Impact of Dietary Arachidonic Acid on Gut Microbiota Composition and Gut–Brain Axis in Male BALB/C Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Diet Design
2.2. Animal Handling
2.3. Fecal Microbiota Analysis
2.4. Immunoblotting Analysis
2.5. RNA Extraction and RT-qPCR Analysis
2.6. Immunohistochemical Analysis
2.7. Statistical Analysis
3. Results
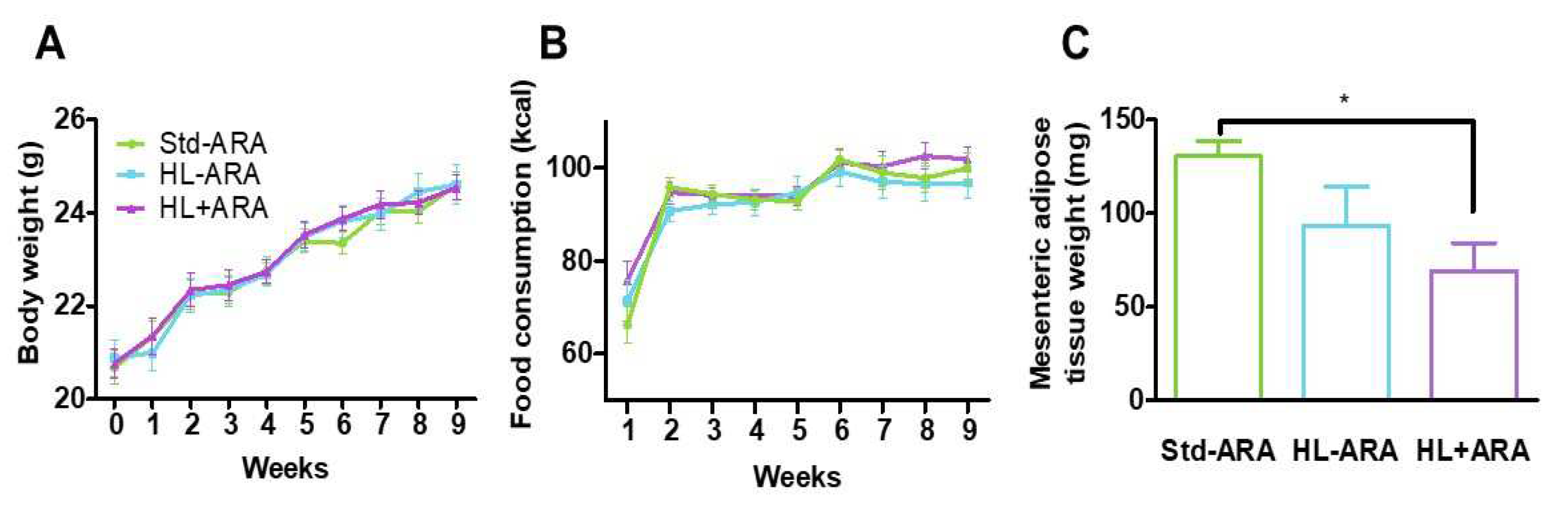
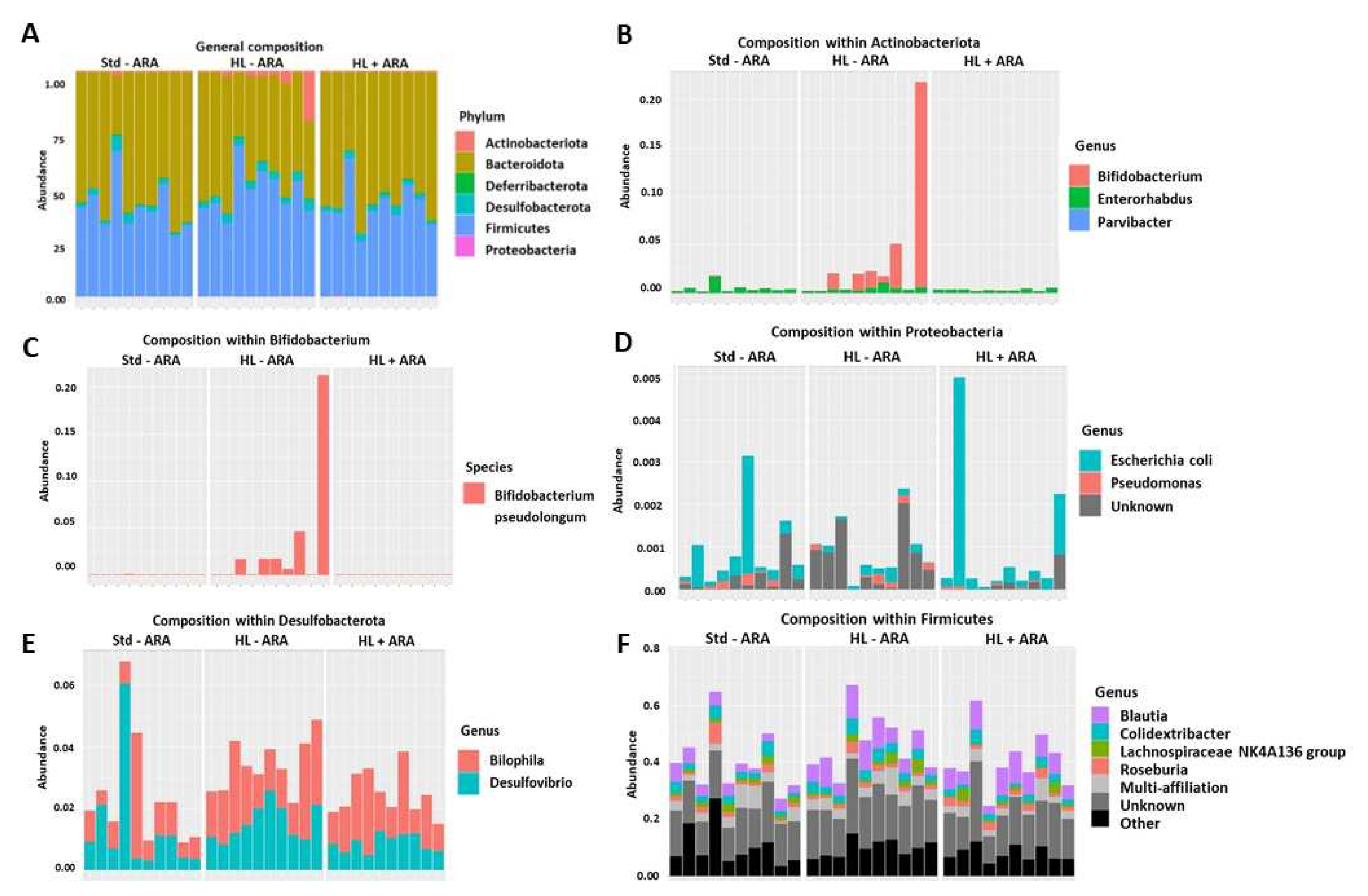
3.1. Impact of Dietary ARA Intake on Murine Growth and Fecal Microbiota
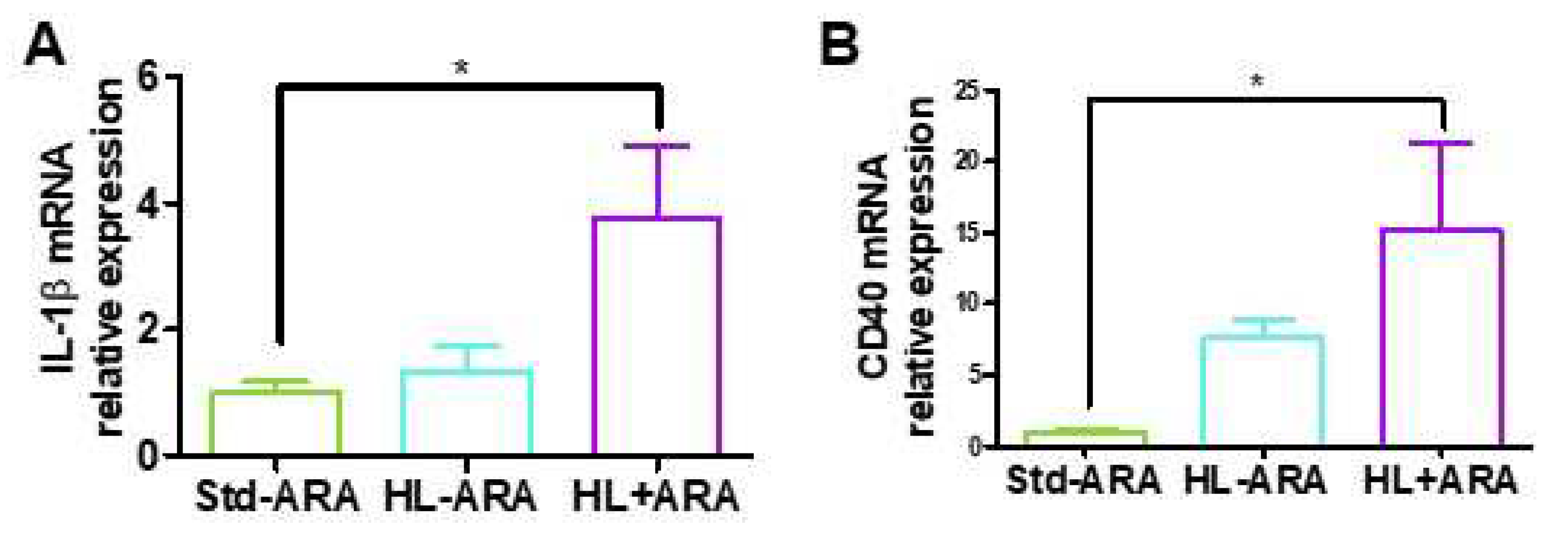
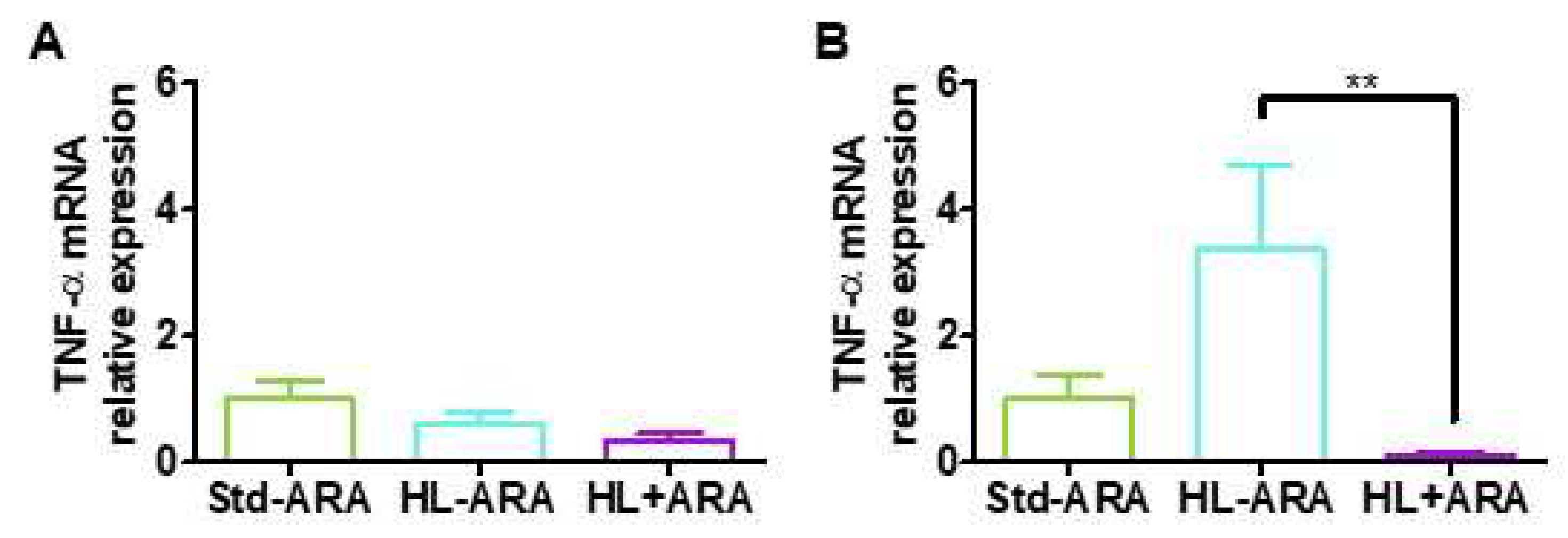
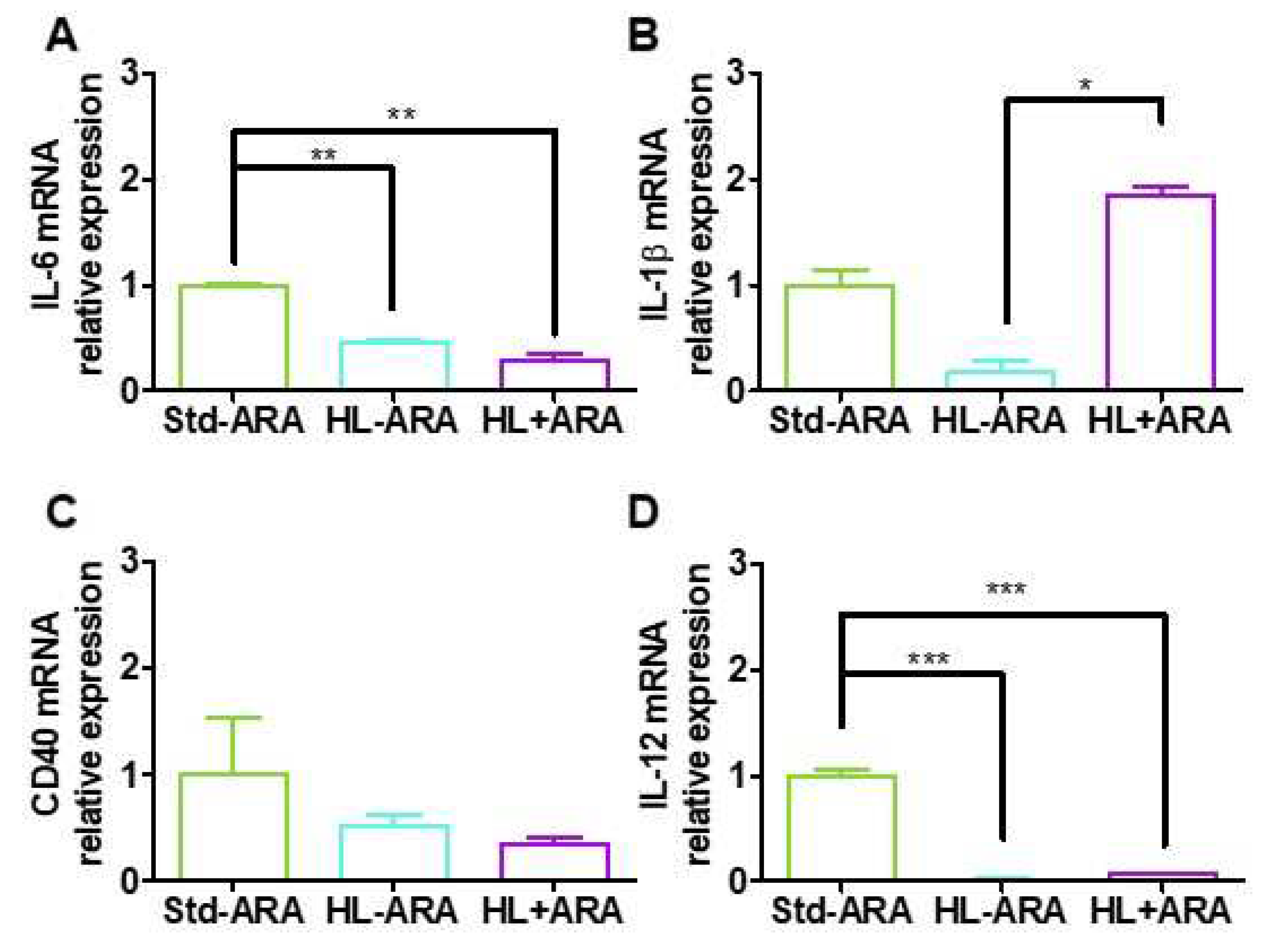
3.2. Impact of Dietary ARA Intake on the Expression Level of Inflammatory Markers in Colon
3.3. Relationship between Low Grade Inflammation in Liver and Adipose Tissues and ARA Intake
3.4. Impact of Dietary ARA on Brain Glial Cells and Pro-Inflammatory Cytokine Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chadaideh, K.S.; Carmody, R.N. Host-microbial interactions in the metabolism of different dietary fats. Cell Metab. 2021, 33, 857–872. [Google Scholar] [CrossRef]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; Wall, R.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C. Health Implications of High Dietary Omega-6 Polyunsaturated Fatty Acids. J. Nutr. Metab. 2012, 2012, 539426. [Google Scholar] [CrossRef]
- Calder, P.C.; Campoy, C.; Eilander, A.; Fleith, M.; Forsyth, S.; Larsson, P.O.; Schelkle, B.; Lohner, S.; Szommer, A.; van de Heijning, B.J.M.; et al. A systematic review of the effects of increasing arachidonic acid intake on PUFA status, metabolism and health-related outcomes in humans. Br. J. Nutr. 2019, 121, 1201–1214. [Google Scholar] [CrossRef]
- Mariamenatu, A.H.; Abdu, E.M. Overconsumption of Omega-6 Polyunsaturated Fatty Acids (PUFAs) versus Deficiency of Omega-3 PUFAs in Modern-Day Diets: The Disturbing Factor for Their “Balanced Antagonistic Metabolic Function” in the Human Body. J. Lipids 2021, 2021, 8848161. [Google Scholar] [CrossRef]
- Forsyth, S.; Gautier, S.; Salem, N., Jr. Global Estimates of Dietary Intake of Docosahexaenoic Acid and Arachidonic Acid in Developing and Developed Countries. Ann. Nutr. Metab. 2016, 68, 258–267. [Google Scholar] [CrossRef]
- Innes, J.K.; Calder, P.C. Omega-6 fatty acids and inflammation. Prostaglandins Leukot. Essent. Fat. Acids 2018, 132, 41–48. [Google Scholar] [CrossRef]
- Watanabe, Y.; Tatsuno, I. Omega-3 polyunsaturated fatty acids for cardiovascular diseases: Present, past and future. Expert Rev. Clin. Pharmacol. 2017, 10, 865–873. [Google Scholar] [CrossRef]
- Serini, S.; Calviello, G. Omega-3 PUFA Responders and Non-Responders and the Prevention of Lipid. Dysmetabolism and Related Diseases. Nutrients 2020, 12, 1363. [Google Scholar] [CrossRef] [PubMed]
- Belkouch, M.; Hachem, M.; Elgot, A.; Lo Van, A.; Picq, M.; Guichardant, M.; Lagarde, M.; Bernoud-Hubac, N. The pleiotropic effects of omega-3 docosahexaenoic acid on the hallmarks of Alzheimer’s disease. J. Nutr. Biochem. 2016, 38, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bigornia, S.J.; Scott, T.M.; Harris, W.S.; Tucker, K.L. Prospective Associations of Erythrocyte Composition and Dietary Intake of n-3 and n-6 PUFA with Measures of Cognitive Function. Nutrients 2018, 10, 1253. [Google Scholar] [CrossRef] [PubMed]
- Freitas-Simoes, T.M.; Cofán, M.; Blasco, M.A.; Soberón, N.; Foronda, M.; Corella, D.; Asensio, E.M.; Serra-Mir, M.; Roth, I.; Calvo, C.; et al. The red blood cell proportion of arachidonic acid relates to shorter leukocyte telomeres in Mediterranean elders: A secondary analysis of a randomized controlled trial. Clin. Nutr. 2019, 38, 958–961. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, P.; Wang, W.; Wang, J.; Zhang, Y.; Jiao, J. Polyunsaturated fatty acids intake, omega-6/omega-3 ratio and mortality: Findings from two independent nationwide cohorts. Clin. Nutr. 2019, 38, 848–855. [Google Scholar] [CrossRef]
