Roseburia intestinalis and Its Metabolite Butyrate Inhibit Colitis and Upregulate TLR5 through the SP3 Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
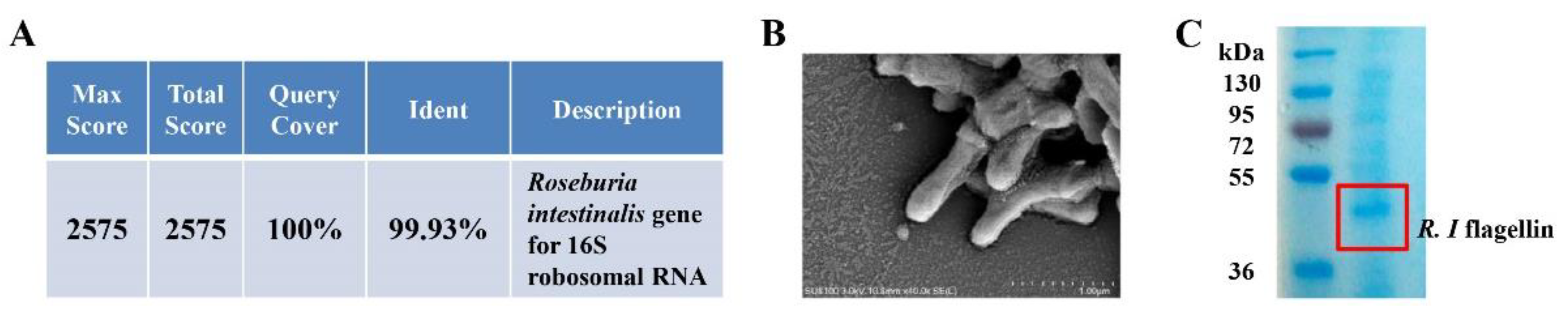
2.1. Preparation of R. intestinalis
2.2. Animals and Groups
2.3. Disease Activity Index
2.4. Extraction of Flagellin
2.5. Cell Culture
2.6. RT–qPCR and Western Blotting
2.7. IHC Staining
2.8. Immunofluorescence
2.9. ELISA
2.10. Cytokine Quantitation
2.11. siRNA Interference
2.12. ChIP–qPCR
2.13. Analysis of Fecal SCFAs
2.14. Statistical Analyses
3. Results
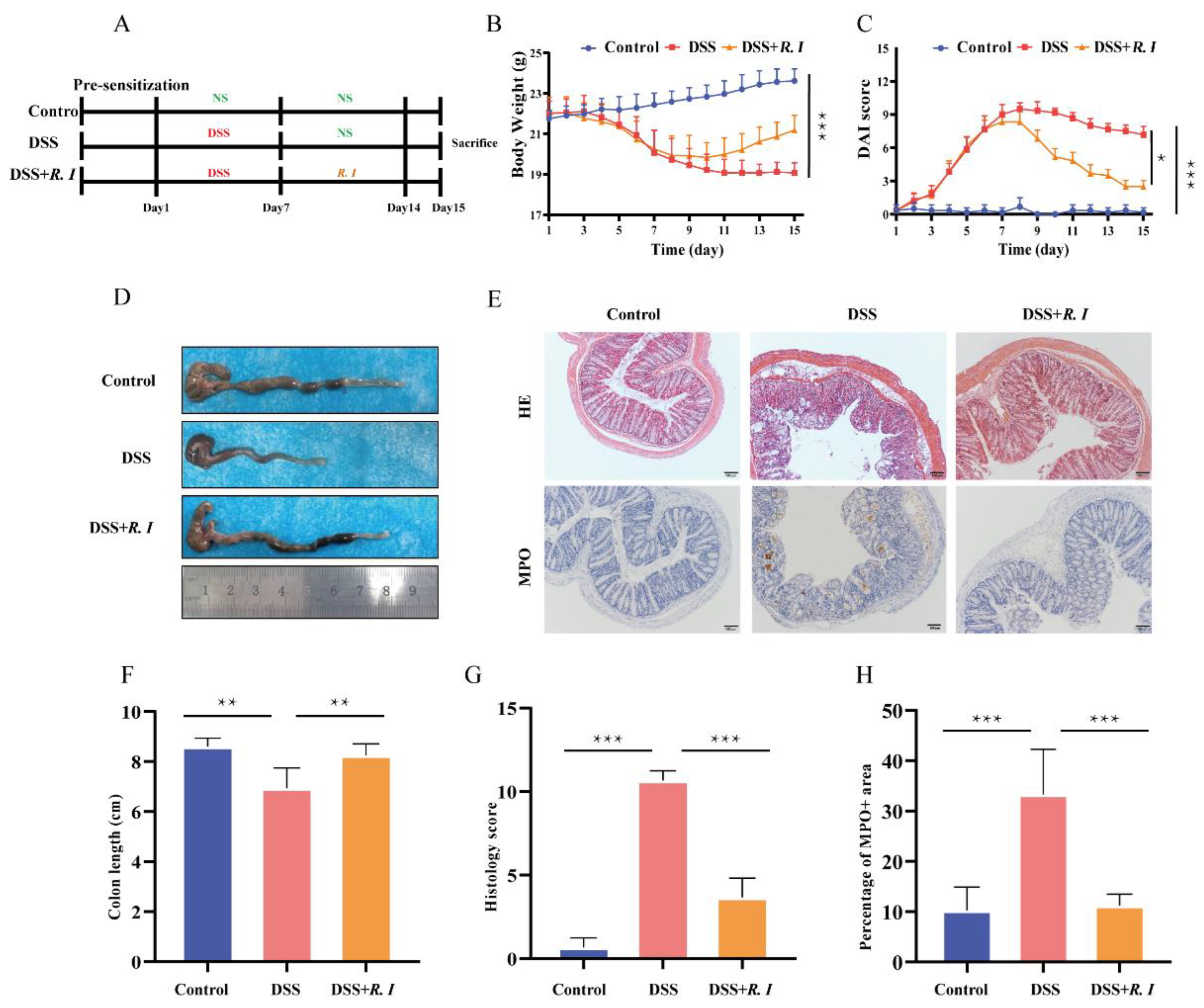
3.1. DSS-Induced Intestinal Inflammation Relieved by R. intestinalis
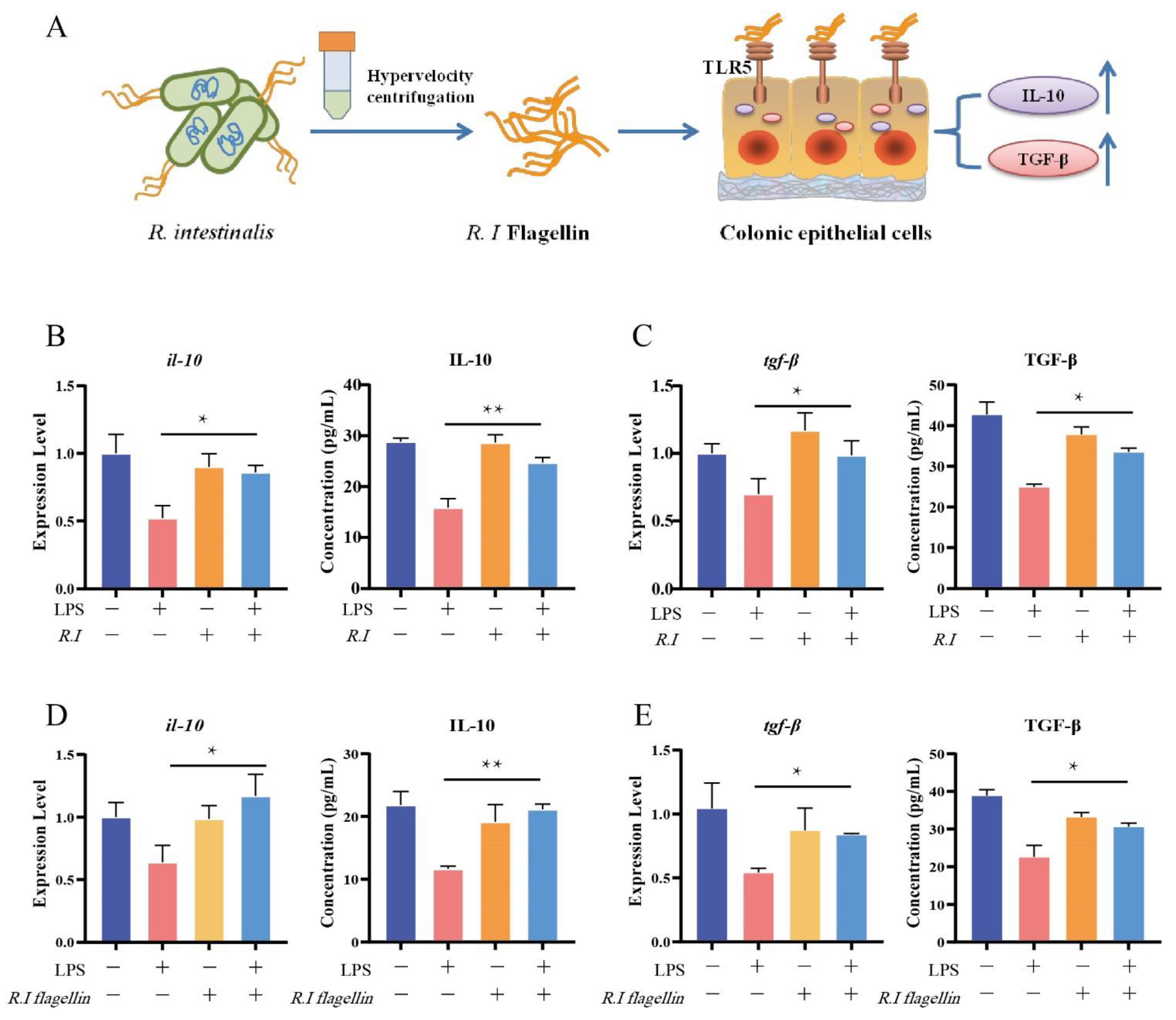
3.2. Flagellin from R. intestinalis Displayed Anti-Inflammatory Effects in HT29 Cells
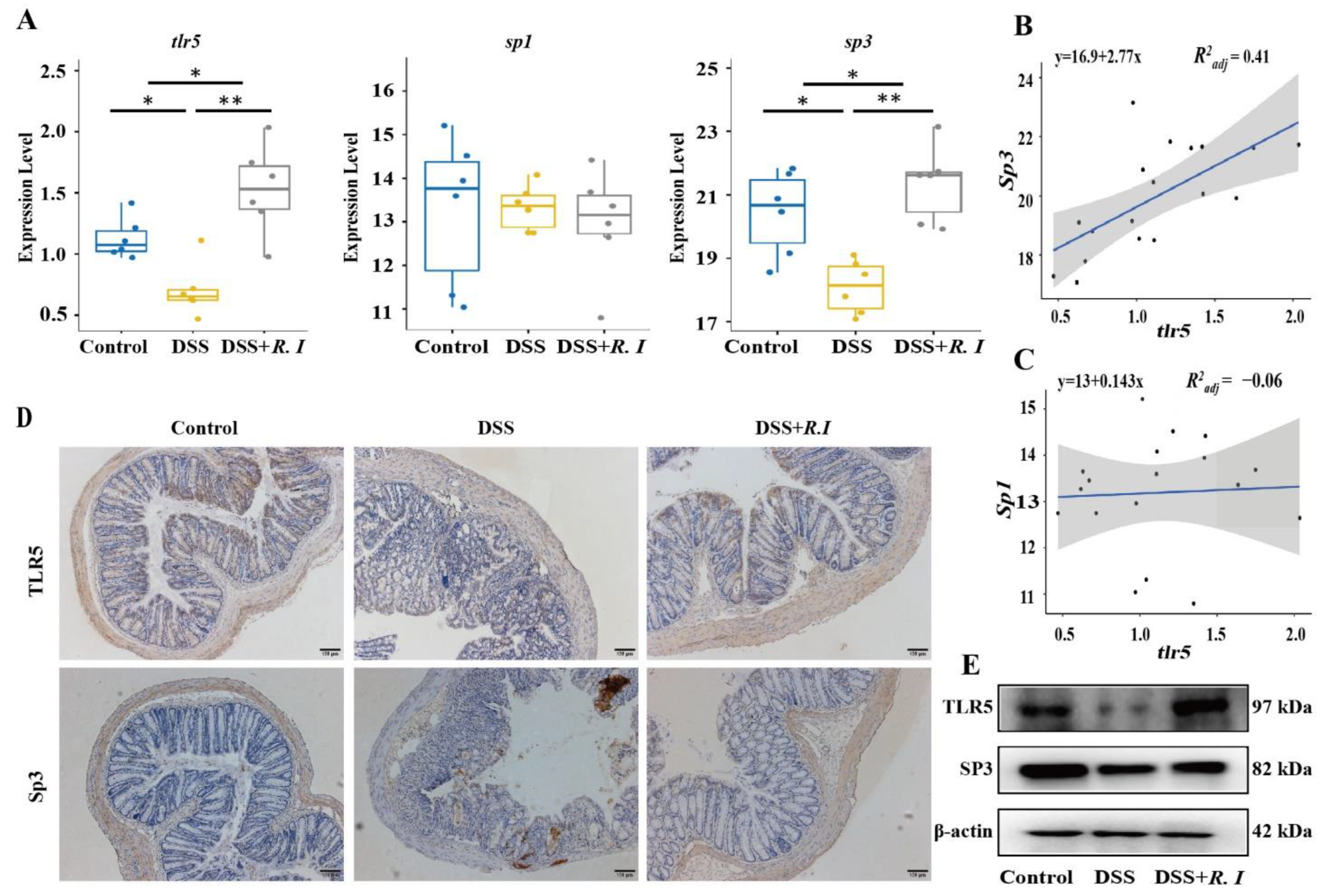
3.3. Determination of TLR5 Expression and Screening of Related Transcription Factors
3.4. R. intestinalis Inhibited the Inflammatory Progression of UC through Its Metabolite Butyrate
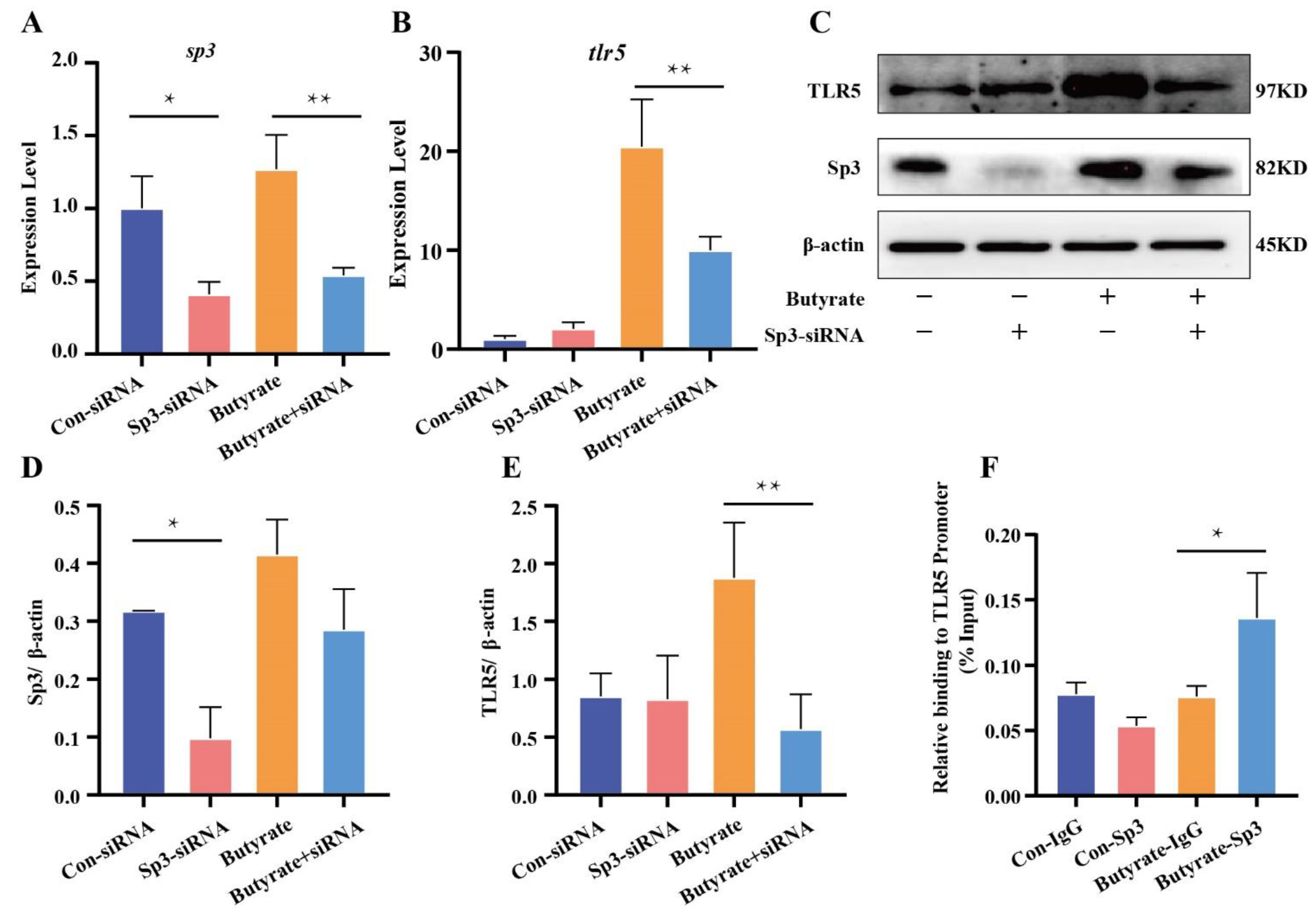
3.5. Butyrate, a Metabolite of R. intestinalis, Promoted TLR5 Expression at the Transcriptional Level
3.6. Butyrate Promoted Sp3 Binding to the Promoter of TLR5
