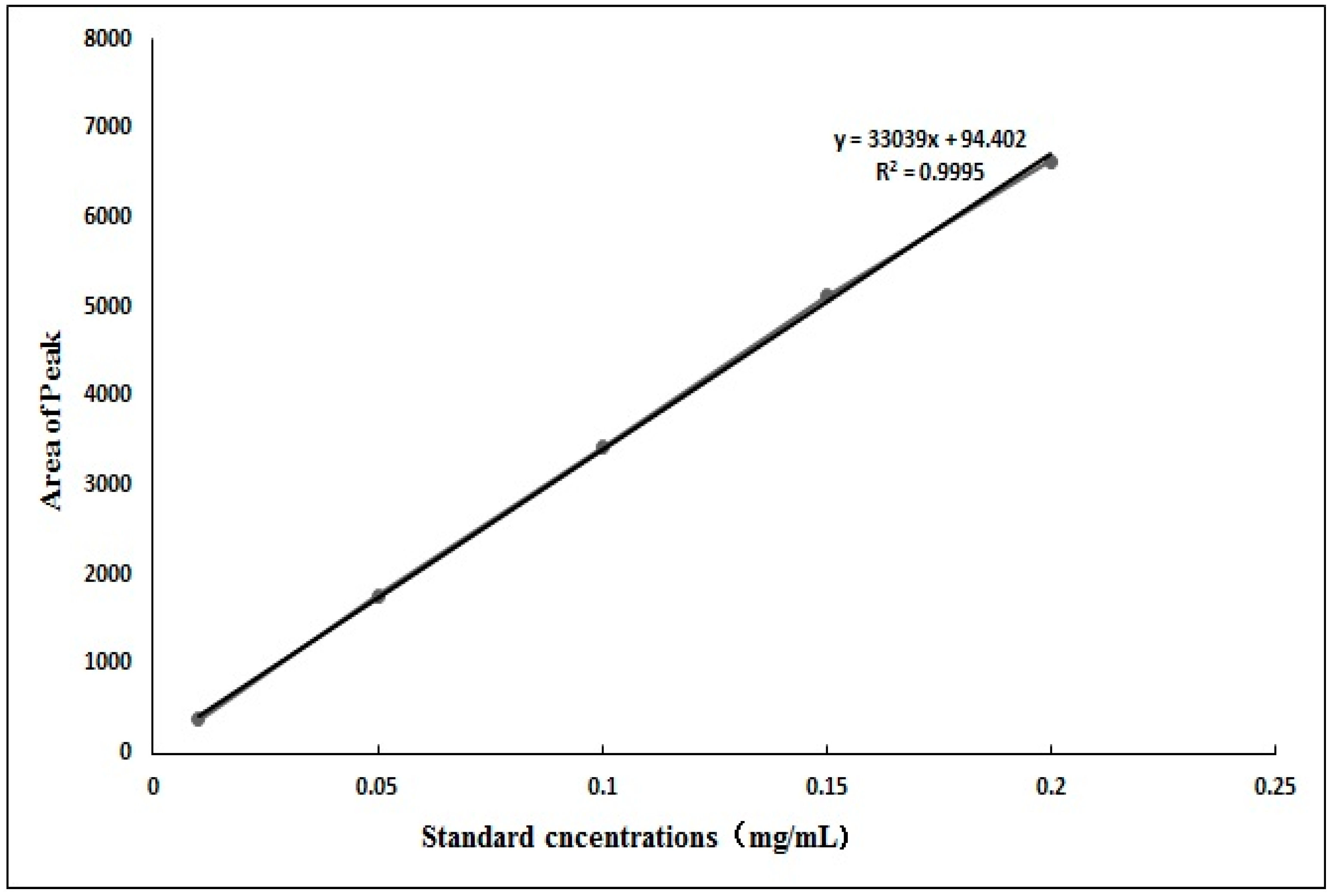
3.2.1. Standard Curve of Cryptochlorogenic Acid
3.2.2. Determination of Cryptochlorogenic Acid in Ageratina adenophora
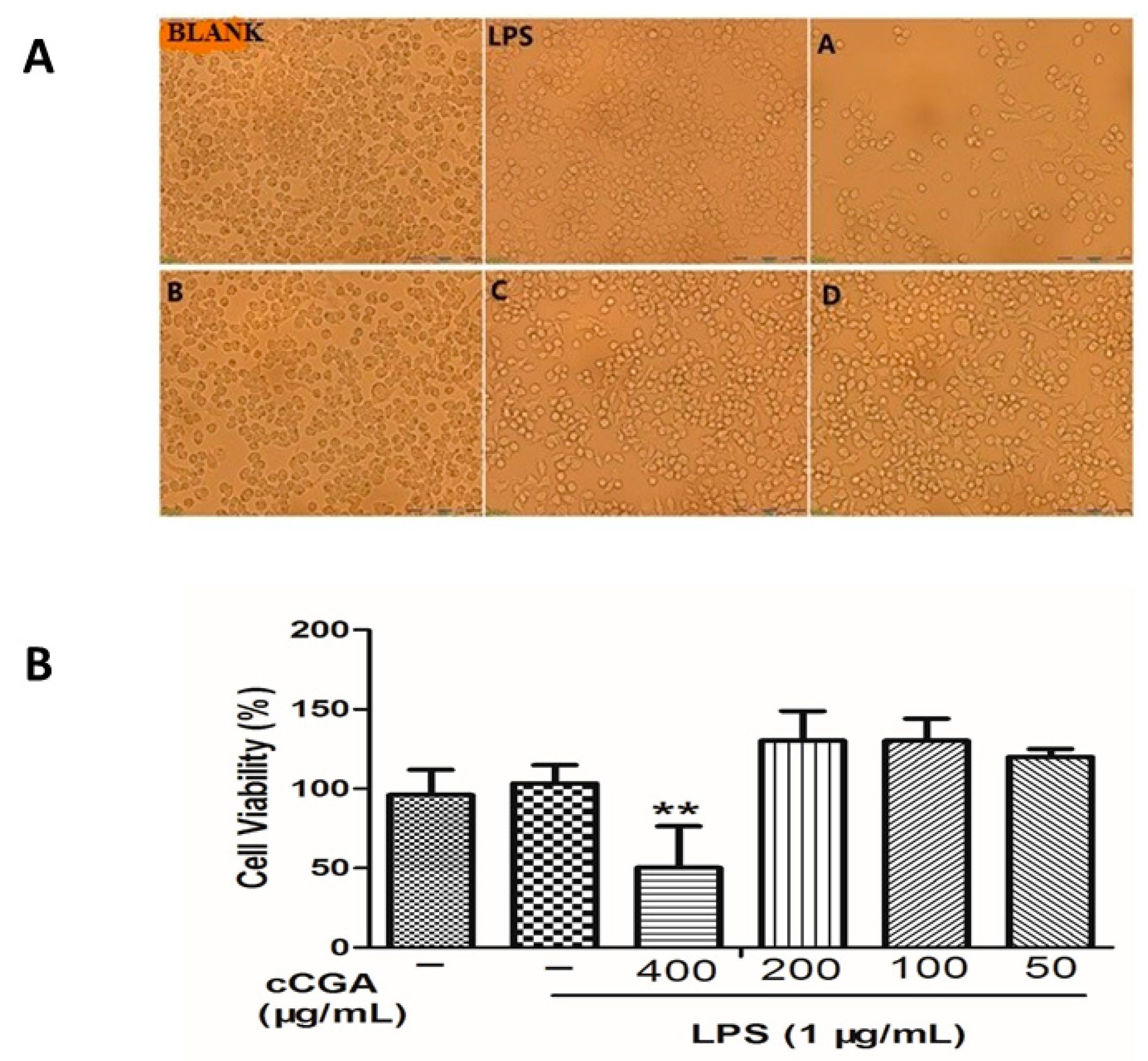
3.3. Effects of CCGA on Cell Morphology and Cytotoxicity
3.4. Effect of CCGA on LPS-Induced Nitric Oxide (NO) and iNOS Production
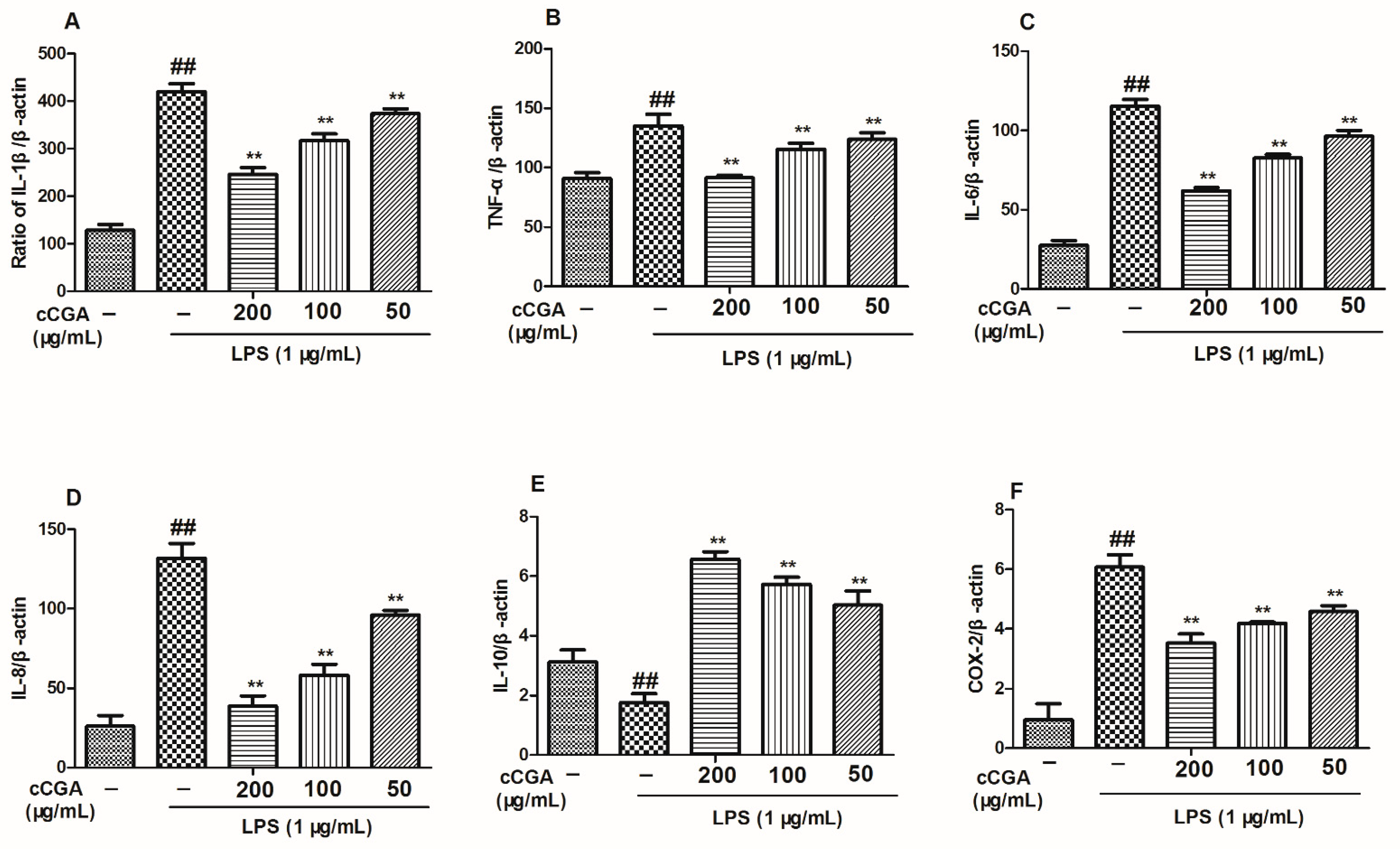
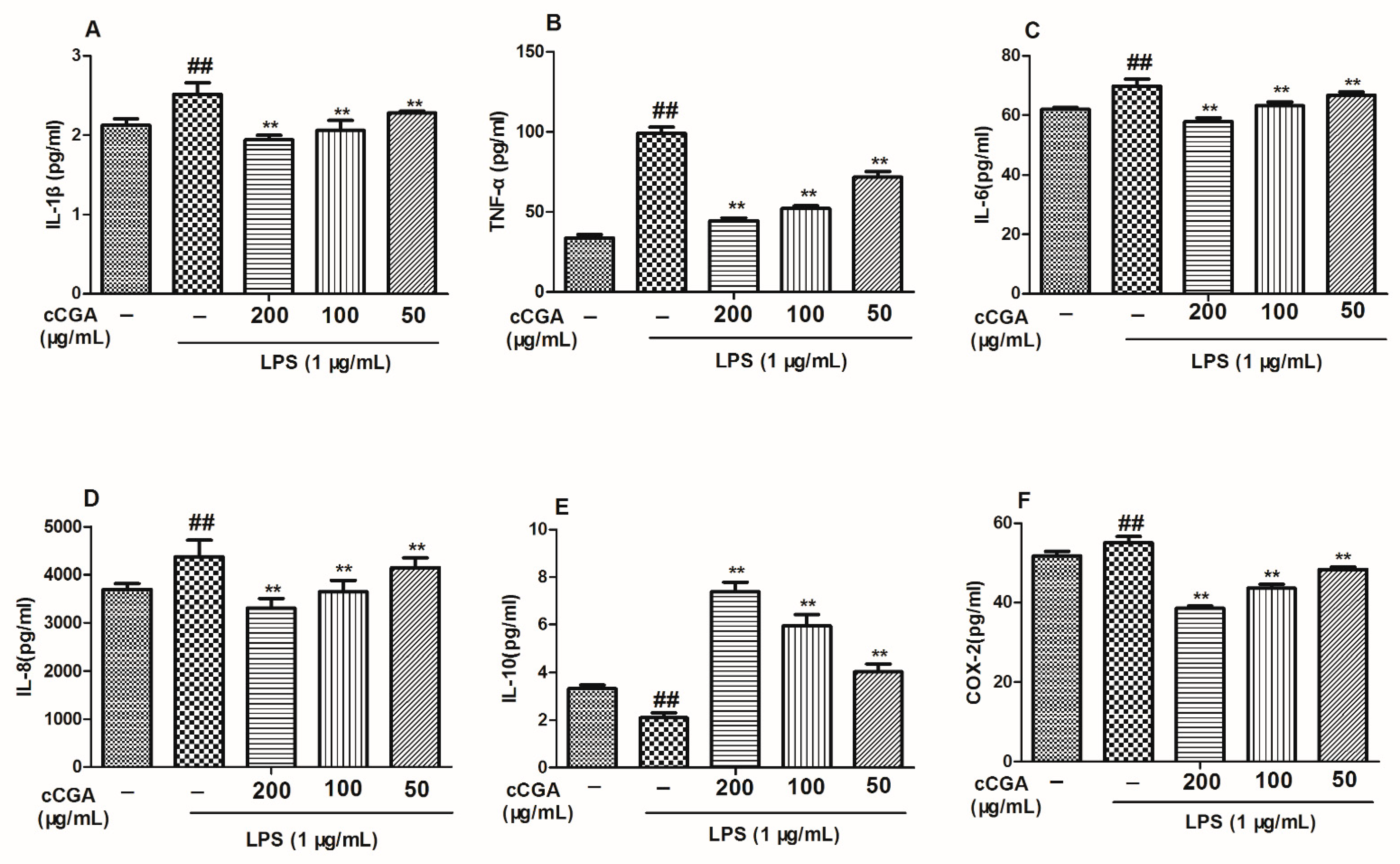
3.5. The Regulatory Effect of CCGA from A. adenophora on Inflammation Related Cytokines in LPS-Induced RAW264.7 Cells
3.6. Effect of CCGA from A. adenophora on COX-2 Expression
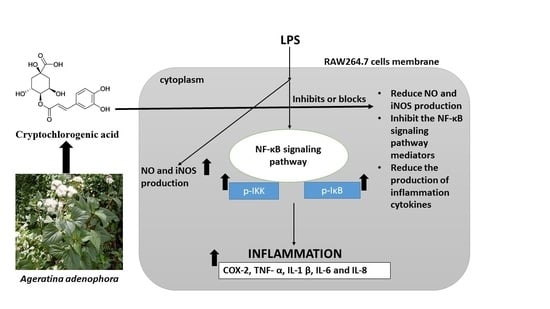
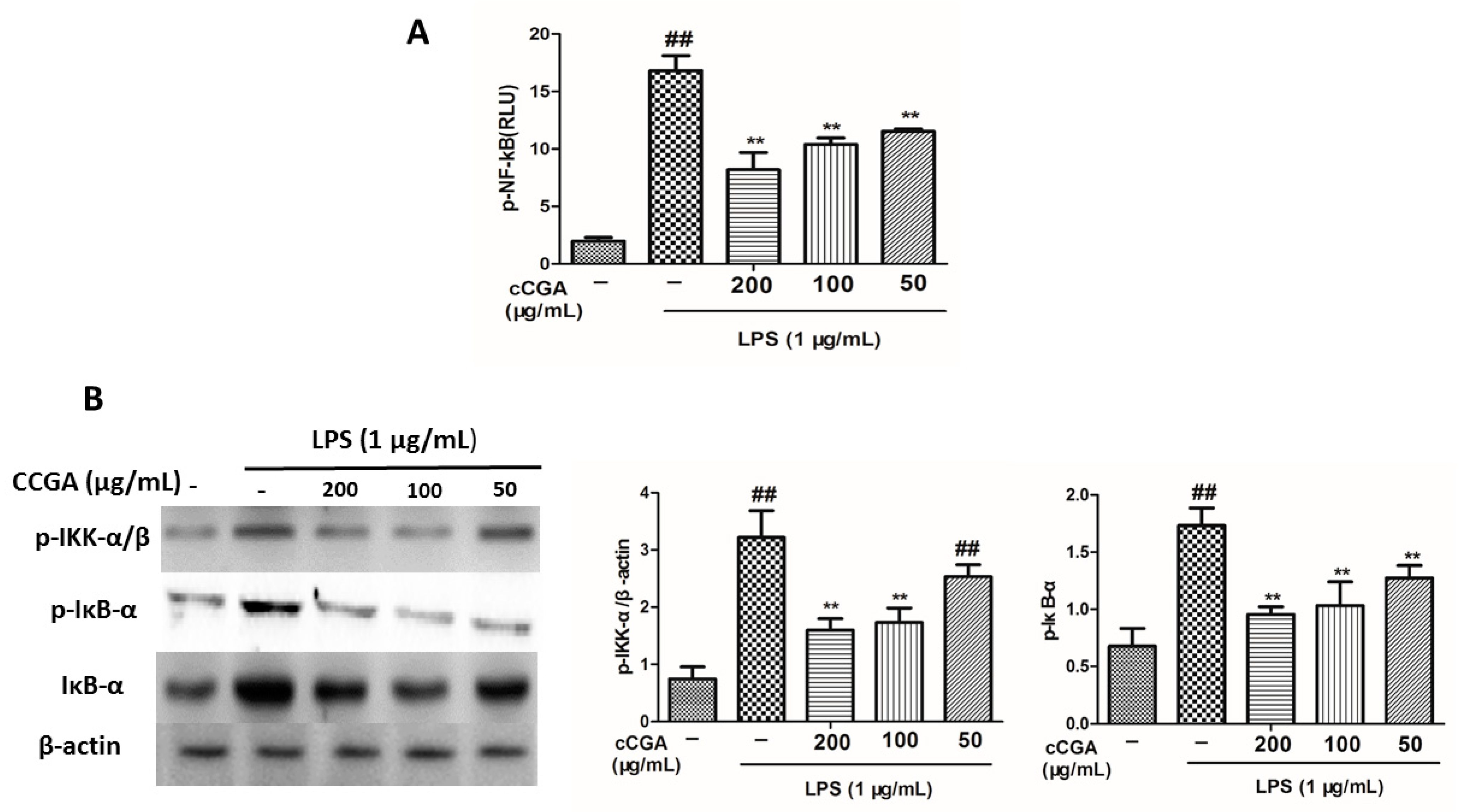
3.7. CCGA Inhibited LPS-Induced NF-kB Activation in RAW 264.7 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serhan, C.N.; Savill, J. Resolution of inflammation: The beginning programs the end. Nat. Immunol. 2005, 6, 1191–1197. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation, metaflammation and immunometabolic disorders. Nature 2017, 542, 177. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Catrysse, L.G.; van Loo, G. Inflammation and the metabolic syndrome: The tissuespecific functions of NF-κB. Trends Cell Biol. 2017, 27, 417–429. [Google Scholar] [CrossRef]
- Eming, S.A.; Krieg, T.; Davidson, J.M. Inflammation in wound repair: Molecular and cellular mechanisms. J. Investig. Dermatol. 2007, 127, 514–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, D.L. Inflammation in Tic Disorders and Obsessive-Compulsive Disorder: Are PANS and PANDAS a Path Forward? J. Child Neurol. 2019, 34, 598–611. [Google Scholar] [CrossRef]
- Kay, J.; Thadhani, E.; Samson, L.; Engelward, B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair 2019, 83, 102673. [Google Scholar] [CrossRef]
- Reinehr, T. Inflammatory markers in children and adolescents with type 2 diabetes mellitus. Clin. Chim. Acta 2019, 496, 100–107. [Google Scholar] [CrossRef]
- McGeer, E.G.; McGeer, P.L. Inflammatory processes in Alzheimer’s disease. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2003, 27, 741–749. [Google Scholar] [CrossRef]