- Lankinen, M.A.; Fauland, A.; Shimizu, B.I.; Ågren, J.; Wheelock, C.E.; Laakso, M.; Schwab, U.; Pihlajamäki, J. Inflammatory response to dietary linoleic acid depends on FADS1 genotype. Am. J. Clin. Nutr. 2019, 109, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Harauma, A.; Sueyasu, T.; Tokuda, H.; Yasuda, H.; Hoshi, Y.; Kaneda, Y.; Rogi, T.; Shibata, H.; Nakamura, M.T.; Moriguchi, T. Changes in behavior and fatty acid composition induced by long-term reduction in murine Δ6-desaturation activity. Prostaglandins Leukot. Essent. Fatty Acids 2020, 155, 102079. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Arenaza, L.; Rios, C.; Plows, J.F.; Berger, P.K.; Alderete, T.L.; Fogel, J.L.; Nayak, K.; Mohamed, P.; Hwang, D.; et al. PNPLA3 Genotype, Arachidonic Acid Intake, and Unsaturated Fat Intake Influences Liver Fibrosis in Hispanic Youth with Obesity. Nutrients 2021, 13, 1621. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.H.; Paris, C.; Magnien, M.; Colin, J.; Pelleïeux, S.; Coste, F.; Escanyé, M.C.; Pillot, T.; Olivier, J.L. Dietary arachidonic acid increases deleterious effects of amyloid-β oligomers on learning abilities and expression of AMPA receptors: Putative role of the ACSL4-cPLA2 balance. Alzheimer’s Res. Ther. 2017, 9, 69–93. [Google Scholar] [CrossRef]
- Mak, I.L.; Lavery, P.; Agellon, S.; Rauch, F.; Murshed, M.; Weiler, H.A. Arachidonic acid exacerbates diet-induced obesity and reduces bone mineral content without impacting bone strength in growing male rats. J. Nutr. Biochem. 2019, 73, 108226. [Google Scholar] [CrossRef]
- Boyd, J.T.; LoCoco, P.M.; Furr, A.R.; Bendele, M.R.; Tram, M.; Li, Q.; Chang, F.M.; Colley, M.E.; Samenuk, G.M.; Arris, D.A.; et al. Elevated dietary ω-6 polyunsaturated fatty acids induce reversible peripheral nerve dysfunction that exacerbates comorbid pain conditions. Nat. Metab. 2021, 3, 762–773. [Google Scholar] [CrossRef]
- Adam, A.C.; Lie, K.K.; Moren, M.; Skjærven, K.H. High dietary arachidonic acid levels induce changes in complex lipids and immune-related eicosanoids and increase levels of oxidised metabolites in zebrafish (Danio rerio). Br. J. Nutr. 2017, 117, 1075–1085. [Google Scholar] [CrossRef]
- Nayak, S.; Khozin-Goldberg, I.; Cohen, G.; Zilberg, D. Dietary Supplementation With ω6 LC-PUFA-Rich Algae Modulates Zebrafish Immune Function and Improves Resistance to Streptococcal Infection. Front. Immunol. 2018, 9, 1960. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Al Ashhab, A.; Zilberg, D.; Khozin-Goldberg, I. Dietary Supplementation with Omega-6 LC-PUFA-Rich Microalgae Regulates Mucosal Immune Response and Promotes Microbial Diversity in the Zebrafish Gut. Biology 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Weeks, T.L.; Hazen, S.L. Gut Microbiota and Cardiovascular Disease. Circ. Res. 2020, 127, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Ussar, S.; Fujisaka, S.; Kahn, C.R. Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol. Metab. 2016, 5, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, V.V.; Barichello De Quevedo, C.E.; Petronilho, F. Microbiota-gut-brain axis in the Alzheimer’s disease pathology—An overview. Neurosci. Res. 2022, 181, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Nuzum, N.D.; Loughman, A.; Szymlek-Gay, E.A.; Teo, W.P.; Hendy, A.M.; Macpherson, H. To the Gut Microbiome and Beyond: The Brain-First or Body-First Hypothesis in Parkinson’s Disease. Front. Microbiol. 2022, 13, 791213. [Google Scholar] [CrossRef]
- Taniya, M.A.; Chung, H.J.; Al Mamun, A.; Alam, S.; Aziz, M.A.; Emon, N.U.; Islam, M.M.; Hong, S.S.; Podder, B.R.; Ara Mimi, A.; et al. Role of Gut Microbiome in Autism Spectrum Disorder and Its Therapeutic Regulation. Front. Cell Infect. Microbiol. 2022, 12, 915701. [Google Scholar] [CrossRef]
