Microbiome Therapeutics for Food Allergy
Abstract
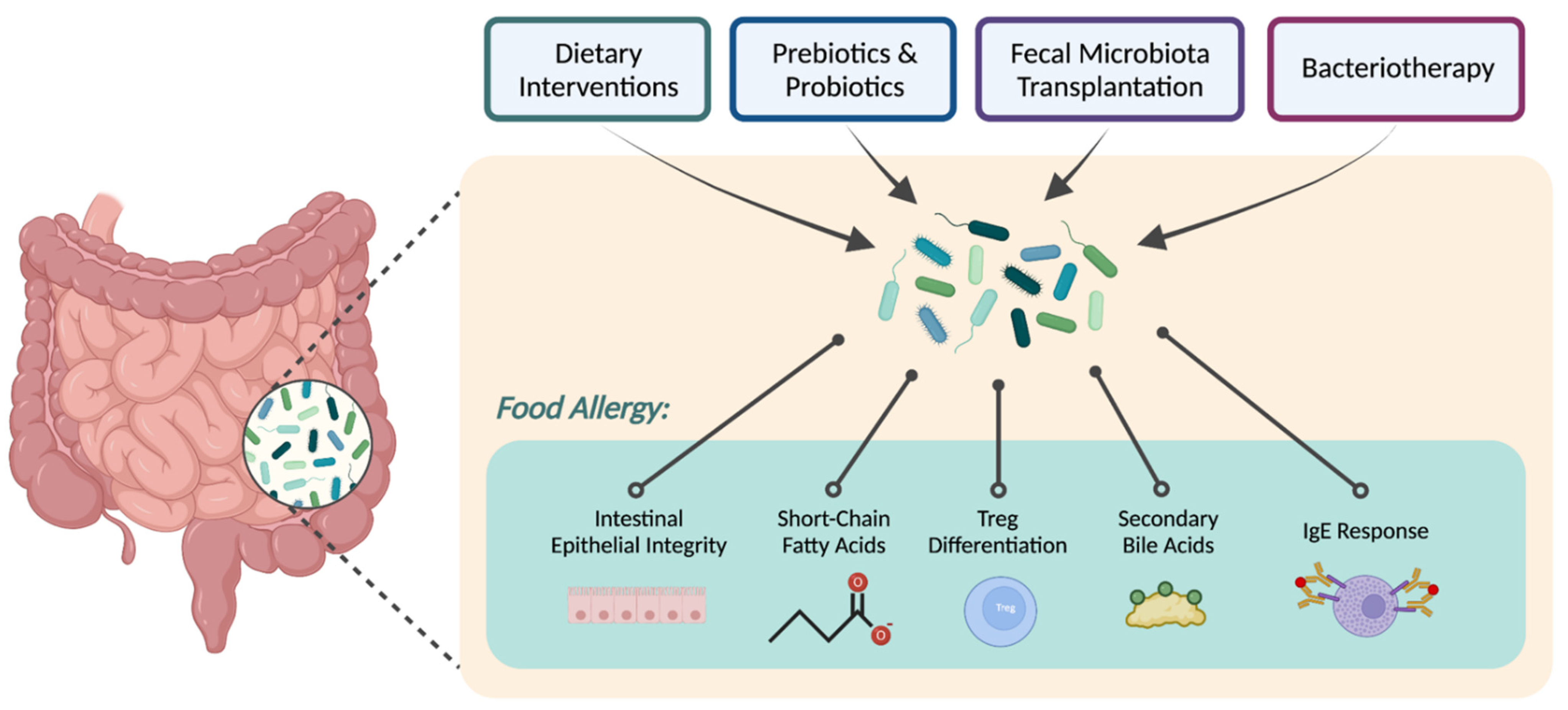
:1. Introduction
1.1. Food Allergy
1.2. Gut Microbiota and Food Allergy
1.3. Early Life Influences on Gut Microbiota and Immune Development
2. Dietary Approaches
3. Metabolites
3.1. Short Chain Fatty Acids
3.2. Secondary Bile Acids
3.3. Sphingolipids
3.4. Amino Acids
4. Targeted Microbial Therapies
4.1. Fecal Microbiota Transplantation
4.2. Bacteriotherapy
4.3. Industry Developments and Ongoing Clinical Trials
| Therapeutic Strategy | Company or Organization | Clinical Trial Phase | Intervention Name | Intervention Description | Study Subjects | Primary Study Outcome | Findings | Trial Identifier | Trial Name |
|---|---|---|---|---|---|---|---|---|---|
| Biotherapeutics | Boston Children’s Hospital | Phase 1 | N/A | Unspecified oral encapsulated microbiota transplantation | Adults 18–40 years with peanut allergy | Presence of fecal microbiota transplantation-related adverse events grade 2 or above | Not yet published | NCT02960074 | Evaluating the Safety and Efficacy of Oral Encapsulated Fecal Microbiota Transplant in Peanut Allergic Patients |
| Pre/Pro/Synbiotic | Royal Children’s Hospital; Murdoch Children’s Research Institute; Prota Therapeutics | Phase 2b/3 | PRT100 | Probiotic Lactobacillus rhamnosus ATCC 53103; peanut oral immunotherapy | Children 1–10 years with peanut allergy | Sustained unresponsiveness to peanut protein by double-blind placebo-controlled food challenge | Sustained unresponsiveness at 12 months in 36/79 (46%) in the probiotic and peanut oral immunotherapy group vs. 42/85 (51%) in the peanut oral immunotherapy group vs. 2/39 (5%) in placebo group [138]. | ACTRN12616000322437 | A multicentre, randomised, controlled trial evaluating the effectiveness of probiotic and peanut oral immunotherapy (PPOIT) in inducing desensitisation or tolerance in children with peanut allergy compared with oral immunotherapy (OIT) alone and with placebo |
| Royal Children’s Hospital | Phase 2b | NCC4007 | Probiotic Lactobacillus rhamnosus CGMCC 1.3724; peanut oral immunotherapy | Children 1–10 years with peanut allergy | Sustained unresponsiveness to peanut protein by double-blind placebo-controlled food challenge | Sustained unresponsiveness after 2 to 5 weeks in 23/28 (82%) in the probiotic and peanut oral immunotherapy group vs. 1/28 (4%) in placebo [139]. Quality-of-life scores increased in the probiotic and peanut oral immunotherapy group (n = 19) but not the placebo group (n = 19), with a strong correlation between quality-of-life scores and frequency/amount of peanuts eaten up to the final endpoint at four years [140]. | ACTRN12608000594325 | Study of effectiveness of probiotics and peanut oral immunotherapy (OIT) in inducing desensitisation or tolerance in children with peanut allergy | |
| National Health and Medical Research Council; Sydney Children’s Hospital Network | N/A | Butyrylated high-amylase maize starch (HAMSB) | Prebiotic dietary fiber; peanut immunotherapy | Children 10–16 years with peanut allergy | Sustained unresponsiveness to peanut protein by double-blind placebo-controlled food challenge | Not yet published | ACTRN12617000914369 | Oral peanut immunotherapy with a modified dietary starch adjuvant for treatment of peanut allergy in children aged 10–16 years | |
| Danone Nutricia Research | N/A | N/A | Amino acid formula with prebiotic oligofructose, prebiotic inulin, and probiotic Bifidobacterium breve M-16V | Infants <13 months with cow’s milk allergy | Cow’s milk tolerance by double-blind placebo-controlled food challenge | No significant difference in cow’s milk tolerance at 12 and 24 months [146]. | NTR3725 | A prospective double blind randomised controlled study to evaluate the immunological benefits and clinical effects of an elimination diet using an amino acid formula (AAF) with an added pre-probiotic blend in infants with Cow’s Milk Allergy (CMA) | |
| Danone Nutricia Research | N/A | N/A | Amino acid formula with prebiotic oligofructose, prebiotic inulin, and probiotic Bifidobacterium breve M-16V | Infants <13 months with suspected cow’s milk allergy | Fecal percentages of bifidobacteria and Eubacterium rectale/Clostridium coccoides | Experimental vs. placebo group had higher median percentage bifidobacteria and lower Eubacterium rectale/Clostridium coccoides at 8 weeks: (35.4% vs. 9.7%; p < 0.001), (9.5% vs. 24.2%; p < 0.001) respectively [144] and at 26 weeks: (47.0% vs. 11.8%; p < 0.001), (13.7% vs. 23.6%; p = 0.003) respectively [145]. | NTR3979 | An Amino Acid based Formula with synbiotics: Effects on gut microbiota diversity and clinical effectiveness in suspected gastrointestinal non-IgE mediated Cow’s Milk Allergy (ASSIGN I) | |
| University of Naples Federico II | N/A | Nutramigen LGG | Lactobacillus GG in extensively hydrolyzed casein formula | Children 1–24 months with cow’s milk allergy | Tolerance to oral food challenge | Group receiving Lactobacillus GG vs. control had more patients achieving tolerance to non-IgE mediated cow’s milk allergy at 6 months (16 vs. 6; p = 0.017), IgE mediated cow’s allergy at 12 months (5 vs. 1; p = 0.046), and non-IgE mediated cow’s milk allergy at 12 months (17 vs. 8; p = 0.006) [68]. | ACTRN12610000566033 | A randomised controlled trial on the effect of extensively hydrolyzed casein formula containing Lactobacillus GG (LGG) vs. extensively hydrolyzed casein formula on time of tolerance acquisition in children with cow’s milk allergy | |