- Fingleton, B. Matrix metalloproteinases as regulators of inflammatory processes. Biochimi. Biophys. (BBA)-Mol. Cell Res. 2017, 1864, 2036–2042. [Google Scholar] [CrossRef]
- Reddy, P.; Lent-Schochet, D.; Ramakrishnan, N.; McLaughlin, M.; Jialal, I. Metabolic syndrome is an inflammatory disorder: A conspiracy between adipose tissue and phagocytes. Clin. Chim. Acta 2019, 496, 35–44. [Google Scholar] [CrossRef]
- Catarro, M.; Serrano, J.L.; Ramos, S.S.; Silvestre, S.; Almeida, P. Nimesulide analogues: From anti-inflammatory to antitumor agents. Bioorg. Chem. 2019, 88, 102966. [Google Scholar] [CrossRef] [PubMed]
- Schumann, R.R.; Pfeil, D.; Lamping, N.; Kirschning, C.; Scherzinger, G.; Schlag, P.; Karawajew, L.; Herrmann, F. Lipopolysaccharide induces the rapid tyrosine phosphorylation of the mitogen-activated protein kinases erk-1 and p38 in cultured human vascular endothelial cells requiring the presence of soluble CD14. Blood 1996, 87, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wu, Y.N.; Wang, H.; Ma, J.Y.; Zhai, S.S.; Duan, J. Dapk1 improves inflammation, oxidative stress and autophagy in LPS-induced acute lung injury via p38MAPK/NF-κB signaling pathway. Mol. Immunol. 2020, 120, 13–22. [Google Scholar] [CrossRef]
- Mitoma, H.; Horiuchi, T.; Hatta, N.; Tsukamoto, H.; Harashima, S.; Kikuchi, Y.; Otsuka, J.; Okamura, S.; Fujita, S.; Harada, M. Infliximab induces potent anti-inflammatory responses by outside-toinside signals through transmembrane TNF-α. Gastroenterology 2005, 128, 376–392. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Nemeth, D.P.; McKim, D.B.; Zhu, L.; DiSabato, D.J.; Berdysz, O.; Gorantla, G.; Oliver, B.; Witcher, K.G.; Wang, Y.; et al. Cell-Type-Specific Interleukin 1 Receptor 1 Signaling in the Brain Regulates Distinct Neuroimmune Activities. Immunity 2019, 50, 317–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; London, N.R.; Gibson, C.C.; Davis, C.T.; Tong, Z.; Sorensen, L.K.; Shi, D.S.; Guo, J.; Smith, M.C.; Grossmann, A.H.; et al. Interleukin receptor activates a MYD88-ARNO-ARF6 cascade to disrupt vascular stability. Nature 2012, 492, 252–255. [Google Scholar] [CrossRef] [Green Version]