- Suda, K.; Matsuda, K. How Microbes Affect Depression: Underlying Mechanisms via the Gut-Brain Axis and the Modulating Role of Probiotics. Int. J. Mol. Sci. 2022, 23, 1172. [Google Scholar] [CrossRef]
- Toledo, A.R.L.; Monroy, G.R.; Salazar, F.E.; Lee, J.Y.; Jain, S.; Yadav, H.; Borlongan, C.V. Gut-Brain Axis as a Pathological and Therapeutic Target for Neurodegenerative Disorders. Int. J. Mol. Sci. 2022, 23, 1184. [Google Scholar] [CrossRef]
- Ahmed, H.; Leyrolle, Q.; Koistinen, V.; Kärkkäinen, O.; Layé, S.; Delzenne, N.; Hanhineva, K. Microbiota-derived metabolites as drivers of gut-brain communication. Gut Microbes 2022, 14, 2102878. [Google Scholar] [CrossRef]
- Sochocka, M.; Donskow-Łysoniewska, K.; Diniz, B.S.; Kurpas, D.; Brzozowska, E.; Leszek, J. The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer’s Disease-a Critical Review. Mol. Neurobiol. 2019, 56, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, X.; Du, H. Inflammatory bowel disease: A potential pathogenic factor of Alzheimer’s disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 2022, 119, 110610. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, T.J.; Haskey, N.; Kwong, E.; Greuel, B.K.; Gates, E.J.; Gibson, D.L.; Klegeris, A. Dietary fats modulate neuroinflammation in mucin 2 knock out mice model of spontaneous colitis. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166336. [Google Scholar] [CrossRef] [PubMed]
- Fairley, A.; Stewart, C.J.; Cassidy, A.; Woodside, J.V.; McEvoy, C.T. Diet Patterns, the Gut Microbiome, and Alzheimer’s Disease. J. Alzheimer’s Dis. 2022, 88, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Wang, Y.; Huang, L.; Shen, M.; Yu, Y.; Yu, Q.; Chen, Y.; Xie, J. Review of the relationships among polysaccharides, gut microbiota, and human health. Food Res. Int. 2021, 140, 109858. [Google Scholar] [CrossRef]
- Ghosh, S.; Molcan, E.; DeCoffe, D.; Dai, C.; Gibson, D.L. Diets rich in n-6 PUFA induce intestinal microbial dysbiosis in aged mice. Br. J. Nutr. 2013, 110, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Selmin, O.I.; Papoutsis, A.J.; Hazan, S.; Smith, C.; Greenfield, N.; Donovan, M.G.; Wren, S.N.; Doetschman, T.C.; Snider, J.M.; Snider, A.J.; et al. n-6 High Fat Diet Induces Gut Microbiome Dysbiosis and Colonic Inflammation. Int. J. Mol. Sci. 2021, 22, 6919. [Google Scholar] [CrossRef]
- Zhuang, P.; Shou, Q.; Lu, Y.; Wang, G.; Qiu, J.; Wang, J.; He, L.; Chen, J.; Jiao, J.; Zhang, Y. Arachidonic acid sex-dependently affects obesity through linking gut microbiota-driven inflammation to hypothalamus-adipose-liver axis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2715–2726. [Google Scholar] [CrossRef]
- Benoit, B.; Laugerette, F.; Plaisancié, P.; Géloën, A.; Bodennec, J.; Estienne, M.; Pineau, G.; Bernalier-Donadille, A.; Vidal, H.; Michalski, M.C. Increasing fat content from 20 to 45 wt% in a complex diet induces lower endotoxemia in parallel with an increased number of intestinal goblet cells in mice. Nutr. Res. 2015, 35, 346–356. [Google Scholar] [CrossRef]
- Atienza, M.; Ziontz, J.; Cantero, J.L. Low-grade inflammation in the relationship between sleep disruption, dysfunctional adiposity, and cognitive decline in aging. Sleep Med. Rev. 2018, 42, 171–183. [Google Scholar] [CrossRef]
- Więckowska-Gacek, A.; Mietelska-Porowska, A.; Wydrych, M.; Wojda, U. Western diet as a trigger of Alzheimer’s disease: From metabolic syndrome and systemic inflammation to neuroinflammation and neurodegeneration. Ageing Res. Rev. 2021, 70, 101397. [Google Scholar] [CrossRef] [PubMed]
- Ackman, R.G. Remarks on official methods employing boron trifluoride in the preparation of methyl esters of the fatty acids of fish oils. J. Am. Oil Chem. Soc. 1998, 75, 541–545. [Google Scholar] [CrossRef]