| University of Naples Federico II | N/A | Nutramigen LGG | Lactobacillus GG in extensively hydrolyzed casein formula | Infants 1–12 months with cow’s milk allergy | Time to tolerance acquisition | Compared to groups receiving other formula types, there more patients who achieved tolerance to cow’s milk allergy in the groups receiving Lactobacillus GG with extensively hydrolyzed casein formula (78.9%; p < 0.05) and extensively hydrolyzed casein formula alone (43.6%; p < 0.05) [142]. | NCT01634490 | Effects of Different Dietary Regimens on Tolerance Acquisition in Children With Cow’s Milk Allergy |
| Therapeutic Strategy | Company or Organization | Clinical Trial Phase | Intervention Name | Intervention Description | Study Subjects | Primary Study Outcome | Trial Identifier | Trial Name |
|---|---|---|---|---|---|---|---|---|
| Biotherapeutics | Massachusetts General Hospital; Vedanta Biosciences | Phase 1/2 | VE416 | Consortium of inactive commensals | Patients 12–55 years with peanut allergy | Number of patients with treatment related adverse events; peanut protein tolerance by double-blind placebo-controlled food challenge | NCT03936998 | VE416 for Treatment of Food Allergy |
| Siolta Therapeutics | Phase 1b/2 | STMC-103H | Live biotherapeutic of unspecified intestinal bacteria | Children 1–6 years; 1–12 months; 0–7 days with immediate family history of allergic disorder | Frequency, type, and severity of adverse events; incidence of atopic dermatitis Secondary outcome: incidence of sensitization to food allergen; incidence of food allergies | NCT05003804 | Allergic Disease Onset Prevention Study (adored) | |
| Evelo Biosciences | Phase 2 | EDP1815 | Prevotella histicola | Adults 18–75 years with atopic dermatitis | EDP1815 efficacy defined as >50% decrease in Eczema Area Severity Index | NCT05121480 | A Study Investigating the Effect of EDP1815 in the Treatment of Mild, Moderate and Severe Atopic Dermatitis | |
| Pre/Pro/Synbiotic | University of Chicago | Phase 1/2 | N/A | Unspecified prebiotic | Children 4–7 years with peanut allergy | Peanut protein tolerance by double-blind placebo-controlled food challenge | NCT05138757 | Pinpoint Trial: Prebiotics IN Peanut Oral ImmunoTherapy |
| Chinese University of Hong Kong | N/A | N/A | Unspecified probiotic; peanut oral immunotherapy | Children 1–17 years with peanut allergy | Sustained unresponsiveness to peanut protein by double-blind placebo-controlled food challenge | NCT05165329 | A Randomised, Controlled Trial Evaluating the Effectiveness of Probiotic and Peanut Oral Immunotherapy (PPOIT) in Inducing Desensitisation or Remission in Chinese Children With Peanut Allergy Compared With Oral Immunotherapy (OIT) Alone and With Placebo | |
| Massachusetts General Hospital; Mead Johnson Nutrition | Phase 2 (Terminated) | N/A | Extensively hydrolyzed casein formula; Lactobacillus GG; Amino acid formula | Infants up to 120 days with suspected cow’s milk allergy | Tolerance to cow’s milk protein | NCT02719405 | Impact of Infant Formula on Resolution of Cow’s Milk Allergy | |
| Johnson & Johnson; Evolve BioSystems | N/A | Evivo | Bifidobacterium longum subspecies infantis strain EVC001 | Healthy infants up to 14 days | Number of subjects with atopic dermatitis Secondary outcome: Percentage of subjects with Bifidobacterium Infantis gut colonization | NCT04662619 | A Study of a Probiotic Food Supplement Containing B. Infantis (EVC001) in Healthy Breastfed Infants at Risk of Developing Atopic Dermatitis | |
| NovoNatum | N/A | BioAmicus Lactobacillus drops | Lactobacillus Reuteri NCIMB 30351 | Children 1–5 months with colic, constipation, diarrhea, or regurgitation | Change in number with infantile colic Secondary outcome: presence of skin or food allergies; stool 16 s RNA sequencing | NCT04262648 | Randomized Placebo-controlled Study of L. Reuteri NCIMB 30351 in GI Functional Disorders and Food Allergy in Newborns | |
| Nutricia UK | N/A | N/A | Hypoallergenic formula containing unspecified synbiotics | Infants <13 months with confirmed or suspected cow’s milk allergy | Healthcare utilization by electronic health records Secondary outcome: Clinical outcomes related to cow’s milk allergy | NCT05046418 | Synbiotics Cohort Study | |
| Vaginal Seeding | National Institute of Allergy and Infectious Diseases; Immune Tolerance Network; Pharmaceutical Product Development; Rho Federal Systems Division | Phase 1 | N/A | Maternal vaginal microbiota | Neonate of adult female 18–45 with first degree relative with atopic disease or food allergies | Presence of sensitization to at least one food allergen | NCT03567707 | Vaginal Microbiome Exposure and Immune Responses in C-section Infants |