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.C. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: Exploring a common ground hypothesis. J. Biomed. Sci. 2018, 25, 79. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Shen, Z.H.; Zhu, C.X.; Quan, Y.S.; Yang, Z.Y.; Wu, S.; Luo, W.W.; Tan, B.; Wang, X.Y. Relationship between intestinal microbiota and ulcerative colitis: Mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018, 24, 5–14. [Google Scholar] [CrossRef]
- van der Lelie, D.; Oka, A.; Taghavi, S.; Umeno, J.; Fan, T.J.; Merrell, K.E.; Watson, S.D.; Ouellette, L.; Liu, B.; Awoniyi, M.; et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat. Commun. 2021, 12, 3105. [Google Scholar] [CrossRef]
- Zhang, H.L.; Li, W.S.; Xu, D.N.; Zheng, W.W.; Liu, Y.; Chen, J.; Qiu, Z.B.; Dorfman, R.G.; Zhang, J.; Liu, J. Mucosa-reparing and microbiota-balancing therapeutic effect of Bacillus subtilis alleviates dextrate sulfate sodium-induced ulcerative colitis in mice. Exp. Ther. Med. 2016, 12, 2554–2562. [Google Scholar] [CrossRef] [Green Version]
- Tamaki, H.; Nakase, H.; Inoue, S.; Kawanami, C.; Itani, T.; Ohana, M.; Kusaka, T.; Uose, S.; Hisatsune, H.; Tojo, M.; et al. Efficacy of probiotic treatment with Bifidobacterium longum 536 for induction of remission in active ulcerative colitis: A randomized, double-blinded, placebo-controlled multicenter trial. Dig. Endosc. 2016, 28, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Zhu, C.; Quan, Y.; Yang, J.; Yuan, W.; Yang, Z.; Wu, S.; Luo, W.; Tan, B.; Wang, X. Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J. Gastroenterol. Hepatol. 2018, 33, 1751–1760. [Google Scholar] [CrossRef]
