Kombuchas from Green and Black Tea Modulate the Gut Microbiota and Improve the Intestinal Health of Wistar Rats Fed a High-Fat High-Fructose Diet
Abstract
1. Introduction
2. Material and Methods
2.1. Kombuchas Preparation
2.2. Animal Study
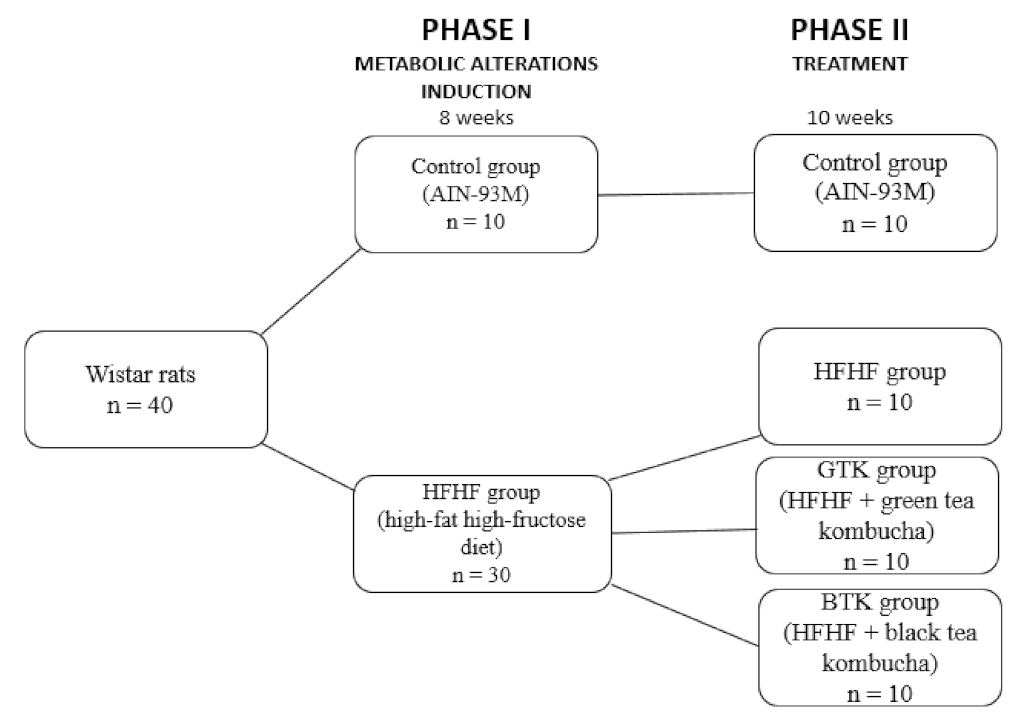
2.2.1. Study Design
2.2.2. Kombuchas Characterization and Dosage
2.2.3. Euthanasia and Samples Collection
2.2.4. Histological Analysis
2.2.5. Intestinal Permeability
2.2.6. Fecal pH and Short-Chain Fatty Acids Analysis
2.2.7. DNA Extraction and Microbiota Profile
2.3. Statistical Analysis
3. Results
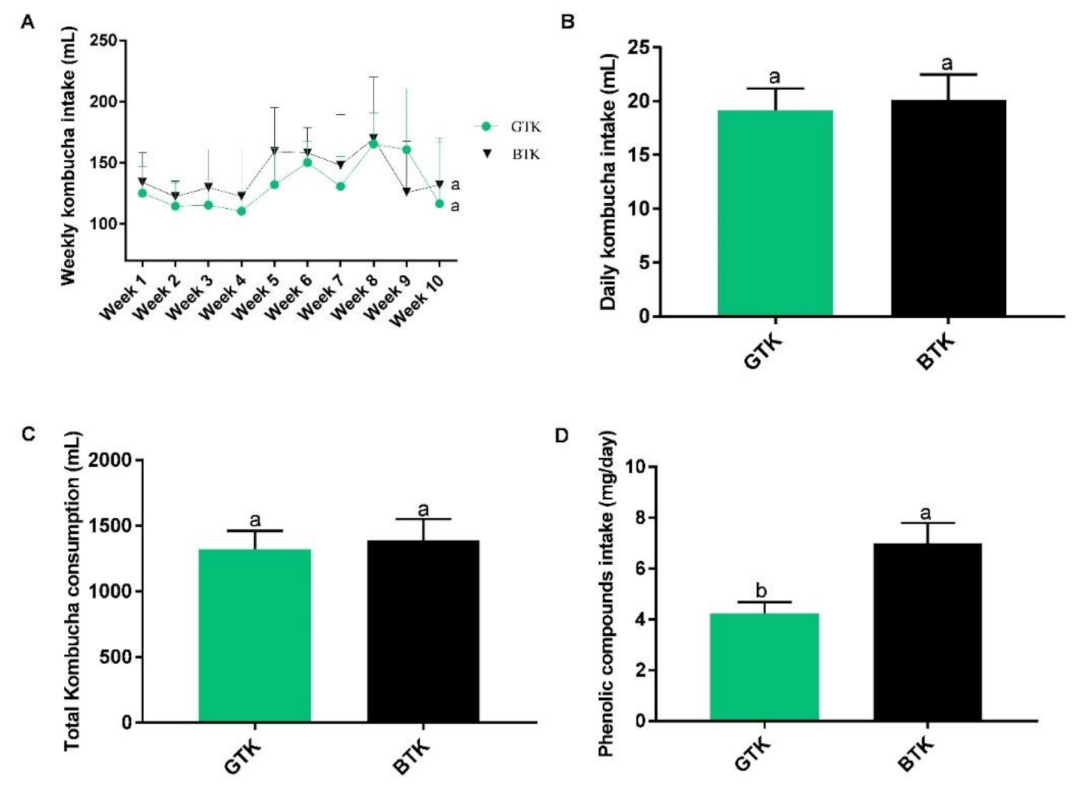
3.1. Kombucha Chemical Characterization and Consumption
3.2. Biometric Parameters
3.3. Bioinformatics Analysis
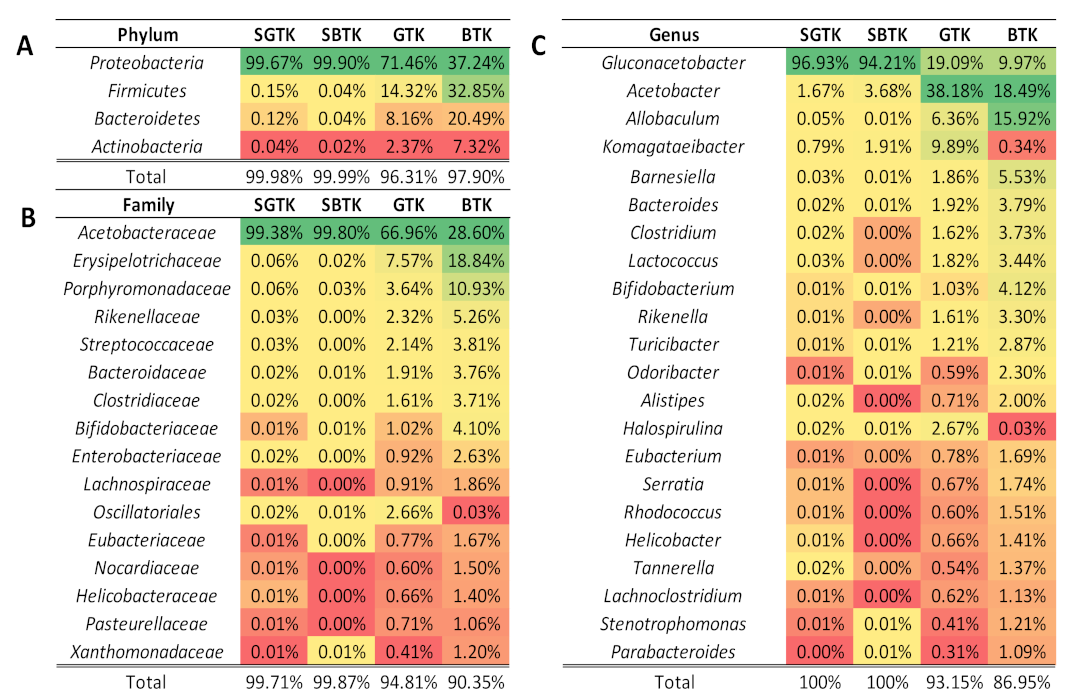
3.4. Microbiota Profiling of GTK and BTK and Their Respective SCOBYs
3.5. Alpha and Beta-Diversity Metrics of Gut Microbiota
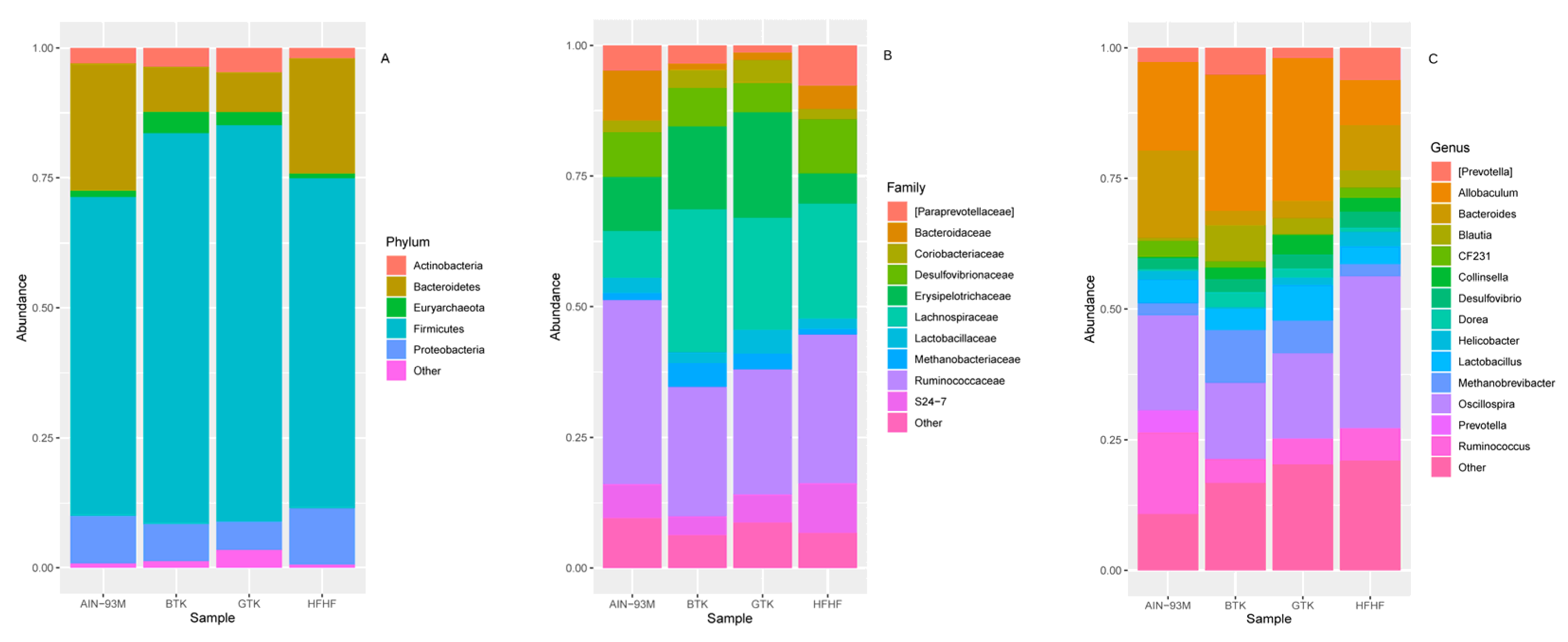
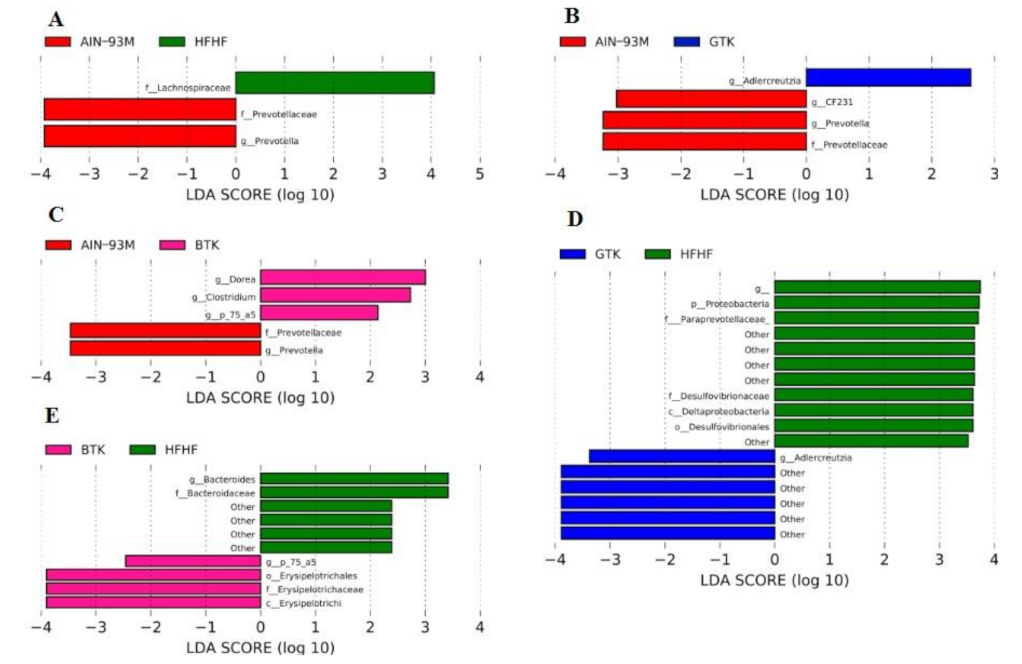
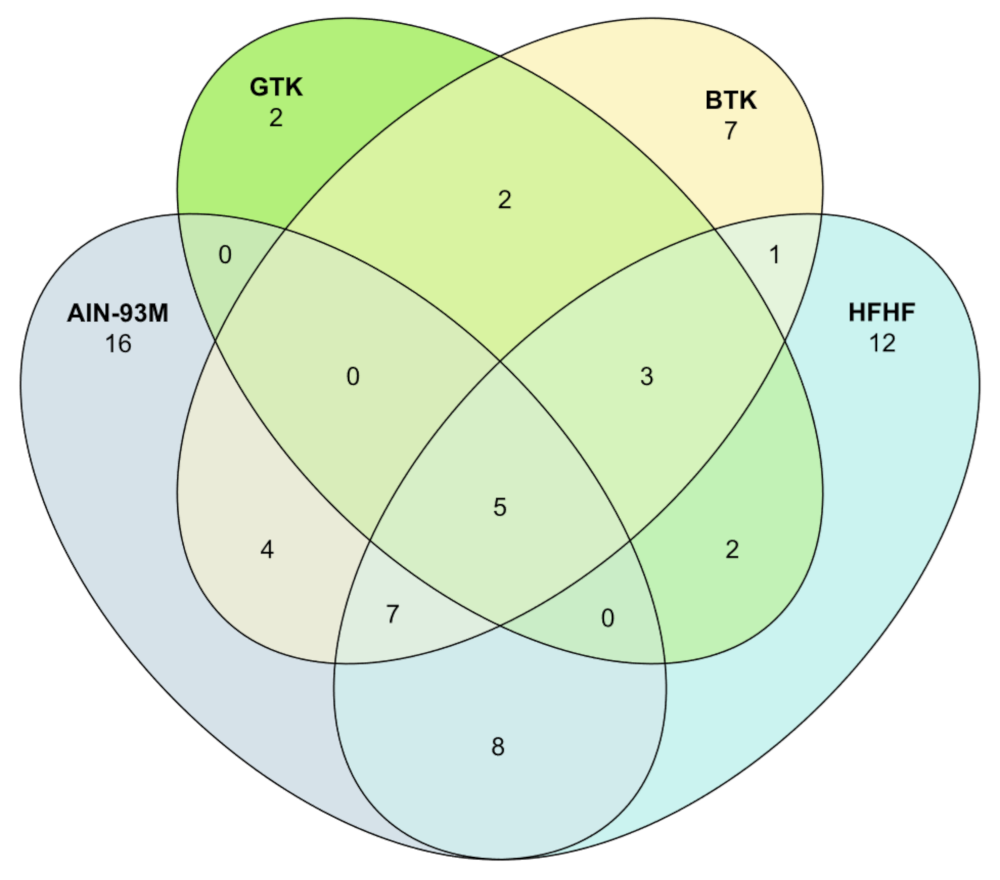
3.6. Taxonomic Assignment and Gut Bacterial Composition
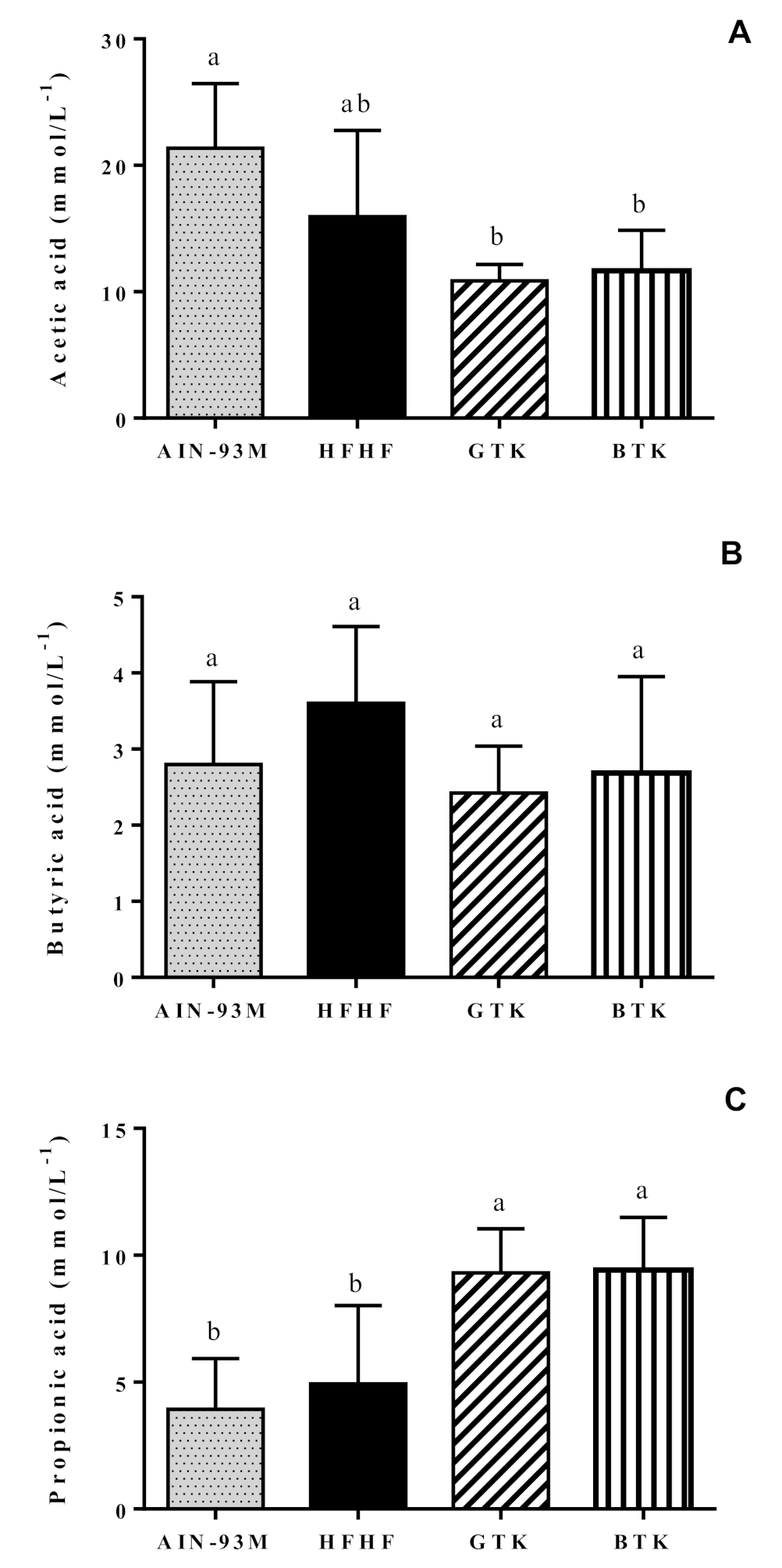
3.7. Fecal pH and Short-Chain Fatty Acids Content
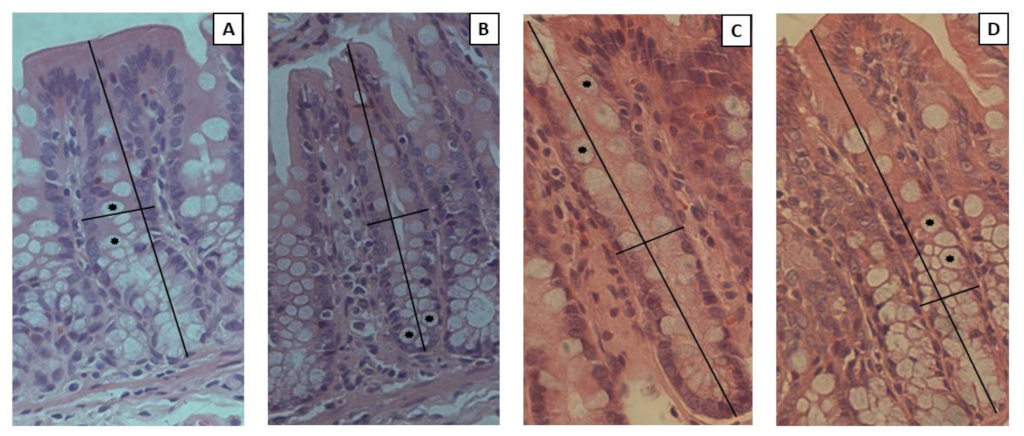
3.8. Intestinal Permeability and Histological Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malesza, I.J.; Malesza, M.; Walkowiak, J.; Mussin, N.; Walkowiak, D.; Aringazina, R.; Bartkowiak-Wieczorek, J.; Mądry, E. High-fat, western-style diet, systemic inflammation, and gut microbiota: A narrative review. Cells 2021, 10, 3164. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Chen, S.Y.; Sheng, L.; Jena, P.K.; Kalanetra, K.M.; Mills, D.A.; Wan, Y.J.Y.; Slupsky, C.M. Long-Term Effects of Western Diet Consumption in Male and Female Mice. Sci. Rep. 2020, 10, 14686. [Google Scholar] [CrossRef] [PubMed]
- Kopp, W. How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 2221–2236. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.Y.; Ha, C.W.Y.; Campbell, C.R.; Mitchell, A.J.; Dinudom, A.; Oscarsson, J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE 2012, 7, e34233. [Google Scholar] [CrossRef]
- Schoeler, M.; Caesar, R. Dietary Lipids, Gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef]
- Li, X.; Guo, J.; Ji, K.; Zhang, P. Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota. Sci. Rep. 2016, 6, 32953. [Google Scholar] [CrossRef]
- Miura, K.; Ishioka, M.; Iijima, K. The roles of the gut microbiota and toll-like receptors in obesity and nonalcoholic fatty liver disease. J. Obes. Metab. Syndr. 2017, 26, 86–96. [Google Scholar] [CrossRef]