| Karolinska Institutet; Uppsala University Linkoeping University Umeå University Örebro University | N/A | N/A | Maternal vaginal and fecal microbiota | Neonate of healthy adult mother | Incidence of immunoglobulin E-associated allergic disease | NCT03928431 | Restoration of Microbiota in Neonates (RoMaNs) |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Platts-Mills, T.A. The allergy epidemics: 1870–2010. J. Allergy Clin. Immunol. 2015, 136, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinakar, C. Anaphylaxis in Children: Current Understanding and Key Issues in Diagnosis and Treatment. Curr. Allergy Asthma Rep. 2012, 12, 641–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaker, M.S.; Wallace, D.V.; Golden, D.B.K.; Oppenheimer, J.; Bernstein, J.A.; Campbell, R.L.; Dinakar, C.; Ellis, A.; Greenhawt, M.; Khan, D.A.; et al. Anaphylaxis-a 2020 practice parameter update, systematic review, and Grading of Recommendations, Assessment, Development and Evaluation (GRADE) analysis. J. Allergy Clin. Immunol. 2020, 145, 1082–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaker, M.; Greenhawt, M. Peanut allergy: Burden of illness. Allergy Asthma Proc. 2019, 40, 290–294. [Google Scholar] [CrossRef]
- Golding, M.A.; Gunnarsson, N.V.; Middelveld, R.; Ahlstedt, S.; Protudjer, J.L. A scoping review of the caregiver burden of pediatric food allergy. Ann. Allergy Asthma Immunol. 2021, 127, 536–547.e3. [Google Scholar] [CrossRef]
- Patel, N.; Herbert, L.; Green, T.D. The emotional, social, and financial burden of food allergies on children and their families. Allergy Asthma Proc. 2017, 38, 88–91. [Google Scholar] [CrossRef]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.M.; Burks, A.W. Food Allergy. N. Engl. J. Med. 2017, 377, 1168–1176. [Google Scholar] [CrossRef]
- Macdougall, J.D.; Burks, A.W.; Kim, E.H. Current Insights into Immunotherapy Approaches for Food Allergy. Immuno. Targets Ther. 2021, 10, 1–8. [Google Scholar] [CrossRef]
- PALISADE Group of Clinical Investigators; Vickery, B.P.; Vereda, A.; Casale, T.B.; Beyer, K.; du Toit, G.; Hourihane, J.O.; Jones, S.M.; Shreffler, W.G.; Marcantonio, A.; et al. AR101 Oral Immunotherapy for Peanut Allergy. N. Engl. J. Med. 2018, 379, 1991–2001. [Google Scholar]
- Badina, L.; Belluzzi, B.; Contorno, S.; Bossini, B.; Benelli, E.; Barbi, E.; Berti, I. Omalizumab effectiveness in patients with a previously failed oral immunotherapy for severe milk allergy. Immun. Inflamm. Dis. 2022, 10, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Albuhairi, S.; Rachid, R. The use of adjunctive therapies during oral immunotherapy: A focus on biologics. J. Food Allergy 2022, 4, 65–70. [Google Scholar] [CrossRef]
- Fiocchi, A.; Vickery, B.P.; Wood, R.A. The use of biologics in food allergy. Clin. Exp. Allergy 2021, 51, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.C.; Jackson, C.M.; Ting, T.; Harizaj, A.; Järvinen, K.M. Predictors and biomarkers of food allergy and sensitization in early childhood. Ann. Allergy Asthma Immunol. 2022, 129, 292–300. [Google Scholar] [CrossRef]
- Brough, H.A.; Lanser, B.J.; Sindher, S.B.; Teng, J.M.C.; Leung, D.Y.M.; Venter, C.; Chan, S.M.; Santos, A.F.; Bahnson, H.T.; Guttman-Yassky, E.; et al. Early intervention and prevention of allergic diseases. Allergy 2022, 77, 416–441. [Google Scholar] [CrossRef]
- Kanchan, K.; Clay, S.; Irizar, H.; Bunyavanich, S.; Mathias, R.A. Current insights into the genetics of food allergy. J. Allergy Clin. Immunol. 2021, 147, 15–28. [Google Scholar] [CrossRef]
- Iweala, O.I.; Nagler, C.R. The Microbiome and Food Allergy. Annu. Rev. Immunol. 2019, 37, 377–403. [Google Scholar] [CrossRef]
- Ober, C.; Sperling, A.I.; von Mutius, E.; Vercelli, D. Immune development and environment: Lessons from Amish and Hutterite children. Curr. Opin. Immunol. 2017, 48, 51–60. [Google Scholar] [CrossRef]
- Lambrecht, B.N.; Hammad, H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol. 2017, 18, 1076–1083. [Google Scholar] [CrossRef]
- Yu, J.E.; Mallapaty, A.; Miller, R.L. It’s not just the food you eat: Environmental factors in the development of food allergies. Environ. Res. 2018, 165, 118–124. [Google Scholar] [CrossRef]
- Du Toit, G.; Katz, Y.; Sasieni, P.; Mesher, D.; Maleki, S.J.; Fisher, H.R.; Fox, A.T.; Turcanu, V.; Amir, T.; Zadik-Mnuhin, G.; et al. Early consumption of peanuts in infancy is associated with a low prevalence of peanut allergy. J. Allergy Clin. Immunol. 2008, 122, 984–991. [Google Scholar] [CrossRef]
- Maslin, K.; Pickett, K.; Ngo, S.; Anderson, W.; Dean, T.; Venter, C. Dietary diversity during infancy and the association with childhood food allergen sensitization. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2022, 33, e13650. [Google Scholar] [CrossRef]
- Hesselmar, B.; Hicke-Roberts, A.; Lundell, A.-C.; Adlerberth, I.; Rudin, A.; Saalman, R.; Wennergren, G.; Wold, A.E. Pet-keeping in early life reduces the risk of allergy in a dose-dependent fashion. PLoS ONE 2018, 13, e0208472. [Google Scholar] [CrossRef] [Green Version]
- Tun, H.M.; Konya, T.; Takaro, T.K.; Brook, J.R.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; et al. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 2017, 5, 40. [Google Scholar] [CrossRef] [Green Version]