- Seo, B.; Jeon, K.; Moon, S.; Lee, K.; Kim, W.K.; Jeong, H.; Cha, K.H.; Lim, M.Y.; Kang, W.; Kweon, M.N.; et al. Roseburia spp. Abundance Associates with Alcohol Consumption in Humans and Its Administration Ameliorates Alcoholic Fatty Liver in Mice. Cell Host Microbe 2020, 27, 25–40.e26. [Google Scholar] [CrossRef] [PubMed]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-Chain Fatty Acids and Their Association with Signalling Pathways in Inflammation, Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Sun, S.; Zhou, Y.; Wang, H.; Yu, Y.; Hu, T.; Yao, Y.; Zhou, C. Bacteroides fragilis restricts colitis-associated cancer via negative regulation of the NLRP3 axis. Cancer Lett. 2021, 523, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ran, X.; Li, B.; Li, Y.; He, D.; Huang, B.; Fu, S.; Liu, J.; Wang, W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine 2018, 30, 317–325. [Google Scholar] [CrossRef] [Green Version]
- Couto, M.R.; Goncalves, P.; Magro, F.; Martel, F. Microbiota-derived butyrate regulates intestinal inflammation: Focus on inflammatory bowel disease. Pharmacol. Res. 2020, 159, 104947. [Google Scholar] [CrossRef]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Cheng, Y.; Ruan, G.; Fan, L.; Tian, Y.; Xiao, Z.; Chen, D.; Wei, Y. New pathway ameliorating ulcerative colitis: Focus on Roseburia intestinalis and the gut-brain axis. Therap. Adv. Gastroenterol 2021, 14, 17562848211004469. [Google Scholar] [CrossRef]