- Lambertz, J.; Weiskirchen, S.; Landert, S.; Weiskirchen, R. Fructose: A dietary sugar in crosstalk with microbiota contributing to the development and progression of non-alcoholic liver disease. Front. Immunol. 2017, 8, 1159. [Google Scholar] [CrossRef]
- Do, M.H.; Lee, E.; Oh, M.J.; Kim, Y.; Park, H.Y. High-glucose or-fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef]
- Kapp, J.M.; Sumner, W. Kombucha: A systematic review of the empirical evidence of human health benefit. Ann. Epidemiol. 2019, 30, 66–70. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbaša, R.V.; Lončar, E.S.; Vitas, J.S.; Sathishkumar, M. A review on kombucha tea-microbiology, composition, fermentation, beneficial effects, toxicity, and tea fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.-P.; Taillandier, P. Understanding kombucha tea fermentation: A review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Greenwalt, C.J.; Steinkraus, K.H.; Ledford, R.A. Kombucha, the fermented tea: Microbiology, composition, and claimed health effects. J. Food Prot. 2000, 63, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Vina, I.; Semjonovs, P.; Linde, R.; Denina, I. Current evidence on physiological activity and expected health effects of kombucha fermented beverage. J. Med. Food 2014, 17, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Watawana, M.I.; Jayawardena, N.; Gunawardhana, C.B.; Waisundara, V.Y. Health, Wellness, and Safety Aspects of the Consumption of Kombucha. J. Chem. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Gaggìa, F.; Baffoni, L.; Galiano, M.; Nielsen, D.S.; Jakobsen, R.R.; Castro-Mejía, J.L.; Bosi, S.; Truzzi, F.; Musumeci, F.; Dinelli, G.; et al. Kombucha beverage from green, black and rooibos teas: A comparative study looking at microbiology, chemistry and antioxidant activity. Nutrients 2018, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.R.; Neto, R.O.; dos Santos D’Almeida, C.T.; do Nascimento, T.P.; Pressete, C.G.; Azevedo, L.; Martino, H.S.D.; Cameron, L.C.; Ferreira, M.S.L.; de Barros, F.A.R. Kombuchas from green and black teas have different phenolic profile, which impacts their antioxidant capacities, antibacterial and antiproliferative activities. Food Res. Int. 2020, 128, 108782. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Troise, A.D.; Vitaglione, P.; Ercolini, D. Different temperatures select distinctive acetic acid bacteria species and promotes organic acids production during kombucha tea fermentation. Food Microbiol. 2018, 73, 11–16. [Google Scholar] [CrossRef]