- Ahmadizar, F.; Vijverberg, S.J.H.; Arets, H.G.M.; de Boer, A.; Lang, J.E.; Garssen, J.; Kraneveld, A.; Maitland-van Der Zee, A.H. Early-life antibiotic exposure increases the risk of developing allergic symptoms later in life: A meta-analysis. Allergy 2018, 73, 971–986. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.K.; Masilamani, M.; Li, X.-M.; Sampson, H.A. The false alarm hypothesis: Food allergy is associated with high dietary advanced glycation end-products and proglycating dietary sugars that mimic alarmins. J. Allergy Clin. Immunol. 2017, 139, 429–437. [Google Scholar] [CrossRef] [Green Version]
- Chernikova, D.; Yuan, I.; Shaker, M. Prevention of allergy with diverse and healthy microbiota: An update. Curr. Opin. Pediatr. 2019, 31, 418–425. [Google Scholar] [CrossRef]
- Stiemsma, L.T.; Michels, K.B. The Role of the Microbiome in the Developmental Origins of Health and Disease. Pediatrics 2018, 141, e20172437. [Google Scholar] [CrossRef] [Green Version]
- Sudo, N.; Sawamura, S.; Tanaka, K.; Aiba, Y.; Kubo, C.; Koga, Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J. Immunol. 1997, 159, 1739–1745. [Google Scholar]
- Abdel-Gadir, A.; Stephen-Victor, E.; Gerber, G.K.; Rivas, M.N.; Wang, S.; Harb, H.; Wang, L.; Li, N.; Crestani, E.; Spielman, S.; et al. Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat. Med. 2019, 25, 1164–1174. [Google Scholar] [CrossRef]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Hong, S.M.C.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Investig. 2021, 131, e141935. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; LeVan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, C.L.; Sitarik, A.R.; Kim, H.; Huffnagle, G.; Fujimura, K.; Yong, G.J.M.; Levin, A.M.; Zoratti, E.; Lynch, S.; Ownby, D.R.; et al. Infant gut bacterial community composition and food-related manifestation of atopy in early childhood. Pediatr. Allergy Immunol. 2022, 33, e13704. [Google Scholar] [CrossRef] [PubMed]
- Rachid, R.; Stephen-Victor, E.; Chatila, T.A. The microbial origins of food allergy. J. Allergy Clin. Immunol. 2021, 147, 808–813. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.-Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef] [Green Version]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; Lieber, A.D.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef] [Green Version]
- Madan, J.C.; Hoen, A.G.; Lundgren, S.N.; Farzan, S.F.; Cottingham, K.; Morrison, H.G.; Sogin, M.L.; Li, H.; Moore, J.; Karagas, M.R. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA Pediatr. 2016, 170, 212–219. [Google Scholar] [CrossRef]
- Coker, M.O.; Hoen, A.G.; Dade, E.; Lundgren, S.; Li, Z.; Wong, A.D.; Zens, M.S.; Palys, T.J.; Morrison, H.G.; Sogin, M.L.; et al. Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: A prospective cohort study. BJOG Int. J. Obstet. Gynaecol. 2019, 127, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Stearns, J.C.; Simioni, J.; Gunn, E.; McDonald, H.; Holloway, A.C.; Thabane, L.; Mousseau, A.; Schertzer, J.D.; Ratcliffe, E.M.; Rossi, L.; et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep. 2017, 7, 16527. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Faden, H.S.; Zhu, L. The Response of the Gut Microbiota to Dietary Changes in the First Two Years of Life. Front. Pharmacol. 2020, 11, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernikova, D.A.; Madan, J.C.; Housman, M.L.; Zain-Ul-Abideen, M.; Lundgren, S.N.; Morrison, H.G.; Sogin, M.L.; Williams, S.; Moore, J.H.; Karagas, M.R.; et al. The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatr. Res. 2018, 84, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Naimi, S.; Viennois, E.; Gewirtz, A.T.; Chassaing, B. Direct impact of commonly used dietary emulsifiers on human gut microbiota. Microbiome 2021, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Muinck, E.J.; Trosvik, P. Individuality and convergence of the infant gut microbiota during the first year of life. Nat. Commun. 2018, 9, 2233. [Google Scholar] [CrossRef] [Green Version]
- Al Nabhani, Z.; Dulauroy, S.; Marques, R.; Cousu, C.; Al Bounny, S.; Déjardin, F.; Sparwasser, T.; Bérard, M.; Cerf-Bensussan, N.; Eberl, G. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity 2019, 50, 1276–1288.e5. [Google Scholar] [CrossRef]
- Du Toit, G.; Roberts, G.; Sayre, P.H.; Bahnson, H.T.; Radulovic, S.; Santos, A.F.; Brough, H.A.; Phippard, D.; Basting, M.; Feeney, M.; et al. Randomized Trial of Peanut Consumption in Infants at Risk for Peanut Allergy. N. Engl. J. Med. 2015, 372, 803–813. [Google Scholar] [CrossRef] [Green Version]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef] [Green Version]
- Bunyavanich, S.; Berin, M.C. Food allergy and the microbiome: Current understandings and future directions. J. Allergy Clin. Immunol. 2019, 144, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898.e11. [Google Scholar] [CrossRef] [PubMed]
- Méndez, C.S.; Bueno, S.M.; Kalergis, A.M. Contribution of Gut Microbiota to Immune Tolerance in Infants. J. Immunol. Res. 2021, 2021, 7823316. [Google Scholar] [CrossRef] [PubMed]
- Cabana, M.D. The Role of Hydrolyzed Formula in Allergy Prevention. Ann. Nutr. Metab. 2017, 70, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Venter, C.; Agostoni, C.; Arshad, S.H.; Ben-Abdallah, M.; Du Toit, G.; Fleischer, D.M.; Greenhawt, M.; Glueck, D.H.; Groetch, M.; Lunjani, N.; et al. Dietary factors during pregnancy and atopic outcomes in childhood: A systematic review from the European Academy of Allergy and Clinical Immunology. Pediatr. Allergy Immunol. 2020, 31, 889–912. [Google Scholar] [CrossRef] [PubMed]