- Godon, J.J.; Zumstein, E.; Dabert, P.; Habouzit, F.; Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997, 63, 2802–2813. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kittana, H.; Gomes-Neto, J.C.; Heck, K.; Geis, A.L.; Segura Muñoz, R.R.; Cody, L.A.; Schmaltz, R.J.; Bindels, L.B.; Sinha, R.; Hostetter, J.M.; et al. Commensal Escherichia coli Strains Can Promote Intestinal Inflammation via Differential Interleukin-6 Production. Front. Immunol. 2018, 9, 20318. [Google Scholar] [CrossRef]
- Sasajima, N.; Ogasawara, T.; Takemura, N.; Fujiwara, R.; Watanabe, J.; Sonoyama, K. Role of intestinal Bifidobacterium pseudolongum in dietary fructo-oligosaccharide inhibition of 2,4-dinitrofluorobenzene-induced contact hypersensitivity in mice. Br. J. Nutr. 2010, 103, 539–548. [Google Scholar] [CrossRef]
- Ogrinc Wagner, A.; Friedrich, V.; Barthels, C.; Marconi, P.; Blutke, A.; Brombacher, F.; Brocker, T. Strain specific maturation of Dendritic cells and production of IL-1β controls CD40-driven colitis. PLoS ONE 2019, 14, e0210998. [Google Scholar] [CrossRef]
- Aloisi, F.; Penna, G.; Polazzi, E.; Minghetti, L.; Adorini, L. CD40-CD154 interaction and IFN-gamma are required for IL-12 but not prostaglandin E2 secretion by microglia during antigen presentation to Th1 cells. J. Immunol. 1999, 162, 1384–1391. [Google Scholar]
- Bo, T.B.; Wen, J.; Zhao, Y.C.; Tian, S.J.; Zhang, X.Y.; Wang, D.H. Bifidobacterium pseudolongum reduces triglycerides by modulating gut microbiota in mice fed high-fat food. J. Steroid Biochem. Mol. Biol. 2020, 198, 105602. [Google Scholar] [CrossRef] [PubMed]
- Ghadimi, D.; Yoness Hassan, M.F.; Fölster-Holst, R.; Röcken, C.; Ebsen, M.; de Vrese, M.; Heller, K.J. Regulation of hepcidin/iron-signalling pathway interactions by commensal bifidobateria plays an important role for the inhibition of metaflammation-related biomarkers. Immunobiology 2020, 225, 151874. [Google Scholar] [CrossRef] [PubMed]
- Gorissen, L.; De Vuyst, L.; Raes, K.; De Smet, S.; Leroy, F. Conjugated linoleic and linolenic acid production kinetics by bifidobacteria differ among strains. Int. J. Food Microbiol. 2012, 155, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Wei, J.; Gao, Y.; Yang, Y.; Li, Y.; Zhu, H.; Su, L.; Su, X.; Zhang, Y.; Yang, R. Reg4 and complement factor D prevent the overgrowth of E. coli in the mouse gut. Commun. Biol. 2020, 3, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, K.E.; Lee, S.A.; Jang, H.M.; Kim, D.H. Interplay Between Human Gut Bacteria Escherichia coli and Lactobacillus mucosae in the occurrence of neuropsychiatric disorders in mice. Front. Immunol. 2020, 11, 273. [Google Scholar] [CrossRef]
- Weng, Y.J.; Jiang, D.X.; Liang, J.; Ye, S.C.; Tan, W.K.; Yu, C.Y.; Zhou, Y. Effects of Pretreatment with Bifidobacterium bifidum Using 16S Ribosomal RNA Gene Sequencing in a Mouse Model of Acute Colitis Induced by Dextran Sulfate Sodium. Med. Sci. Monit. 2021, 27, e928478. [Google Scholar] [CrossRef]
- Das, U.N. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: A review. J. Adv. Res. 2018, 11, 57–66. [Google Scholar] [CrossRef]
- Naoe, S.; Tsugawa, H.; Takahashi, M.; Ikeda, K.; Arita, M. Characterization of Lipid Profiles after Dietary Intake of Polyunsaturated Fatty Acids Using Integrated Untargeted and Targeted Lipidomics. Metabolites 2019, 9, 241. [Google Scholar] [CrossRef]
- Jo, J.K.; Seo, S.H.; Park, S.E.; Kim, H.W.; Kim, E.J.; Kim, J.S.; Pyo, J.Y.; Cho, K.M.; Kwon, S.J.; Park, D.H.; et al. Gut Microbiome and Metabolome Profiles Associated with High-Fat Diet in Mice. Metabolites 2021, 11, 482. [Google Scholar] [CrossRef]
- Bolte, L.A.; Vich Vila, A.; Imhann, F.; Collij, V.; Gacesa, R.; Peters, V.; Wijmenga, C.; Kurilshikov, A.; Campmans-Kuijpers, M.J.E.; Fu, J.; et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021, 70, 1287–1298. [Google Scholar] [CrossRef]