- Petersen, A.; Pedersen, B. The role of IL-6 in mediating the anti-inflammatory. J. Physiol. Pharmacol. 2006, 57, 43–51. [Google Scholar]
- Singh, J.K.; Simões, B.M.; Howell, S.J.; Farnie, G.; Clarke, R.B. Recent advances reveal IL-8 signaling as a potential key to targeting breast cancer stem cells. Breast Cancer Res. 2013, 15, 210. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Yao, Y.; Yang, J.; Zhengxie, J.; Li, X.; Hu, S.; Zhang, A.; Dong, J.; Zhang, C.; Gan, G. COX-2-PGE2 signaling pathway con-tributes to hippocampal neuronal injury and cognitive impairment in PTZ-kindled epilepsy mice. Int. Immunopharmacol. 2020, 87, 106801. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.K.; Sung, S.H.; Kim, Y.C.; Kim, S.G. Inhibition of lipopolysaccharide inducible nitric oxide synthase, TNF-alpha and COX-2 expression by sauchinone effects on I-kappaBalpha phosphorylation, C/EBP and AP-1 activation. Br. J. Pharmacol. 2003, 139, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Spiller, F.; Oliveira Formiga, R.; Fernandes da Silva Coimbra, J.; Alves-Filho, J.C.; Cunha, T.M.; Cunha, F.Q. Targeting nitric oxide as a key modulator of sepsis, arthritis and pain. Nitric Oxide 2019, 89, 32–40. [Google Scholar] [CrossRef]
- Ally, A.; Powell, I.; Ally, M.M.; Chaitoff, K.; Nauli, S.M. Role of neuronal nitric oxide synthase on cardiovascular functions in physiological and pathophysiological states. Nitric Oxide 2020, 102, 52–73. [Google Scholar] [CrossRef]
- Ying, X.; Yu, K.; Chen, X.; Chen, H.; Hong, J.; Cheng, S.; Peng, L. Piperine inhibits LPS induced expression of inflammatory mediators in RAW 264.7 cells. Cell Immunol. 2013, 285, 49–54. [Google Scholar] [CrossRef]
- Detmers, P.A.; Hernandez, M.; Mudgett, J.; Hassing, H.; Burton, C.; Mundt, S.; Chun, S.; Fletcher, D.; Card, D.J.; Lisnock, J.; et al. Deficiency in inducible nitric oxide synthase results in reduced atherosclerosis in apolipoprotein E-deficient mic. J. Immunol. 2000, 16, 3430–3435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korhonen, R.; Lahti, A.; Kankaanranta, H.; Moilanen, E. Nitric oxide production and signaling in inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 471–479. [Google Scholar] [CrossRef]