- de Noronha, M.C.; Cardoso, R.R.; dos Santos D’Almeida, C.T.; do Carmo, M.A.V.; Azevedo, L.; Maltarollo, V.G.; Júnior, J.I.R.; Eller, M.R.; Cameron, L.C.; Ferreira, M.S.L.; et al. Black tea kombucha: Physicochemical, microbiological and comprehensive phenolic profile changes during fermentation, and antimalarial activity. Food Chem. 2022, 384, 132515. [Google Scholar] [CrossRef]
- Arıkan, M.; Mitchell, A.L.; Finn, R.D.; Gürel, F. Microbial composition of kombucha determined using amplicon sequencing and shotgun metagenomics. J. Food Sci. 2020, 85, 455–464. [Google Scholar] [CrossRef]
- Harrison, K.; Curtin, C.; Arıkan, M.; Mitchell, A.L.; Finn, R.D.; Gürel, F. Microbial composition of scoby starter cultures used by commercial kombucha brewers in north america. J. Food Sci. 2021, 9, 1060. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kim, I.; Mannaa, M.; Kim, J.; Wang, S.; Park, I.; Kim, J.; Seo, Y.S. Effect of kombucha on gut-microbiota in mouse having non-alcoholic fatty liver disease. Food Sci. Biotechnol. 2019, 28, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Feng, Z.; Sang, X.; Chen, W.; Zhang, X.; Xiao, J.; Chen, Y.; Chen, Q.; Yang, M.; Su, J. Kombucha ameliorates LPS-induced sepsis in a mouse model. Food Funct. 2021, 12, 10263–10280. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.A.d.C.; Vilela, D.L.d.S.; Fraiz, G.M.; Lopes, I.L.; Coelho, A.I.M.; Castro, L.C.V.; Martin, J.G.P. Effect of kombucha intake on the gut microbiota and obesity-related comorbidities: A systematic review. Crit. Rev. Food Sci. Nutr. 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.R.; Moreira, L.d.P.D.; Costa, M.A.d.C.; Toledo, R.C.L.; Grancieri, M.; Nascimento, T.P.d.; Ferreira, M.S.L.; da Matta, S.L.P.; Eller, M.R.; Martino, H.S.D.; et al. Kombuchas from green and black teas reduce oxidative stress, liver steatosis and inflammation, and improve glucose metabolism in wistar rats fed a high-fat high-fructose diet. Food Funct. 2021, 12, 10813–10827. [Google Scholar] [CrossRef]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C. AIN-93 Purified diets for laboratory rodents: Final report of the american institute of nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef]
- Martinez, O.D.M.; Theodoro, J.M.V.; Grancieri, M.; Toledo, R.C.L.; Queiroz, V.A.V.; de Barros, F.A.R.; Martino, H.S.D. Dry heated whole sorghum flour (BRS 305) with high tannin and resistant starch improves glucose metabolism, modulates adiposity, and reduces liver steatosis and lipogenesis in wistar rats fed with a high-fat high-fructose diet. J. Cereal Sci. 2021, 99. [Google Scholar] [CrossRef]
- Zenebon, O.; Pascuet, N.S.; Tiglea, P. Métodos fíSicos-Quimicos Para Análise de Alimentos; Instituto Adolfo Lutz: São Paulo, Brazil, 2008; pp. 1–1020. [Google Scholar]