- Ozato, N.; Saito, S.; Yamaguchi, T.; Katashima, M.; Tokuda, I.; Sawada, K.; Katsuragi, Y.; Kakuta, M.; Imoto, S.; Ihara, K.; et al. Blautia genus associated with visceral fat accumulation in adults 20–76 years of age. NPJ Biofilms Microbiomes 2019, 5, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Dahl, W.J.; Rivero Mendoza, D.; Lambert, J.M. Diet, nutrients and the microbiome. Prog. Mol. Biol. Transl. Sci. 2020, 171, 237–263. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Lamas, B.; Pham, H.P.; Michel, M.L.; Rainteau, D.; Bridonneau, C.; da Costa, G.; van Hylckama Vlieg, J.; Sovran, B.; Chamignon, C.; et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 2018, 9, 2802–2817. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Membrez, M.; Morin-Rivron, D.; Siddharth, J.; Chou, C.J.; Henry, H.; Bruce, S.; Metairon, S.; Raymond, F.; Betrisey, B.; et al. Transcriptomics-driven lipidomics (TDL) identifies the microbiome-regulated targets of ileal lipid metabolism. NPJ Syst. Biol. Appl. 2017, 3, 33–48. [Google Scholar] [CrossRef]
- Ramakers, J.D.; Mensink, R.P.; Verstege, M.I.; te Velde, A.A.; Plat, J. An arachidonic acid-enriched diet does not result in more colonic inflammation as compared with fish oil- or oleic acid-enriched diets in mice with experimental colitis. Br. J. Nutr. 2008, 100, 347–354. [Google Scholar] [CrossRef]
- Tateishi, N.; Kakutani, S.; Kawashima, H.; Shibata, H.; Morita, I. Dietary supplementation of arachidonic acid increases arachidonic acid and lipoxin A4 contents in colon, but does not affect severity or prostaglandin E2 content in murine colitis model. Lipids Health Dis. 2014, 13, 30–40. [Google Scholar] [CrossRef]
- Charles-Messance, H.; Mitchelson, K.A.J.; De Marco Castro, E.; Sheedy, F.J.; Roche, H.M. Regulating metabolic inflammation by nutritional modulation. J. Allergy Clin. Immunol. 2020, 146, 706–720. [Google Scholar] [CrossRef]
- Aarts, S.; Reiche, M.; den Toom, M.; Gijbels, M.; Beckers, L.; Gerdes, N.; Lutgens, E. Depletion of CD40 on CD11c+ cells worsens the metabolic syndrome and ameliorates hepatic inflammation during NASH. Sci. Rep. 2019, 9, 14702–14713. [Google Scholar] [CrossRef]
- Polese, L.; Angriman, I.; Cecchetto, A.; Norberto, L.; Scarpa, M.; Ruffolo, C.; Barollo, M.; Sommariva, A.; D’Amico, D.F. The role of CD40 in ulcerative colitis: Histochemical analysis and clinical correlation. Eur. J. Gastroenterol. Hepatol. 2002, 14, 237–241. [Google Scholar] [CrossRef]
- Kamińska, B.; Roszko-Kirpsza, I.; Landowski, P.; Szlagatys-Sidorkiewicz, A.; Guzińska-Ustymowicz, K.; Maciorkowska, E. Evaluation of CD40 and CD80 receptors in the colonic mucosal membrane of children with inflammatory bowel disease. Ann. Agric. Environ. Med. 2015, 22, 695–699. [Google Scholar] [CrossRef]
- Elson, C.O.; Cong, Y. Understanding immune-microbial homeostasis in intestine. Immunol. Res. 2002, 26, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, E.; Ogita, T.; Miyamoto, J.; Kawamoto, S.; Morita, H.; Ohno, H.; Suzuki, T.; Tanabe, S. Bifidobacterium longum alleviates dextran sulfate sodium-induced colitis by suppressing IL-17A response: Involvement of intestinal epithelial costimulatory molecules. PLoS ONE 2013, 8, e79735. [Google Scholar] [CrossRef] [PubMed]
- Mion, F.; D’Incà, F.; Danelli, L.; Toffoletto, B.; Guarnotta, C.; Frossi, B.; Burocchi, A.; Rigoni, A.; Gerdes, N.; Lutgens, E.; et al. Mast cells control the expansion and differentiation of IL-10-competent B cells. J. Immunol. 2014, 193, 4568–4579. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, S.; Sharma, K.K.A. Gut-organ axis: A microbial outreach and networking. Lett. Appl. Microbiol. 2021, 72, 636–668. [Google Scholar] [CrossRef]
- Cintra, D.E.; Ropelle, E.R.; Moraes, J.C.; Pauli, J.R.; Morari, J.; Souza, C.T.; Grimaldin, R.; Stahl, M.; Carvalheira, J.B.; Saad, M.J.; et al. Unsaturated fatty acids revert diet-induced hypothalamic inflammation in obesity. PLoS ONE 2012, 7, e30571. [Google Scholar] [CrossRef]