- Murtas, D.; Piras, F.; Minerba, L.; Ugalde, J.; Piga, M.; Maxia, C.; Perra, M.T.; Sirigu, P. Nuclear factor-κB expression is predictive of overall survival in patients with cutaneous melanoma. Oncol. Lett. 2010, 1, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Talalay, P. Relevance of anti-inflammatory and antioxidant activities of exemestane and synergism with sulforaphane for disease prevention. Proc. Natl. Acad. Sci. USA 2013, 110, 19065–19070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babu, P.V.; Liu, D.; Gilbert, E.R. Recent advances in understanding the anti-diabetic actions of dietary flavonoids. J. Nutr. Biochem. 2013, 24, 1777–1789. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Goel, N. Phenolic acids: Natural versatile molecules with promising therapeutic applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Itagaki, S.; Kurokawa, T.; Ogura, J.; Kobayashi, M.; Hirano, T.; Iseki, K. In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. Int. J. Pharm. 2011, 403, 136–138. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Zhu, S.; Ma, C.; Wang, Z. Antibacterial activity and mechanism of action of chlorogenic acid. J. Food Sci. 2011, 76, M398–M403. [Google Scholar] [CrossRef]
- Dos Santos, M.D.; Almeida, M.C.; Lopes, N.P.; De Souza, G.E.P. Evaluation of the antiinflammatory, analgesic and antipyretic activities of the natural polyphenol chlorogenic acid. Biol. Pharm. Bull. 2006, 29, 2236–2240. [Google Scholar] [CrossRef] [Green Version]
- Barahuie, F.; Hussein, M.Z.; Arulselvan, P.; Fakurazi, S.; Zainal, Z. Controlled in vitro release of the anticancer drug chlorogenic acid using magnesium/aluminiumlayered double hydroxide as a nanomatrix. Sci. Adv. Mater. 2016, 8, 501–513. [Google Scholar] [CrossRef]
- Hwang, S.J.; Kim, Y.W.; Park, Y.; Lee, H.J.; Kim, K.W. Anti-inflammatory effects of chlorogenic acid in lipopolysaccharide-stimulated raw 264.7 cells. Inflamm. Res. 2014, 63, 81–90. [Google Scholar] [CrossRef]
- Zhao, X.L.; Yu, L.; Zhang, S.D.; Ping, K.; Ni, H.Y.; Qin, X.Y.; Zhao, C.J.; Wang, W.; Efferth, T.; Fu, Y.J. Cryptochlorogenic acid attenu-ates LPS-induced inflam;matory response and oxidative stress via upregulation of the Nrf2/HO-1 signaling pathway in RAW 264.7 macrophages. Int. Immunopharmacol. 2020, 83, 106436. [Google Scholar] [CrossRef]