- Jayabalan, R.; Chen, P.N.; Hsieh, Y.S.; Prabhakaran, K.; Pitchai, P.; Marimuthu, S.; Thangaraj, P.; Swaminathan, K.; Yun, S.E. Effect of solvent fractions of kombucha tea on viability and invasiveness of cancer cells-characterization of Dimethyl 2-(2-Hydroxy-2-Methoxypropylidine) malonate and vitexin. Indian J. Biotechnol. 2011, 10, 75–82. [Google Scholar]
- Azevedo, L.; de Araujo Ribeiro, P.F.; de Carvalho Oliveira, J.A.; Correia, M.G.; Ramos, F.M.; de Oliveira, E.B.; Barros, F.; Stringheta, P.C. Camu-Camu (Myrciaria Dubia) from commercial cultivation has higher levels of bioactive compounds than native cultivation (amazon forest) and presents antimutagenic effects in vivo. J. Sci. Food Agric. 2019, 99, 624–631. [Google Scholar] [CrossRef]
- De Sá, L.R.V.; De Oliveira, M.A.L.; Cammarota, M.C.; Matos, A.; Ferreira-Leitão, V.S. Simultaneous analysis of carbohydrates and volatile fatty acids by hplc for monitoring fermentative biohydrogen production. Int. J. Hydrog. Energy 2011, 36, 15177–15186. [Google Scholar] [CrossRef]
- Song, P.; Zhang, R.; Wang, X.; He, P.; Tan, L.; Ma, X. Dietary grape-seed procyanidins decreased postweaning diarrhea by modulating intestinal permeability and suppressing oxidative stress in rats. J. Agric. Food Chem. 2011, 59, 6227–6232. [Google Scholar] [CrossRef] [PubMed]
- Grancieri, M.; Costa, N.M.B.; Tostes, M.d.G.V.; de Oliveira, D.S.; de Carvalho Nunes, L.; de Nadai Marcon, L.; Veridiano, T.A.; Viana, M.L. Yacon flour (smallanthus sonchifolius) attenuates intestinal morbidity in rats with colon cancer. J. Funct. Foods 2017, 37, 666–675. [Google Scholar] [CrossRef]
- Siegfried, R.; Ruckemann, H.; Stumpf, G. Method for the determination of organic acids in silage by high performance liquid chromatography. Landwirt 1984, 37, 298–304. [Google Scholar]
- Stevenson, D.M.; Weimer, P.J. Dominance of prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S RRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef]
- Campbell, L.; Yu, R.; Li, F.; Zhou, Q.; Chen, D.; Qi, C.; Yin, Y.; Sun, J. Diabetes, Metabolic syndrome and obesity: Targets and therapy dovepress modulation of fat metabolism and gut microbiota by resveratrol on high-fat diet-induced obese mice. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Ormerod, K.L.; Wood, D.L.A.; Lachner, N.; Gellatly, S.L.; Daly, J.N.; Parsons, J.D.; Dal’Molin, C.G.O.; Palfreyman, R.W.; Nielsen, L.K.; Cooper, M.A.; et al. Genomic characterization of the uncultured bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 2016, 4, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, T.A.F.; Rogero, M.M.; Hassimotto, N.M.A.; Lajolo, F.M. The two-way polyphenols-microbiota interactions and their effects on obesity and related metabolic diseases. Front. Nutr. 2019, 6, 188. [Google Scholar] [CrossRef]