- Wernstedt Asterholm, I.; Tao, C.; Morley, T.S.; Wang, Q.A.; Delgado-Lopez, F.; Wang, Z.V.; Scherer, P.E. Benoit. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell. Metab. 2014, 20, 103–118. [Google Scholar] [CrossRef]
- Bobbo, V.C.; Engel, D.F.; Jara, C.P.; Mendes, N.F.; Haddad-Tovolli, R.; Prado, T.P.; Sidarta-Oliveira, D.; Morari, J.; Velloso, L.A.; Araujo, E.P. Interleukin-6 actions in the hypothalamus protects against obesity and is involved in the regulation of neurogenesis. J. Neuroinflammation 2021, 18, 192–209. [Google Scholar] [CrossRef]
- Lin, H.; Hikawa, N.; Takenaka, T.; Ishikawa, Y. Interleukin-12 promotes neurite outgrowth in mouse sympathetic superior cervical ganglion neurons. Neurosci. Lett. 2000, 278, 129–132. [Google Scholar] [CrossRef]
- Eede, P.; Obst, J.; Benke, E.; Yvon-Durocher, G.; Richard, B.C.; Gimber, N.; Schmoranzer, J.; Böddrich, A.; Wanker, E.E.; Prokop, S.; et al. Interleukin-12/23 deficiency differentially affects pathology in male and female Alzheimer’s disease-like mice. EMBO Rep. 2020, 21, e48530. [Google Scholar] [CrossRef]
- Calder, P.C. Functional Roles of fatty acids and their effects on human health. J. Parenter. Enter. Nutr. 2015, 39, 18S–32S. [Google Scholar] [CrossRef]


| Std-ARA | HL-ARA | HL + ARA | |
|---|---|---|---|
| Proteins (% w/w) | 14 | 14 | 14 |
| Carbohydrates (% w/w) | 71 | 61 | 61 |
| Lipids (% w/w) | 5 | 15 | 15 |
| Others (% w/w) | 10 | 10 | 10 |
| Energy (kcal/100 g) 1 | 390 | 437 | 437 |
| Fatty Acids | Std-ARA | HL-ARA | HL + ARA |
|---|---|---|---|
| C10:0 to C15:0 | 1.00 | 0.96 | 1.25 |
| C16:0 palmitic acid | 17.25 | 17.11 | 17.50 |
| C16:1 palmitoleic acid | 1.54 | 1.58 | 1.48 |
| C17:0 3-hydroxyheptadecanoic acid | 0.15 | 0.14 | 0.18 |
| C18:0 stearic acid | 8.60 | 8.63 | 9.16 |
| C18:1 ω-9 oleic acid | 31.84 | 31.65 | 32.07 |
| C18:2 ω-6 linoleic acid | 29.83 | 29.87 | 23.23 |
| C18:3 ω-6 γ-linolenic acid | 1.11 | 1.12 | 1.18 |
| C18:3 ω-3 α-linolenic acid | 7.42 | 7.48 | 5.33 |
| C18:4 ω-3 stearidonic acid | 0.57 | 0.54 | 0.52 |
| C20:1 ω-9 11-eicosenoic acid | 0.31 | 0.31 | 0.41 |
| C20:4 ω-6 arachidonic acid | 0.21 | 0.25 | 6.99 |
| Ʃ saturated fatty acids | 27 | 26.8 | 27.1 |
| Ʃ mono-unsaturated fatty acids | 33.7 | 33.5 | 34 |
| Ʃ ω-6 PUFA | 31.2 | 31.3 | 31.4 |
| Ʃ ω-3 PUFA | 8 | 8 | 5.9 |
| ω-6/ω-3 PUFA ratio | 3.9 | 3.9 | 5.3 |
| Gene | Primer Sequence (5’–3’) | |
|---|---|---|
| GAPDH | Forward: | AATTCAACGGCACAGTCAAGGC |
| Reverse: | CGTGGTTCACACCCATCACAAA | |
| IL-1β | Forward: | AGGCCACAGGTATTTTGTCGT |
| Reverse: | TGTCCAGATGAGAGCATCCAG | |
| IL-6 | Forward: | TAGTCCTTCCTACCCCAATTTCC |
| Reverse: | TTGGTCCTTAGCCACTCCTTC | |
| TNF-α | Forward: | AGCCCACGTCGTAGCAAAC |
| Reverse: | GATAGCAAATCGGCTGACGG | |
| IL-12 | Forward: | ACCTGTGACACGCCTGAAG |
| Reverse: | CTCAGAGTCTCGCCTCCTTTG | |
| Adiponectin | Forward: | GACGACACCAAAAGGGCTCA |
| Reverse: | AGGTGAAGAGAACGGCCTTG | |
| CD40 | Forward: | TTGTTGACAGCGGTCCATCT |
| Reverse: | TCACGACAGGAATGACCAGC | |
| Claudin 1 | Forward: | GCCATCTACGAGGGACTGTG |
| Reverse: | CACTAATGTCGCCAGACCTGAA | |
| Occludin | Forward: | TGAATGGGTCACCGAGGGAG |
| Reverse: | AGATAAGCGAACCTGCCGA | |
| Comparison between HL-ARA and HL + ARA Groups | ||||
|---|---|---|---|---|
| Pathways | Description | Results | p-Values (Corrected) | Difference between Means |
| PWY-5676 | Acetyl-CoA fermentation to butanoate, organic acid synthesis, energetic metabolism | HL-ARA > HL + ARA | 0.0096 | −0.0933 |
| PWY-6471 | Peptidoglycan biosynthesis IV (Enterococcus faecium), bacterial wall synthesis | HL-ARA > HL + ARA | 0.0148 | −0.0778 |
| P124-PWY | Bifidobacterium shunt, degradation of complex carbohydrates, organic acid synthesis | HL-ARA > HL + ARA | 0.0245 | −0.0726 |
| UDPNAGSYN-PWY | UDP-N-acetyl-D-glucosamine biosynthesis I, bacterial wall synthesis | HL-ARA > HL + ARA | 0.0443 | −0.0693 |
| P4-PWY | Superpathway of L-lysine, L-threonine and L-methionine biosynthesis I | HL-ARA > HL + ARA | 0.0062 | −0.0690 |