- Hold, G.L.; Schwiertz, A.; Aminov, R.I.; Blaut, M.; Flint, H.J. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 2003, 69, 4320–4324. [Google Scholar] [CrossRef] [Green Version]
- Nie, K.; Ma, K.; Luo, W.; Shen, Z.; Yang, Z.; Xiao, M.; Tong, T.; Yang, Y.; Wang, X. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front. Cell. Infect. Microbiol. 2021, 11, 757718. [Google Scholar] [CrossRef]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020, 469, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Kim, B.G.; Kim, J.H.; Chun, J.; Im, J.P.; Kim, J.S. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int. Immunopharmacol. 2017, 51, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 2014, 104, 15.25.11–15.25.14. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Dasgupta, N.; Ta, A.; Das, S. Physiological TLR5 expression in the intestine is regulated by differential DNA binding of Sp1/Sp3 through simultaneous Sp1 dephosphorylation and Sp3 phosphorylation by two different PKC isoforms. Nucleic Acids Res. 2016, 44, 5658–5672. [Google Scholar] [CrossRef] [Green Version]
- Davie, J.R. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003, 133, 2485S–2493S. [Google Scholar] [CrossRef]
- Xu, H.; McCoy, A.; Li, J.; Zhao, Y.; Ghishan, F.K. Sodium butyrate stimulates NHE8 expression via its role on activating NHE8 basal promoter activity. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G500–G505. [Google Scholar] [CrossRef] [Green Version]