- Venter, C.; Meyer, R.W.; Greenhawt, M.; Pali-Schöll, I.; Nwaru, B.; Roduit, C.; Untersmayr, E.; Adel-Patient, K.; Agache, I.; Agostoni, C.; et al. Role of dietary fiber in promoting immune health—An EAACI position paper. Allergy 2022, 77, 3185–3198. [Google Scholar] [CrossRef]
- de Silva, D.; Halken, S.; Singh, C.; Muraro, A.; Angier, E.; Arasi, S.; Arshad, H.; Beyer, K.; Boyle, R.; du Toit, G.; et al. Preventing food allergy in infancy and childhood: Systematic review of randomised controlled trials. Pediatr. Allergy Immunol. 2020, 31, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Halken, S.; Muraro, A.; de Silva, D.; Khaleva, E.; Angier, E.; Arasi, S.; Arshad, H.; Bahnson, H.T.; Beyer, K.; Boyle, R.; et al. EAACI guideline: Preventing the development of food allergy in infants and young children (2020 update). Pediatr. Allergy Immunol. 2021. [Google Scholar] [CrossRef]
- Urashima, M.; Mezawa, H.; Okuyama, M.; Urashima, T.; Hirano, D.; Gocho, N.; Tachimoto, H. Primary Prevention of Cow’s Milk Sensitization and Food Allergy by Avoiding Supplementation With Cow’s Milk Formula at Birth: A Randomized Clinical Trial. JAMA Pediatr. 2019, 173, 1137–1145. [Google Scholar] [CrossRef]
- Perkin, M.R.; Logan, K.; Tseng, A.; Raji, B.; Ayis, S.; Peacock, J.; Brough, H.; Marrs, T.; Radulovic, S.; Craven, J.; et al. Randomized Trial of Introduction of Allergenic Foods in Breast-Fed Infants. N. Engl. J. Med. 2016, 374, 1733–1743. [Google Scholar] [CrossRef] [Green Version]
- Heine, R.G. Food Allergy Prevention and Treatment by Targeted Nutrition. Ann. Nutr. Metab. 2018, 72, 33–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, C.; Palumbo, M.P.; Glueck, D.H.; Sauder, K.A.; O’Mahony, L.; Fleischer, D.M.; Ben-Abdallah, M.; Ringham, B.M.; Dabelea, D. The maternal diet index in pregnancy is associated with offspring allergic diseases: The Healthy Start study. Allergy 2021, 77, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef]
- Martin, V.J.; Shreffler, W.G.; Yuan, Q. Presumed Allergic Proctocolitis Resolves with Probiotic Monotherapy: A Report of 4 Cases. Am. J. Case Rep. 2016, 17, 621–624. [Google Scholar] [CrossRef] [Green Version]
- Tan-Lim, C.S.C.; Esteban-Ipac, N.A.R. Probiotics as treatment for food allergies among pediatric patients: A meta-analysis. World Allergy Organ J. 2018, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Sestito, S.; D’Auria, E.; Baldassarre, M.E.; Salvatore, S.; Tallarico, V.; Stefanelli, E.; Tarsitano, F.; Concolino, D.; Pensabene, L. The Role of Prebiotics and Probiotics in Prevention of Allergic Diseases in Infants. Front. Pediatr. 2020, 8, 583946. [Google Scholar] [CrossRef]
- Cukrowska, B.; Ceregra, A.; Maciorkowska, E.; Surowska, B.; Zegadło-Mylik, M.A.; Konopka, E.; Trojanowska, I.; Zakrzewska, M.; Bierła, J.B.; Zakrzewski, M.; et al. The Effectiveness of Probiotic Lactobacillus rhamnosus and Lactobacillus casei Strains in Children with Atopic Dermatitis and Cow’s Milk Protein Allergy: A Multicenter, Randomized, Double Blind, Placebo Controlled Study. Nutrients 2021, 13, 1169. [Google Scholar] [CrossRef]
- Berni Canani, R.; Nocerino, R.; Terrin, G.; Coruzzo, A.; Cosenza, L.; Leone, L.; Troncone, R. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: A randomized trial. J. Allergy Clin. Immunol. 2012, 129, 580–582.e1–e5. [Google Scholar] [CrossRef]
- Roggero, P.; Liotto, N.; Pozzi, C.; Braga, D.; Troisi, J.; Menis, C.; Giannì, M.L.; Canani, R.B.; Paparo, L.; Nocerino, R.; et al. Analysis of immune, microbiota and metabolome maturation in infants in a clinical trial of Lactobacillus paracasei CBA L74-fermented formula. Nat. Commun. 2020, 11, 2703. [Google Scholar] [CrossRef]
- McKenzie, C.; Tan, J.; Macia, L.; Mackay, C.R. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev. 2017, 278, 277–295. [Google Scholar] [CrossRef]
- Di Costanzo, M.; De Paulis, N.; Biasucci, G. Butyrate: A Link between Early Life Nutrition and Gut Microbiome in the Development of Food Allergy. Life 2021, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Macfarlane, G. Collaborative JPEN-Clinical Nutrition Scientific Publications Role of intestinal bacteria in nutrient metabolism. J. Parenter. Enter. Nutr. 1997, 21, 357–365. [Google Scholar] [CrossRef]
- Topping, D.L.; Clifton, P.M. Short-Chain Fatty Acids and Human Colonic Function: Roles of Resistant Starch and Nonstarch Polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Lee, S.W.; Hong, S. Regulation of Allergic Immune Responses by Microbial Metabolites. Immune Netw. 2018, 18, e15. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G Protein-coupled Receptors GPR41 and GPR43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [Green Version]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Karaki, S.-I.; Mitsui, R.; Hayashi, H.; Kato, I.; Sugiya, H.; Iwanaga, T.; Furness, J.B.; Kuwahara, A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006, 324, 353–360. [Google Scholar] [CrossRef]
- Karaki, S.-I.; Tazoe, H.; Hayashi, H.; Kashiwabara, H.; Tooyama, K.; Suzuki, Y.; Kuwahara, A. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. Histochem. J. 2008, 39, 135–142. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016, 15, 2809–2824. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef] [Green Version]