- Ouyang, C.; Liu, X.; Yan, D.; Li, Y.; Wang, Q.; Cao, A.; Guo, M. Immunotoxicity assessment of cadinene sesquiterpenes from Eupatorium adenophorum in mice. J. Integr. Agric. 2016, 15, 2319–2325. [Google Scholar] [CrossRef] [Green Version]
- Okyere, S.K.; Wen, J.; Cui, Y.; Xie, L.; Gao, P.; Wang, J.; Wang, S.; Hu, Y. Toxic mechanisms and pharmacological properties of euptox A, a toxic monomer from A. adenophora. Fitoterapia 2021, 155, 105032. [Google Scholar] [CrossRef]
- Ren, Z.; Gao, P.; Okyere, S.K.; Cui, Y.; Wen, J.; Jing, B.; Deng, J.; Hu, Y. Ageratina adenophora inhibits spleen immune function in rats via the loss of the FRC Network and Th1-Th2 Cell Ratio elevation. Toxins 2021, 13, 309. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Okyere, S.K.; Gao, P.; Wen, J.; Cao, S.; Wang, Y.; Deng, J.; Hu, Y. Ageratina adenophora Disrupts the Intestinal Structure and Immune Barrier Integrity in Rats. Toxins 2021, 13, 651. [Google Scholar] [CrossRef]
- Okyere, S.K.; Mo, Q.; Pei, G.; Ren, Z.; Deng, J.; Hu, Y. Euptox A induces G0/G1 arrest and apoptosis of hepatocyte via ROS, mitochondrial dysfunction and caspases-dependent path- ways in vivo. J. Toxicol. Sci. 2020, 45, 661–671. [Google Scholar] [CrossRef]
- Ren, Z.; Okyere, S.K.; Wen, J.; Xie, L.; Cui, Y.; Wang, S.; Wang, J.; Cao, S.; Shen, L.; Ma, X.; et al. An Overview: The Toxicity of Ageratina adenophora on Animals and Its Possible Interventions. Int. J. Mol. Sci. 2021, 22, 11581. [Google Scholar] [CrossRef] [PubMed]
- Okyere, S.K.; Wen, J.; Cui, Y.; Xie, L.; Gao, P.; Zhang, M.; Wang, J.; Wang, S.; Ran, Y.; Ren, Z.; et al. Bacillus toyonensis SAU-19 and SAU-20 isolated from Ageratina adenophora alleviates the intestinal structure and integrity damage associated with gut dysbiosis in mice fed high fat mice. Front. Microbiol. 2021; in press. [Google Scholar] [CrossRef]
- Okyere, S.K.; Xie, L.; Wen, J.; Ran, Y.; Ren, Z.; Deng, J.; Hu, Y. Bacillus toyonensis SAU-19 Ameliorates Hepatic Insulin Resistance in High-Fat Diet/Streptozocin-Induced Diabetic Mice. Nutrients 2021, 13, 4512. [Google Scholar] [CrossRef] [PubMed]