| P441-PWY | Superpathway of N-acetylneuraminate degradation, degradation of complex carbohydrates, organic acid synthesis | HL-ARA > HL + ARA | 0.0171 | −0.0652 |
| PWY-5347 | Superpathway of L-methionine biosynthesis (trans-sulfuration) | HL-ARA > HL + ARA | 0.0252 | −0.0649 |
| MET-SAM-PWY | Superpathway of S-adenosyl-L-methionine biosynthesis | HL-ARA > HL + ARA | 0.0349 | −0.0528 |
| OANTIGEN-PWY | O-antigen building blocks biosynthesis (E. coli), bacterial wall synthesis | HL-ARA > HL + ARA | 0.0458 | −0.0421 |
| HOMOSER-METSYN-PWY | L-methionine biosynthesis I | HL-ARA > HL + ARA | 0.0403 | −0.0379 |
| PWY-5304 | Superpathway of sulfur oxidation (Acidianus ambivalens) | HL-ARA > HL + ARA | 0.0401 | −0.0349 |
| PWY-7013 | L-1,2-propanediol degradation | HL-ARA > HL + ARA | 0.0115 | −0.0338 |
| PWY0-1479 | tRNA processing, protein synthesis | HL-ARA > HL + ARA | 0.0203 | −0.0323 |
| PWY-181 | Photorespiration | HL-ARA > HL + ARA | 0.0053 | −0.0293 |
| PWY-5005 | Biotin biosynthesis II | HL-ARA > HL + ARA | 0.0252 | −0.0110 |
| P341-PWY | Glycolysis V (Pyrococcus), anaerobic carbohydrate metabolism | HL-ARA > HL + ARA | 0.0475 | −0.0042 |
| LEU-DEG2-PWY | L-leucine degradation I | HL-ARA > HL + ARA | 0.0153 | −0.0032 |
| PWY-5741 | Ethylmalonyl-CoA pathway | HL-ARA > HL + ARA | 0.0263 | −0.0010 |
| PWY-5747 | 2-methylcitrate cycle II | HL-ARA > HL + ARA | 0.0476 | −0.0001 |
| PWY0-42 | 2-methylcitrate cycle I | HL-ARA > HL + ARA | 0.0492 | −0.0001 |
| PWY-4984 | Urea cycle | HL-ARA < HL + ARA | 0.0488 | 0.0111 |
| HEME-BIOSYNTHESIS-II | Heme biosynthesis I (aerobic) | HL-ARA < HL + ARA | 0.0459 | 0.0201 |
| GALACTUROCAT-PWY | D-galacturonate degradation I, carbohydrate metabolism | HL-ARA < HL + ARA | 0.0429 | 0.0226 |
| RHAMCAT-PWY | L-rhamnose degradation I, carbohydrate metabolism | HL-ARA < HL + ARA | 0.0120 | 0.0239 |
| PWY-7323 | Superpathway of GDP-mannose-derived O-antigen building blocks biosynthesis, bacterial wall synthesis | HL-ARA < HL + ARA | 0.0449 | 0.0320 |
| PANTOSYN-PWY | Pantothenate and coenzyme A biosynthesis I | HL-ARA < HL + ARA | 0.0314 | 0.0389 |
| PWY-6897 | Thiamin salvage II, vitamin B1 synthesis | HL-ARA < HL + ARA | 0.0149 | 0.0398 |
| PWY0-845 | Superpathway of pyridoxal 5’-phosphate biosynthesis and salvage, vitamin B6 synthesis | HL-ARA < HL + ARA | 0.0354 | 0.0414 |
| PWY-6892 | Thiazole biosynthesis I (E. coli), vitamin B1 synthesis | HL-ARA < HL + ARA | 0.0128 | 0.0423 |
| PWY-6507 | 4-deoxy-L-threo-hex-4-enopyranuronate degradation, complex carbohydrate degradation | HL-ARA < HL + ARA | 0.0437 | 0.0437 |
| PYRIDOXSYN-PWY | Pyridoxal 5’-phosphate biosynthesis I, vitamin B6 synthesis | HL-ARA < HL + ARA | 0.0341 | 0.0472 |
| PYRIDNUCSYN-PWY | NAD biosynthesis I (from aspartate) | HL-ARA < HL + ARA | 0.0395 | 0.0476 |
| PANTO-PWY | Phosphopantothenate biosynthesis I | HL-ARA < HL + ARA | 0.0324 | 0.0511 |
| PWY-7539 | 6-hydroxymethyl-dihydropterin diphosphate biosynthesis III (Chlamydia), folate synthesis | HL-ARA < HL + ARA | 0.0480 | 0.0534 |
| RIBOSYN2-PWY | Flavin biosynthesis I (bacteria and plants) | HL-ARA < HL + ARA | 0.0203 | 0.0537 |
| THISYN-PWY | Superpathway of thiamin diphosphate biosynthesis I, vitamin B1 synthesis | HL-ARA < HL + ARA | 0.0038 | 0.0592 |
| FOLSYN-PWY | Superpathway of tetrahydrofolate biosynthesis and salvage, folate synthesis | HL-ARA < HL + ARA | 0.0092 | 0.0669 |
| PWY-6612 | Superpathway of tetrahydrofolate biosynthesis, folate synthesis | HL-ARA < HL + ARA | 0.0071 | 0.0692 |
| Comparison between Std-ARA and HL-ARA groups | ||||
| ARGSYNBSUB-PWY | L-arginine biosynthesis II (acetyl cycle) | Std-ARA > HL-ARA | 0.0071 | −0.0887 |
| PWY-5505 | L-glutamate and L-glutamine biosynthesis | Std-ARA > HL-ARA | 0.0155 | −0.0809 |
| PWY4FS-7 | Phosphatidylglycerol biosynthesis I (plastidic) | Std-ARA > HL-ARA | 0.0426 | −0.0663 |