- Waldecker, M.; Kautenburger, T.; Daumann, H.; Busch, C.; Schrenk, D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 2008, 19, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Burger-van Paassen, N.; Vincent, A.; Puiman, P.J.; Van Der Sluis, M.; Bouma, J.; Boehm, G.; van Goudoever, J.B.; Van Seuningen, I.; Renes, I.B. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: Implications for epithelial protection. Biochem. J. 2009, 420, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Goverse, G.; Molenaar, R.; Macia, L.; Tan, J.; Erkelens, M.N.; Konijn, T.; Knippenberg, M.; Cook, E.C.L.; Hanekamp, D.; Veldhoen, M.; et al. Diet-Derived Short Chain Fatty Acids Stimulate Intestinal Epithelial Cells To Induce Mucosal Tolerogenic Dendritic Cells. J. Immunol. 2017, 198, 2172–2181. [Google Scholar] [CrossRef] [Green Version]
- Paparo, L.; Nocerino, R.; Ciaglia, E.; Di Scala, C.; De Caro, C.; Russo, R.; Trinchese, G.; Aitoro, R.; Amoroso, A.; Bruno, C.; et al. Butyrate as a bioactive human milk protective component against food allergy. Allergy 2021, 76, 1398–1415. [Google Scholar] [CrossRef]
- Thio, C.L.-P.; Chi, P.-Y.; Lai, A.C.-Y.; Chang, Y.-J. Regulation of type 2 innate lymphoid cell–dependent airway hyperreactivity by butyrate. J. Allergy Clin. Immunol. 2018, 142, 1867–1883.e12. [Google Scholar] [CrossRef] [Green Version]
- Sandin, A.; Bråbäck, L.; Norin, E.; Björkstén, B. Faecal short chain fatty acid pattern and allergy in early childhood. Acta Paediatr. 2009, 98, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Cait, A.; Cardenas, E.; Dimitriu, P.A.; Amenyogbe, N.; Dai, D.; Cait, J.; Sbihi, H.; Stiemsma, L.; Subbarao, P.; Mandhane, P.J.; et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J. Allergy Clin. Immunol. 2019, 144, 1638–1647.e3. [Google Scholar] [CrossRef] [Green Version]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Chan, J.C.Y.; Yap, G.C.; Huang, C.-H.; Kioh, D.Y.Q.; Tham, E.H.; Loo, E.X.L.; Shek, L.P.C.; Karnani, N.; Goh, A.; et al. Evaluation of Stool Short Chain Fatty Acids Profiles in the First Year of Life With Childhood Atopy-Related Outcomes. Front. Allergy 2022, 3, 873168. [Google Scholar] [CrossRef]
- Danielewicz, H. Breastfeeding and Allergy Effect Modified by Genetic, Environmental, Dietary, and Immunological Factors. Nutrients 2022, 14, 3011. [Google Scholar] [CrossRef]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Paparo, L.; Di Scala, C.; Cosenza, L.; Della Gatta, G.; Calignano, A.; De Caro, C.; Laiola, M.; et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci. Rep. 2018, 8, 12500. [Google Scholar] [CrossRef] [Green Version]
- Berni Canani, R.; Sangwan, N.; Stefka, A.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous Bile Acids Are Ligands for the Nuclear Receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Van Best, N.; Rolle-Kampczyk, U.; Schaap, F.G.; Basic, M.; Damink, S.W.M.O.; Bleich, A.; Savelkoul, P.H.M.; Von Bergen, M.; Penders, J.; Hornef, M.W. Bile acids drive the newborn’s gut microbiota maturation. Nat. Commun. 2020, 11, 3692. [Google Scholar] [CrossRef] [PubMed]
- Yoneno, K.; Hisamatsu, T.; Shimamura, K.; Kamada, N.; Ichikawa, R.; Kitazume, M.T.; Mori, M.; Uo, M.; Namikawa, Y.; Matsuoka, K.; et al. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology 2013, 139, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.-B.; Guo, C.-J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef]
- Wu, R.; Yuan, X.; Li, X.; Ma, N.; Jiang, H.; Tang, H.; Xu, G.; Liu, Z.; Zhang, Z. The bile acid-activated retinoic acid response in dendritic cells is involved in food allergen sensitization. Allergy 2022, 77, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Lee-Sarwar, K.; Kelly, R.S.; Lasky-Su, J.; Moody, D.B.; Mola, A.R.; Cheng, T.-Y.; Comstock, L.E.; Zeiger, R.; O’Connor, G.T.; Sandel, M.T.; et al. Intestinal microbial-derived sphingolipids are inversely associated with childhood food allergy. J. Allergy Clin. Immunol. 2018, 142, 335–338.e9. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Kim, E.G.; Kim, M.; Kim, S.Y.; Kim, Y.H.; Sohn, M.H.; Kim, K.W. Metabolomic profiling revealed altered lipid metabolite levels in childhood food allergy. J. Allergy Clin. Immunol. 2022, 149, 1722–1731.e9. [Google Scholar] [CrossRef] [PubMed]
- Obeso, D.; Contreras, N.; Dolores-Hernández, M.; Carrillo, T.; Barbas, C.; Escribese, M.M.; Villaseñor, A.; Barber, D. Development of a Novel Targeted Metabolomic LC-QqQ-MS Method in Allergic Inflammation. Metabolites 2022, 12, 592. [Google Scholar] [CrossRef]
- Johnson, E.L.; Heaver, S.L.; Waters, J.L.; Kim, B.I.; Bretin, A.; Goodman, A.L.; Gewirtz, A.T.; Worgall, T.S.; Ley, R.E. Sphingolipids produced by gut bacteria enter host metabolic pathways impacting ceramide levels. Nat. Commun. 2020, 11, 2471. [Google Scholar] [CrossRef]
- Crestani, E.; Harb, H.; Charbonnier, L.-M.; Leirer, J.; Motsinger-Reif, A.; Rachid, R.; Phipatanakul, W.; Kaddurah-Daouk, R.; Chatila, T.A. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J. Allergy Clin. Immunol. 2020, 145, 897–906. [Google Scholar] [CrossRef] [Green Version]