- Fava, F.; Rizzetto, L.; Tuohy, K.M. Gut microbiota and health: Connecting actors across the metabolic system. Proc. Nutr. Soc. 2019, 78, 177–188. [Google Scholar] [CrossRef]
- Gowd, V.; Karim, N.; Shishir, M.R.I.; Xie, L.; Chen, W. Dietary polyphenols to combat the metabolic diseases via altering gut microbiota. Trends Food Sci. Technol. 2019, 93, 81–93. [Google Scholar] [CrossRef]
- Sheng, S.; Chen, J.; Zhang, Y.; Qin, Q.; Li, W.; Yan, S.; Wang, Y.; Li, T.; Gao, X.; Tang, L.; et al. Structural and functional alterations of gut microbiota in males with hyperuricemia and high levels of liver enzymes. Front. Med. 2021, 8, 779994. [Google Scholar] [CrossRef]
- Caponio, G.R.; Lippolis, T.; Tutino, V.; Gigante, I.; De Nunzio, V.; Milella, R.A.; Gasparro, M.; Notarnicola, M. Nutraceuticals: Focus on anti-inflammatory, anti-cancer, antioxidant properties in gastrointestinal tract. Antioxidants 2022, 11, 1274. [Google Scholar] [CrossRef]
- Hurst, H.; Harborne, J. Plant polyphenols—XVI: Identification of flavonoids by reductive cleavage. Phytochemistry 1967, 6, 1111–1118. [Google Scholar] [CrossRef]
- Rastmanesh, R. High Polyphenol, Low Probiotic diet for weight loss because of intestinal microbiota interaction. Chem. Biol. Interact. 2011, 189, 1–8. [Google Scholar] [CrossRef]
- Caponio, G.R.; Cofano, M.; Lippolis, T.; Gigante, I.; De Nunzio, V.; Difonzo, G.; Noviello, M.; Tarricone, L.; Gambacorta, G.; Giannelli, G.; et al. Anti-proliferative and pro-apoptotic effects of digested aglianico grape pomace extract in human colorectal cancer cells. Molecules 2022, 27, 6791. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Guo, L.; Zhang, Y.; Fan, S.; Gu, M.; Lu, Y.; Jiang, D.; Li, Y.; Huang, C.; Zhou, Z. Extracts of pomelo peels prevent high-fat diet-induced metabolic disorders in C57BL/6 mice through activating the PPARα and GLUT4 pathway. PLoS ONE 2013, 8, e77915. [Google Scholar] [CrossRef] [PubMed]
- Jakubczyk, K.; Kałduńska, J.; Kochman, J.; Janda, K. Chemical profile and antioxidant activity of the kombucha beverage derived from white, green, black and red tea. Antioxidants 2020, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.J.; Borges, M.d.F.; Rosa, M.d.F.; Castro-Gómez, R.J.H.; Spinosa, W.A. Acetic acid bacteria in the food industry: Systematics, characteristics and applications. Food Technol. Biotechnol. 2018, 56, 139–151. [Google Scholar] [CrossRef]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93, 1–16. [Google Scholar] [CrossRef]