- Ruigrok, E.; Gibbons, S.; Wapenaar, K. Cross-correlation beamforming. J. Seismol. 2017, 21, 495–508. [Google Scholar] [CrossRef] [Green Version]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef]
- White, N.R.; Mulligan, P.; King, P.J.; Sanderson, I.R. Sodium butyrate-mediated Sp3 acetylation represses human insulin-like growth factor binding protein-3 expression in intestinal epithelial cells. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 134–141. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Bloom, S.M.; Bijanki, V.N.; Nava, G.M.; Sun, L.; Malvin, N.P.; Donermeyer, D.L.; Dunne, W.M., Jr.; Allen, P.M.; Stappenbeck, T.S. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 2011, 9, 390–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, R.; Xue, X.; Zhang, L.; Yang, X.; Zhao, L.; Zhang, C. Strain-Specific Anti-inflammatory Properties of Two Akkermansia muciniphila Strains on Chronic Colitis in Mice. Front. Cell. Infect. Microbiol. 2019, 9, 239. [Google Scholar] [CrossRef]
- Niederman, R.; Buyle-Bodin, Y.; Lu, B.Y.; Robinson, P.; Naleway, C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 1997, 76, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.; Rodrigues, H.G.; Hatanaka, E.; Hebeda, C.B.; Farsky, S.H.; Curi, R. Short-chain fatty acids stimulate the migration of neutrophils to inflammatory sites. Clin. Sci. Lond. 2009, 117, 331–338. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Sina, C.; Kemper, C.; Derer, S. The intestinal complement system in inflammatory bowel disease: Shaping intestinal barrier function. Semin. Immunol. 2018, 37, 66–73. [Google Scholar] [CrossRef]
- Burgueno, J.F.; Abreu, M.T. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 263–278. [Google Scholar] [CrossRef]
- Meijer, K.; de Vos, P.; Priebe, M.G. Butyrate and other short-chain fatty acids as modulators of immunity: What relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 715–721. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Hernandez-Oliveras, A.; Izquierdo-Torres, E.; Meneses-Morales, I.; Rodriguez, G.; Zarain-Herzberg, A.; Santiago-Garcia, J. Histone deacetylase inhibitors promote ATP2A3 gene expression in hepatocellular carcinoma cells: p300 as a transcriptional regulator. Int. J. Biochem. Cell Biol 2019, 113, 8–16. [Google Scholar] [CrossRef] [PubMed]


| Gene | Sequences (5′-3′) |
|---|---|
| Human gapdh | TGTGGGCATCAATGGATTTGG |
| ACACCATGTATTCCGGGTCAAT | |
| Human il-10 | GAACCAAGACCCAGACATCAAG |
| GCATTCTTCACCTGCTCCAC | |
| Human tgf β | CTAATGGTGGAAACCCACAACG |
| CAGCAACAATTCCTGGCGAT | |
| Human tlr5 | CAACTTGCCTGGGAAACTGA |
| GATGGCTAAATACTCCTGGTGG | |
| Human sp3 | CTGTAAAGAAGGTGGTGGAAGAG |
| AGAATGCCAACGCAGATGAG | |
| Mouse gapdh | TGAAGCAGGCATCTGAGGG |
| CGAAGGTGGAAGAGTGGGAG | |