- Bai, A.; Moss, A.; Kokkotou, E.; Usheva, A.; Sun, X.; Cheifetz, A.; Zheng, Y.; Longhi, M.S.; Gao, W.; Wu, Y.; et al. CD39 and CD161 modulate Th17 responses in Crohn’s disease. J. Immunol. 2014, 193, 3366–3377. [Google Scholar] [CrossRef] [Green Version]
- Takemura, A.; Ohto, N.; Kuwahara, H.; Mizuno, M. Sphingoid base in pineapple glucosylceramide suppresses experimental allergy by binding leukocyte mono-immunoglobulin-like receptor 3. J. Sci. Food Agric. 2022, 102, 2704–2709. [Google Scholar] [CrossRef]
- Dai, Z.-L.; Wu, G.; Zhu, W.-Y. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768–1786. [Google Scholar] [CrossRef] [Green Version]
- Blachier, F.; Mariotti, F.; Huneau, J.F.; Tomé, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007, 33, 547–562. [Google Scholar] [CrossRef]
- Sorensen, K.; Cawood, A.; Gibson, G.; Cooke, L.; Stratton, R. Amino Acid Formula Containing Synbiotics in Infants with Cow’s Milk Protein Allergy: A Systematic Review and Meta-Analysis. Nutrients 2021, 13, 935. [Google Scholar] [CrossRef] [PubMed]
- Gold, M.S.; Quinn, P.J.; Campbell, D.E.; Peake, J.; Smart, J.; Robinson, M.; O’Sullivan, M.; Vogt, J.K.; Pedersen, H.K.; Liu, X.; et al. Effects of an Amino Acid-Based Formula Supplemented with Two Human Milk Oligosaccharides on Growth, Tolerability, Safety, and Gut Microbiome in Infants with Cow’s Milk Protein Allergy. Nutrients 2022, 14, 2297. [Google Scholar] [CrossRef]
- Nocerino, R.; Coppola, S.; Carucci, L.; Paparo, L.; Severina, A.F.D.G.D.S.; Berni, R. Canani Body growth assessment in children with IgE-mediated cow’s milk protein allergy fed with a new amino acid-based formula. Front. Allergy 2022, 3, 977589. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.P.; Singh, U.P.; Rouse, M.; Zhang, J.; Chatterjee, S.; Nagarkatti, P.S.; Nagarkatti, M. Dietary Indoles Suppress Delayed-Type Hypersensitivity by Inducing a Switch from Proinflammatory Th17 Cells to Anti-Inflammatory Regulatory T Cells through Regulation of MicroRNA. J. Immunol. 2016, 196, 1108–1122. [Google Scholar] [CrossRef] [Green Version]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, M.; Viera-Hutchins, L.; Garcia-Lloret, M.; Rivas, M.N.; Wise, P.; McGhee, S.A.; Chatila, Z.K.; Daher, N.; Sioutas, C.; Chatila, T.A. Vehicular exhaust particles promote allergic airway inflammation through an aryl hydrocarbon receptor–notch signaling cascade. J. Allergy Clin. Immunol. 2015, 136, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Ye, Y.; Ji, J.; Sun, J.; Wang, J.-S.; Sun, X. Untargeted Metabolomic Profiling Reveals Changes in Gut Microbiota and Mechanisms of Its Regulation of Allergy in OVA-Sensitive BALB/c Mice. J. Agric. Food Chem. 2022, 70, 3344–3356. [Google Scholar] [CrossRef]
- Larke, J.A.; Kuhn-Riordon, K.; Taft, D.H.; Sohn, K.; Iqbal, S.; Underwood, M.A.; Mills, D.A.; Slupsky, C.M. Preterm Infant Fecal Microbiota and Metabolite Profiles Are Modulated in a Probiotic Specific Manner. J. Craniofacial Surg. 2022, 75, 535–542. [Google Scholar] [CrossRef]
- Kepert, I.; Fonseca, J.; Müller, C.; Milger, K.; Hochwind, K.; Kostric, M.; Fedoseeva, M.; Ohnmacht, C.; Dehmel, S.; Nathan, P.; et al. D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 2017, 139, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Yan, J.; Xiang, Q.; Wang, F.; Dai, H.; Huang, K.; Fang, L.; Yao, H.; Wang, L.; Zhang, W. Fructooligosaccharides protect against OVA-induced food allergy in mice by regulating the Th17/Treg cell balance using tryptophan metabolites. Food Funct. 2021, 12, 3191–3205. [Google Scholar] [CrossRef]
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.W.M.; Tijssen, J.G.P.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youngster, I.; Russell, G.H.; Pindar, C.; Ziv-Baran, T.; Sauk, J.; Hohmann, E.L. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA 2014, 312, 1772–1778. [Google Scholar] [CrossRef] [PubMed]
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P.; et al. Treating Clostridium difficile Infection With Fecal Microbiota Transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, B.; Prioult, G.; Hacini-Rachinel, F.; Moine, D.; Bruttin, A.; Ngom-Bru, C.; Labellie, C.; Nicolis, I.; Berger, B.; Mercenier, A.; et al. Infant gut microbiota is protective against cow’s milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol. Ecol. 2012, 79, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Noval Rivas, M.; Burton, O.T.; Wise, P.; Zhang, Y.-Q.; Hobson, S.A.; Lloret, M.G.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J.; et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, S.; Yang, X.; Huazeng, B.; Cheng, Q. Influences of non-IgE-mediated cow’s milk protein allergy-associated gut microbial dysbiosis on regulatory T cell-mediated intestinal immune tolerance and homeostasis. Microb. Pathog. 2021, 158, 105020. [Google Scholar] [CrossRef]
- Chen, P.-J.; Nakano, T.; Lai, C.-Y.; Chang, K.-C.; Chen, C.-L.; Goto, S. Daily full spectrum light exposure prevents food allergy-like allergic diarrhea by modulating vitamin D3 and microbiota composition. NPJ Biofilms Microbiomes 2021, 7, 41. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Bloom, P.P.; Soto, M.T.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.-B.; Hohmann, E.L. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef]