| PWY4FS-8 | Phosphatidylglycerol biosynthesis II (non-plastidic) | Std-ARA > HL-ARA | 0.0426 | −0.0663 |
| PWY-7315 | dTDP-N-acetylthomosamine biosynthesis, bacterial wall synthesis | Std-ARA > HL-ARA | 0.0021 | −0.0566 |
| ARGSYN-PWY | L-arginine biosynthesis I (via L-ornithine) | Std-ARA > HL-ARA | 0.0121 | −0.0558 |
| PWY-7400 | L-arginine biosynthesis IV (archaebacteria) | Std-ARA > HL-ARA | 0.0117 | −0.0552 |
| OANTIGEN-PWY | O-antigen building blocks biosynthesis (E. coli), bacterial wall synthesis | Std-ARA > HL-ARA | 0.0196 | −0.0547 |
| PWY-5188 | Tetrapyrrole biosynthesis I (from glutamate), heme synthesis | Std-ARA > HL-ARA | 0.0029 | −0.0536 |
| PWY-5189 | Tetrapyrrole biosynthesis II (from glycine), heme synthesis | Std-ARA > HL-ARA | 0.0036 | −0.0500 |
| TRPSYN-PWY | L-tryptophan biosynthesis | Std-ARA > HL-ARA | 0.0126 | −0.0484 |
| PWY-5104 | L-isoleucine biosynthesis IV | Std-ARA > HL-ARA | 0.0401 | −0.0399 |
| PWY0-1479 | tRNA processing, protein synthesis | Std-ARA > HL-ARA | 0.0338 | −0.0368 |
| PWY-7371 | 1,4-dihydroxy-6-naphthoate biosynthesis II, metaquinone synthesis | Std-ARA < HL-ARA | 0.0411 | 0.0423 |
| PWY-5659 | GDP-mannose biosynthesis | Std-ARA < HL-ARA | 0.0314 | 0.0524 |
| PANTOSYN-PWY | Pantothenate and coenzyme A biosynthesis I | Std-ARA < HL-ARA | 0.0375 | 0.0526 |
| FASYN-ELONG-PWY | Fatty acid elongation, saturated lipid synthesis | Std-ARA < HL-ARA | 0.0362 | 0.0654 |
| PWY-7199 | Pyrimidine deoxyribonucleosides salvage, nucleotide synthesis | Std-ARA < HL-ARA | 0.0047 | 0.0701 |
| PWY-5695 | Urate biosynthesis/inosine 5’-phosphate degradation | Std-ARA < HL-ARA | 0.0320 | 0.0743 |
| PWY-6700 | Queuosine biosynthesis, RNA synthesis | Std-ARA < HL-ARA | 0.0295 | 0.0983 |
| Comparison between Std-ARA and HL + ARA groups | ||||
| P23-PWY | Reductive TCA cycle I, lipids, carbohydrates, protein synthesis | Std-ARA > HL + ARA | 0.0292 | −0.0618 |
| TRPSYN-PWY | L-tryptophan biosynthesis | Std-ARA > HL + ARA | 0.0081 | −0.0510 |
| PWY-5188 | Retrapyrrole biosynthesis I (from glutamate), heme synthesis | Std-ARA > HL + ARA | 0.0142 | −0.0440 |
| PWY-5189 | Tetrapyrrole biosynthesis II (from glycine), heme synthesis | Std-ARA > HL + ARA | 0.0137 | −0.0428 |
| PWY-6121 | 5-aminoimidazole ribonucleotide biosynthesis I | Std-ARA < HL + ARA | 0.0334 | 0.0155 |
| GLYCOLYSIS | Glycolysis I (from glucose 6-phosphate) | Std-ARA < HL + ARA | 0.0325 | 0.0391 |
| PWY-7234 | Inosine-5’-phosphate biosynthesis III, guanosine nucleotide synthesis | Std-ARA < HL + ARA | 0.0279 | 0.0694 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinchaud, K.; Hafeez, Z.; Auger, S.; Chatel, J.-M.; Chadi, S.; Langella, P.; Paoli, J.; Dary-Mourot, A.; Maguin-Gaté, K.; Olivier, J.L. Impact of Dietary Arachidonic Acid on Gut Microbiota Composition and Gut–Brain Axis in Male BALB/C Mice. Nutrients 2022, 14, 5338. https://doi.org/10.3390/nu14245338
Pinchaud K, Hafeez Z, Auger S, Chatel J-M, Chadi S, Langella P, Paoli J, Dary-Mourot A, Maguin-Gaté K, Olivier JL. Impact of Dietary Arachidonic Acid on Gut Microbiota Composition and Gut–Brain Axis in Male BALB/C Mice. Nutrients. 2022; 14(24):5338. https://doi.org/10.3390/nu14245338
Chicago/Turabian StylePinchaud, Katleen, Zeeshan Hafeez, Sandrine Auger, Jean-Marc Chatel, Sead Chadi, Philippe Langella, Justine Paoli, Annie Dary-Mourot, Katy Maguin-Gaté, and Jean Luc Olivier. 2022. "Impact of Dietary Arachidonic Acid on Gut Microbiota Composition and Gut–Brain Axis in Male BALB/C Mice" Nutrients 14, no. 24: 5338. https://doi.org/10.3390/nu14245338
APA StylePinchaud, K., Hafeez, Z., Auger, S., Chatel, J.-M., Chadi, S., Langella, P., Paoli, J., Dary-Mourot, A., Maguin-Gaté, K., & Olivier, J. L. (2022). Impact of Dietary Arachidonic Acid on Gut Microbiota Composition and Gut–Brain Axis in Male BALB/C Mice. Nutrients, 14(24), 5338. https://doi.org/10.3390/nu14245338