| Mouse sp3 | GGGAAGGACATACTGCCCAC |
| TGCTTTTGTGTTGGAACTCTTGT | |
| Mouse sp1 | CGGTCCGGGTTCGCTTG |
| ACCCCCATTATTGCCACCAA | |
| Mouse tlr5 | GGCAAGGTTCAGCATCTTCAA |
| GGCAAGGTTCAGCATCTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruan, G.; Chen, M.; Chen, L.; Xu, F.; Xiao, Z.; Yi, A.; Tian, Y.; Ping, Y.; Lv, L.; Cheng, Y.; et al. Roseburia intestinalis and Its Metabolite Butyrate Inhibit Colitis and Upregulate TLR5 through the SP3 Signaling Pathway. Nutrients 2022, 14, 3041. https://doi.org/10.3390/nu14153041
Ruan G, Chen M, Chen L, Xu F, Xiao Z, Yi A, Tian Y, Ping Y, Lv L, Cheng Y, et al. Roseburia intestinalis and Its Metabolite Butyrate Inhibit Colitis and Upregulate TLR5 through the SP3 Signaling Pathway. Nutrients. 2022; 14(15):3041. https://doi.org/10.3390/nu14153041
Chicago/Turabian StyleRuan, Guangcong, Minjia Chen, Lu Chen, Fenghua Xu, Zhifeng Xiao, Ailin Yi, Yuting Tian, Yi Ping, Linling Lv, Yi Cheng, and et al. 2022. "Roseburia intestinalis and Its Metabolite Butyrate Inhibit Colitis and Upregulate TLR5 through the SP3 Signaling Pathway" Nutrients 14, no. 15: 3041. https://doi.org/10.3390/nu14153041
APA StyleRuan, G., Chen, M., Chen, L., Xu, F., Xiao, Z., Yi, A., Tian, Y., Ping, Y., Lv, L., Cheng, Y., & Wei, Y. (2022). Roseburia intestinalis and Its Metabolite Butyrate Inhibit Colitis and Upregulate TLR5 through the SP3 Signaling Pathway. Nutrients, 14(15), 3041. https://doi.org/10.3390/nu14153041