- Maldonado, R.F.; Sá-Correia, I.; Valvano, M.A.; Gasmi, A.; Mujawdiya, P.K.; Pivina, L.; Doşa, A.; Semenova, Y.; Benahmed, A.G.; Bjørklund, G. Lipopolysaccharide modification in gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016, 28, 827–839. [Google Scholar] [CrossRef]
- Gasmi, A.; Mujawdiya, P.K.; Pivina, L.; Doşa, A.; Semenova, Y.; Benahmed, A.G.; Bjørklund, G. Relationship between gut microbiota, gut hyperpermeability and obesity. Curr. Med. Chem. 2020, 28, 827–839. [Google Scholar] [CrossRef]
- Kang, C.; Wang, B.; Kaliannan, K.; Wang, X.; Lang, H.; Hui, S.; Huang, L.; Zhang, Y.; Zhou, M.; Chen, M.; et al. Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. MBio 2017, 8. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.M.; Disciglio, V.; Franco, I.; Sorino, P.; Bonfiglio, C.; Bianco, A.; Campanella, A.; Lippolis, T.; Pesole, P.L.; Polignano, M.; et al. A Low glycemic index mediterranean diet combined with aerobic physical activity rearranges the gut microbiota signature in NAFLD patients. Nutrients 2022, 14, 1773. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Zheng, R.D.; Sun, X.Q.; Ding, W.J.; Wang, X.Y.; Fan, J.G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2017, 16, 375–381. [Google Scholar] [CrossRef]
- Zeng, H.; Larson, K.J.; Cheng, W.H.; Bukowski, M.R.; Safratowich, B.D.; Liu, Z.; Hakkak, R. Advanced liver steatosis accompanies an increase in hepatic inflammation, colonic, secondary bile acids and Lactobacillaceae/Lachnospiraceae bacteria in C57BL/6 mice fed a high-fat diet. J. Nutr. Biochem. 2020, 78, 108336. [Google Scholar] [CrossRef] [PubMed]
- Vojinovic, D.; Radjabzadeh, D.; Kurilshikov, A.; Amin, N.; Wijmenga, C.; Franke, L.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat. Commun. 2019, 10, 5813. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Precup, G.; Vodnar, D.C. Gut Prevotella as a possible biomarker of diet and its Eubiotic versus dysbiotic roles: A Comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef]
- Ke, S.; Fang, S.; He, M.; Huang, X.; Yang, H.; Yang, B.; Chen, C.; Huang, L. Age-Based dynamic changes of phylogenetic composition and interaction networks of health pig gut microbiome feeding in a uniformed condition. BMC Vet. Res. 2019, 15, 1–13. [Google Scholar] [CrossRef]
- Illescas, O.; Rodríguez-Sosa, M.; Gariboldi, M. Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies. Nutrients 2021, 13, 2234. [Google Scholar] [CrossRef]