- Poudel, R.; Neupane, N.P.; Mukeri, I.H.; Alok, S.; Verma, A. An updated review on invasive nature, phytochemical evaluation, & pharmacological activity of Ageratina adenophora. Int. J. Pharm. Sci. Res. 2020, 119, 2510–2520. [Google Scholar]
- Parthasarathy, M.; Evan Prince, S. The potential effect of phytochemicals and herbal plant remedies for treating drug-induced hepatotoxicity: A review. Mol. Biol. Rep. 2021, 48, 4767–4788. [Google Scholar] [CrossRef]
- Forni, C.; Facchiano, F.; Bartoli, M.; Pieretti, S.; Facchiano, A.; D’Arcangelo, D.; Norelli, S.; Valle, G.; Nisini, R.; Beninati, S.; et al. Beneficial Role of Phytochemicals on Oxidative Stress and Age-Related Diseases. Biomed. Res. Int. 2019, 2019, 8748253. [Google Scholar] [CrossRef] [Green Version]
- George, B.P.; Chandran, R.; Abrahamse, H. Role of Phytochemicals in Cancer Chemoprevention: Insights. Antioxidants 2021, 10, 1455. [Google Scholar] [CrossRef]
- Aktan, F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004, 75, 639–653. [Google Scholar] [CrossRef]
- Cirino, G.; Distrutti, E.; Wallace, J.L. Nitric oxide and inflammation. Inflamm. Allergy Drug Targets 2006, 5, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Howard, L.; Brownmiller, C.; Gu, I.; Lee, S.O.; Mauromoustakos, A. Inhibitory effects of cranberry polyphenol and volatile extracts on nitric oxide production in LPS activated RAW 264.7 macrophages. Food Funct. 2019, 10, 7091–7102. [Google Scholar] [CrossRef] [PubMed]
- Bigagli, E.; Cinci, L.; Paccosi, S.; Parenti, A.; D’Ambrosio, M.; Luceri., C. Nutritionally relevant concentrations of resveratrol and hydroxytyrosol mitigate oxidative burst of human granulocytes and monocytes and the production of pro-inflammatory mediators in LPS-stimulated RAW 264.7 macrophages. Int. Immunopharmacol. 2017, 43, 147–155. [Google Scholar] [CrossRef]
- Hung, Y.-L.; Miyazaki, H.; Fang, S.-H.; Li, C.-Y.; Suzuki, K. The Structural Characteristics of Green Tea Polyphenols on Lipopolysaccharide-Stimulated RAW Cells. J. Nutr. Biol. 2018, 2, 151–157. [Google Scholar] [CrossRef]