- Park, S.Y.; Seo, G.S. Fecal Microbiota Transplantation: Is It Safe? Clin. Endosc. 2021, 54, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, M.; Langer, R.; Traverso, G. Microbial therapeutics: New opportunities for drug delivery. J. Exp. Med. 2019, 216, 1005–1009. [Google Scholar] [CrossRef]
- Wargo, J.A. Modulating gut microbes. Science 2020, 369, 1302–1303. [Google Scholar] [CrossRef] [PubMed]
- Nagler, C.R. Drugging the microbiome. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Dai, D.L.; Boutin, R.C.; Sbihi, H.; Sears, M.R.; Moraes, T.J.; Becker, A.B.; Azad, M.B.; Mandhane, P.J.; Subbarao, P.; et al. A rich meconium metabolome in human infants is associated with early-life gut microbiota composition and reduced allergic sensitization. Cell Rep. Med. 2021, 2, 100260. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Korenori, Y.; Washio, M.; Kobayashi, T.; Momoda, R.; Kiyohara, C.; Kuroda, A.; Saito, Y.; Sonomoto, K.; Nakayama, J. Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Loke, P.; Orsini, F.; Lozinsky, A.C.; Gold, M.; O’Sullivan, M.D.; Quinn, P.; Lloyd, M.; Ashley, S.E.; Pitkin, S.; Axelrad, C.; et al. Probiotic peanut oral immunotherapy versus oral immunotherapy and placebo in children with peanut allergy in Australia (PPOIT-003): A multicentre, randomised, phase 2b trial. Lancet Child Adolesc. Health 2022, 6, 171–184. [Google Scholar] [CrossRef]
- Tang, M.L.K.; Ponsonby, A.-L.; Orsini, F.; Tey, D.; Robinson, M.; Su, E.L.; Licciardi, P.; Burks, W.; Donath, S. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J. Allergy Clin. Immunol. 2015, 135, 737–744.e8. [Google Scholar] [CrossRef]
- Galvin, A.D.; Lloyd, M.; Hsiao, K.-C.; Tang, M.L. Long-term benefit of probiotic peanut oral immunotherapy on quality of life in a randomized trial. J. Allergy Clin. Immunol. Pract. 2021, 9, 4493–4495.e1. [Google Scholar] [CrossRef]
- Kalliomäki, M.; Salminen, S.; Arvilommi, H.; Kero, P.; Koskinen, P.; Isolauri, E. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 2001, 357, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; Nocerino, R.; Terrin, G.; Frediani, T.; Lucarelli, S.; Cosenza, L.; Passariello, A.; Leone, L.; Granata, V.; Di Costanzo, M.; et al. Formula selection for management of children with cow’s milk allergy influences the rate of acquisition of tolerance: A prospective multicenter study. J. Pediatr. 2013, 163, 771–777.e1. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.; Isolauri, E.; He, F.; Hashimoto, H.; Benno, Y.; Salminen, S. Differences in Bifidobacterium flora composition in allergic and healthy infants. J. Allergy Clin. Immunol. 2001, 108, 144–145. [Google Scholar] [CrossRef] [PubMed]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr. Res. 2018, 83, 677–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, A.T.; ASSIGN study group; Wopereis, H.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Candy, D.C.A.; Shah, N.; et al. A specific synbiotic-containing amino acid-based formula in dietary management of cow’s milk allergy: A randomized controlled trial. Clin. Transl. Allergy 2019, 9, 5. [Google Scholar] [CrossRef]
- Chatchatee, P.; Nowak-Wegrzyn, A.; Lange, L.; Benjaponpitak, S.; Chong, K.W.; Sangsupawanich, P.; van Ampting, M.T.J.; Oude Nijhuis, M.M.; Harthoorn, L.F.; Langford, J.E.; et al. Tolerance development in cow’s milk–allergic infants receiving amino acid–based formula: A randomized controlled trial. J. Allergy Clin. Immunol. 2022, 149, 650–658.e5. [Google Scholar] [CrossRef]
- Banzon, T.M.; Kelly, M.S.; Bartnikas, L.M.; Sheehan, W.J.; Cunningham, A.; Harb, H.; Crestani, E.; Valeri, L.; Greco, K.F.; Chatila, T.A.; et al. Atopic Dermatitis Mediates the Association Between an IL4RA Variant and Food Allergy in School-Aged Children. J. Allergy Clin. Immunol. Pract. 2022, 10, 2117–2124.e4. [Google Scholar] [CrossRef]
- Vuillermin, P.J.; O’Hely, M.; Collier, F.; Allen, K.J.; Tang, M.L.K.; Harrison, L.C.; Carlin, J.B.; Saffery, R.; Ranganathan, S.; Sly, P.D.; et al. Maternal carriage of Prevotella during pregnancy associates with protection against food allergy in the offspring. Nat. Commun. 2020, 11, 1452. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef] [Green Version]
- McCauley, K.E.; Rackaityte, E.; LaMere, B.; Fadrosh, D.W.; Fujimura, K.E.; Panzer, A.R.; Lin, D.L.; Lynch, K.V.; Halkias, J.; Mendoza, V.F.; et al. Heritable vaginal bacteria influence immune tolerance and relate to early-life markers of allergic sensitization in infancy. Cell Rep. Med. 2022, 3, 100713. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernikova, D.A.; Zhao, M.Y.; Jacobs, J.P. Microbiome Therapeutics for Food Allergy. Nutrients 2022, 14, 5155. https://doi.org/10.3390/nu14235155
Chernikova DA, Zhao MY, Jacobs JP. Microbiome Therapeutics for Food Allergy. Nutrients. 2022; 14(23):5155. https://doi.org/10.3390/nu14235155
Chicago/Turabian StyleChernikova, Diana A., Matthew Y. Zhao, and Jonathan P. Jacobs. 2022. "Microbiome Therapeutics for Food Allergy" Nutrients 14, no. 23: 5155. https://doi.org/10.3390/nu14235155
APA StyleChernikova, D. A., Zhao, M. Y., & Jacobs, J. P. (2022). Microbiome Therapeutics for Food Allergy. Nutrients, 14(23), 5155. https://doi.org/10.3390/nu14235155