- van der Hee, B.; Wells, J.M. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Dahiya, D.K.; Renuka; Puniya, M.; Shandilya, U.K.; Dhewa, T.; Kumar, N.; Kumar, S.; Puniya, A.K.; Shukla, P. Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: A review. Front. Microbiol. 2017, 8, 563. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; MacKay, C.R. Diet, gut microbiota and immune responses. Nat. Immunol. 2011, 12, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Van Hul, M.; Cani, P.D. Targeting carbohydrates and polyphenols for a healthy microbiome and healthy weight. Curr. Nutr. Rep. 2019, 8, 307–316. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Gachhui, R.; Sil, P.C. Effect of kombucha, a fermented black tea in attenuating oxidative stress mediated tissue damage in alloxan induced diabetic rats. Food Chem. Toxicol. 2013, 60, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, N.; Tan, H.Y.; Li, S.; Zhang, C.; Feng, Y. Function of Akkermansia Muciniphila in obesity: Interactions with lipid metabolism, immune response and gut systems. Front. Microbiol. 2020, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Choi, Y.; Kim, D.K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.S.; Jee, Y.K.; Gho, Y.S.; et al. Akkermansia Muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450-11. [Google Scholar] [CrossRef]
- Konikoff, T.; Gophna, U. Oscillospira: A central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef]

| Green Tea Kombucha | Black Tea Kombucha | p-Value | |
|---|---|---|---|
| Chemical composition | |||
| Sucrose (g/L) | 19.30 ± 2.73 b | 34.98 ± 1.42 a | 0.0382 |
| Glucose (g/L) | 3.19 ± 0.15 a | 2.45 ± 0.96 a | 0.4690 |
| Fructose (g/L) | 0.15 ± 0.01 a | 0.05 ± 0.02 a | 0.0583 |
| Ethanol (g/L) | 7.23 ± 0.03 a | 4.91 ± 0.35 a | 0.0653 |
| Theaflavin (g/L) | 0.28 ± 0.03 b | 1.51 ± 0.06 a | 0.0066 |
| Thearubigin (g/L) | 13.30 ± 0.67 b | 19.99 ± 0.10 a | 0.0416 |
| pH | 3.2 ± 0.1 b | 3.5 ± 0.1 a | 0.0078 |
| Total acidity (% w/v) | 0.36 ± 0.01 a | 0.32 ± 0.01 b | 0.0100 |
| Organic acids | |||
| Acetic acid (g/L) | 3.22 ± 0.39 a | 2.78 ± 0.16 a | 0.3336 |
| Glucuronic acid (g/L) | 1.17 ± 0.06 a | 0.47 ± 0.02 b | 0.0323 |
| Lactic acid (g/L) | 0.01 ± 0.00 a | 0.02 ± 0.00 a | 0.2604 |
| Microbiological characterization | |||
| Acetic bacteria (log CFU/mL) | 6.0 ± 0.30 a | 5.30 ± 0.10 a | 0.1071 |
| Lactic bacteria (log CFU/mL) | 6.50 ± 0.20 a | 5.90 ± 0.60 a | 0.3959 |
| Yeast (log CFU/mL) | 6.30 ± 0.40 a | 5.50 ± 0.10 a | 0.1690 |
| Features | AIN93-M (n = 10) | HFHF (n = 10) | GTK (n = 9) | BTK (n = 10) |
|---|---|---|---|---|
| Body composition | ||||
| Initial weight (g) | 349.90 ± 30.71 a | 366.90 ± 36.90 a | 370.40 ± 36.20 a | 364.60 ± 36.05 a |
| Final weight (g) | 415.00 ± 34.50 a | 438.10 ± 66.65 a | 415.30 ± 37.07 a | 409.60 ± 50.08 a |
| Weight gain (g) | 65.00 ± 22.70 a | 71.25 ± 38.10 a | 44.90 ± 30.69 a | 44.90 ± 23.14 a |