- Li, C.-Y.; Suzuki, K.; Hung, Y.-L.; Yang, M.-S.; Yu, C.-P.; Lin, S.-P.; Hou, Y.-C.; Fang, S.-H. Aloe metabolites prevent LPS-induced sepsis and inflammatory response by inhibiting mitogen-activated protein kinase activation. Am. J. Chin. Med. 2017, 45, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Yeom, M.; Kim, J.H.; Min, J.H.; Hwang, M.K.; Jung, H.S.; Sohn, Y. Xanthii fructus inhibits inflammatory responses in LPS-stimulated RAW 264.7 macrophages through suppressing NF-κB and JNK/p38 MAPK. J. Ethnopharmacol. 2015, 176, 394–401. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, S.Y.; Park, Y.L.; Myung, D.S.; Rew, J.S.; Joo, Y.E. Chlorogenic acid suppresses lipopolysaccharide-induced nitric oxide and interleukin-1β expression by inhibiting JAK2/STAT3 activation in RAW264.7 cells. Mol. Med. Rep. 2017, 16, 9224–9232. [Google Scholar] [CrossRef] [Green Version]
- Kunnumakkara, A.B.; Shabnam, B.; Girisa, S.; Harsha, C.; Banik, K.; Devi, T.B.; Choudhury, R.; Sahu, H.; Parama, D.; Sailo, B.L.; et al. Inflammation, NF-κB, and Chronic Diseases: How are They Linked? Crit. Rev. Immunol. 2020, 40, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Endale, M.; Park, S.C.; Kim, S.; Kim, S.H.; Yang, Y.; Cho, J.Y.; Rhee, M.H. Quercetin disrupts tyrosine-phosphorylated phosphatidylinositol 3-kinase and myeloid differentiation factor-88 association, and inhibits MAPK/AP-1 and IKK/NF-κB-induced inflammatory mediators production in RAW 264.7 cells. Immunobiology 2013, 218, 1452–1467. [Google Scholar] [CrossRef]
- Cheng, S.C.; Huang, W.C.; Pang, J.H.; Wu, Y.H.; Cheng, C.Y. Quercetin Inhibits the Production of IL-1β-Induced Inflammatory Cytokines and Chemokines in ARPE-19 Cells via the MAPK and NF-κB Signaling Pathways. Int. J. Mol. Sci. 2019, 20, 2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontoriero, M.; Fiume, G.; Vecchio, E.; de Laurentiis, A.; Albano, F.; Iaccino, E.; Mimmi, S.; Pisano, A.; Agosti, V.; Giovannone, E.; et al. Activation of NF-κB in B cell receptor signaling through Bruton’s tyrosine kinase-dependent phosphorylation of IκB-α. J. Mol. Med. 2019, 97, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Xu, S.; Qian, Y.; Xiao, Q. Resveratrol regulates microglia M1/M2 polarization via PGC-1α in conditions of neuroinflammatory injury. Brain Behav. Immun. 