| BMI (g/cm2) | 0.68 ± 0.08 a | 0.61 ± 0.04 a | 0.61 ± 0.08 a | 0.61 ±0.08 a |
| Cecum weight (empty) (g) | 1.01 ± 0.23 a | 0.97 ± 0.13 a | 0.96 ± 0.18 a | 0.96 ± 0.04 a |
| Cecum weight (full) (g) | 5.09 ± 1.15 a | 3.92 ± 0.89 b | 3.62 ± 0.75 b | 4.03 ± 0.80 ab |
| Cecum weight:body weight ratio | 1.23 ± 0.28 a | 0.91 ± 0.24 ab | 0.87 ± 0.17 b | 0.98 ± 0.21 ab |
| Intestinal Permeability | ||||
| Lactulose:mannitol ratio | 1.51 ± 0.57 a | 1.56 ± 0.78 a | 1.62 ± 0.94 a | 2.17 ± 1.08 a |
| Histological Features | ||||
| Crypt depth (µM) | 179.10 ± 43.20 a | 223.10 ± 40.69 a | 221.20 ± 24.92 a | 209.30 ± 40.83 a |
| Crypt width (µM) | 19.51 ± 2.66 a | 18.85 ± 4.16 a | 21.03 ± 1.46 a | 19.46 ± 2.68 a |
| Number of goblet cells (units) | 18.60 ± 3.27 a | 16.59 ± 4.71 a | 17.94 ± 2.93 a | 22.38 ± 5.51 a |
| Fecal pH | 9.01 ± 0.40 a | 9.17 ± 0.25 a | 9.27 ± 0.07 a | 9.13 ± 0.13 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, M.A.d.C.; Dias Moreira, L.d.P.; Duarte, V.d.S.; Cardoso, R.R.; São José, V.P.B.d.; Silva, B.P.d.; Grancieri, M.; Corich, V.; Giacomini, A.; Bressan, J.; et al. Kombuchas from Green and Black Tea Modulate the Gut Microbiota and Improve the Intestinal Health of Wistar Rats Fed a High-Fat High-Fructose Diet. Nutrients 2022, 14, 5234. https://doi.org/10.3390/nu14245234
Costa MAdC, Dias Moreira LdP, Duarte VdS, Cardoso RR, São José VPBd, Silva BPd, Grancieri M, Corich V, Giacomini A, Bressan J, et al. Kombuchas from Green and Black Tea Modulate the Gut Microbiota and Improve the Intestinal Health of Wistar Rats Fed a High-Fat High-Fructose Diet. Nutrients. 2022; 14(24):5234. https://doi.org/10.3390/nu14245234
Chicago/Turabian StyleCosta, Mirian Aparecida de Campos, Luiza de Paula Dias Moreira, Vinícius da Silva Duarte, Rodrigo Rezende Cardoso, Vinícius Parzanini Brilhante de São José, Bárbara Pereira da Silva, Mariana Grancieri, Viviana Corich, Alessio Giacomini, Josefina Bressan, and et al. 2022. "Kombuchas from Green and Black Tea Modulate the Gut Microbiota and Improve the Intestinal Health of Wistar Rats Fed a High-Fat High-Fructose Diet" Nutrients 14, no. 24: 5234. https://doi.org/10.3390/nu14245234
APA StyleCosta, M. A. d. C., Dias Moreira, L. d. P., Duarte, V. d. S., Cardoso, R. R., São José, V. P. B. d., Silva, B. P. d., Grancieri, M., Corich, V., Giacomini, A., Bressan, J., Martino, H. S. D., & Barros, F. A. R. d. (2022). Kombuchas from Green and Black Tea Modulate the Gut Microbiota and Improve the Intestinal Health of Wistar Rats Fed a High-Fat High-Fructose Diet. Nutrients, 14(24), 5234. https://doi.org/10.3390/nu14245234