2017, 64, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Serreli, G.; Deiana, M. Extra Virgin Olive Oil Polyphenols: Modulation of Cellular Pathways Related to Oxidant Species and Inflammation in Aging. Cells 2020, 9, 478. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Sun, J.; Mohammadtursun, N.; Wu, J.; Dong, J.; Li, L. Curcumin inhibits cigarette smoke-induced inflammation via modulating the PPARγ-NF-κB signaling pathway. Food Funct. 2019, 10, 7983–7994. [Google Scholar] [CrossRef]


| Genes | Primer Sequence (5′→3′) | References |
|---|---|---|
| TNF-α | F- AGCACAGAAAGCATGATCCG R- ATGAGAGGGAGGCCATT | [10] |
| IL-1β | F- GTTCCCCAACTGGTACATCA R- CCATACTTTAGGAAGACACGG | [11] |
| IL-6 | F- CTTCTTGGGACTGATGCTGGT G R- CGCTGGCTTTGTCTTTCTTGTTA | [12] |
| IL-8 | F- CAAGGCTGGTCCATGCTCC R- TGCTATCACTTCCTTTCTGTTG | [11] |
| IL-10 | F- GCTCTTACTGACTGGCATGAG R- CGCAGCTCTAGGAGCATGTG | [13] |
| INOS | F- TCCTACACCACACCAAAC R- CTCCAATCTCTGCCTATCC | [10] |
| COX | F- TGCTGTACAAGCAGTGGCAA R- GCAGCCATTTCCTTCTCTCC | [10] |
| β-actin | F- CGGTTGGCCTTAGGGTTCAGGGGGG R- GTGGGCCGCTCTAGGCACCA | [12] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Okyere, S.K.; Hu, L.; Wen, J.; Ren, Z.; Deng, J.; Hu, Y. Anti-Inflammatory Activity and Mechanism of Cryptochlorogenic Acid from Ageratina adenophora. Nutrients 2022, 14, 439. https://doi.org/10.3390/nu14030439
Ma X, Okyere SK, Hu L, Wen J, Ren Z, Deng J, Hu Y. Anti-Inflammatory Activity and Mechanism of Cryptochlorogenic Acid from Ageratina adenophora. Nutrients. 2022; 14(3):439. https://doi.org/10.3390/nu14030439
Chicago/Turabian StyleMa, Xiaoping, Samuel Kumi Okyere, Liwen Hu, Juan Wen, Zhihua Ren, Junliang Deng, and Yanchun Hu. 2022. "Anti-Inflammatory Activity and Mechanism of Cryptochlorogenic Acid from Ageratina adenophora" Nutrients 14, no. 3: 439. https://doi.org/10.3390/nu14030439
APA StyleMa, X., Okyere, S. K., Hu, L., Wen, J., Ren, Z., Deng, J., & Hu, Y. (2022). Anti-Inflammatory Activity and Mechanism of Cryptochlorogenic Acid from Ageratina adenophora. Nutrients, 14(3), 439. https://doi.org/10.3390/nu14030439