GC1f Vitamin D Binding Protein Isoform as a Marker of Severity in Autism Spectrum Disorders
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. Vitamin D Binding Protein SNPs Genotyping
2.3. Statistical Analysis
3. Results
3.1. DBP rs2282679, rs7041, and rs4588 Genotype Distribution in ASD Children and Healthy Controls
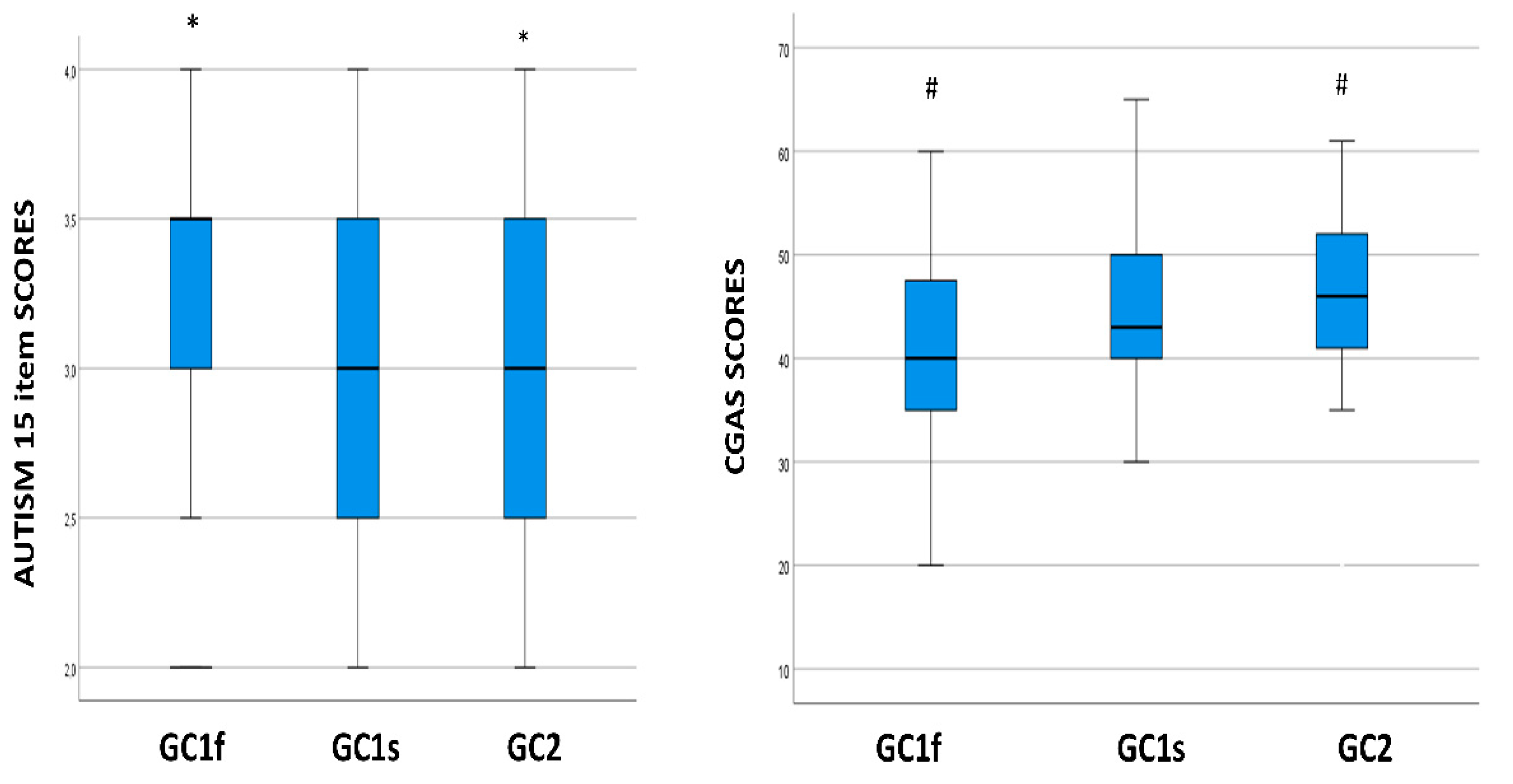
3.2. GC Isoform and Phenotype Distribution in ASD Children and Healthy Controls
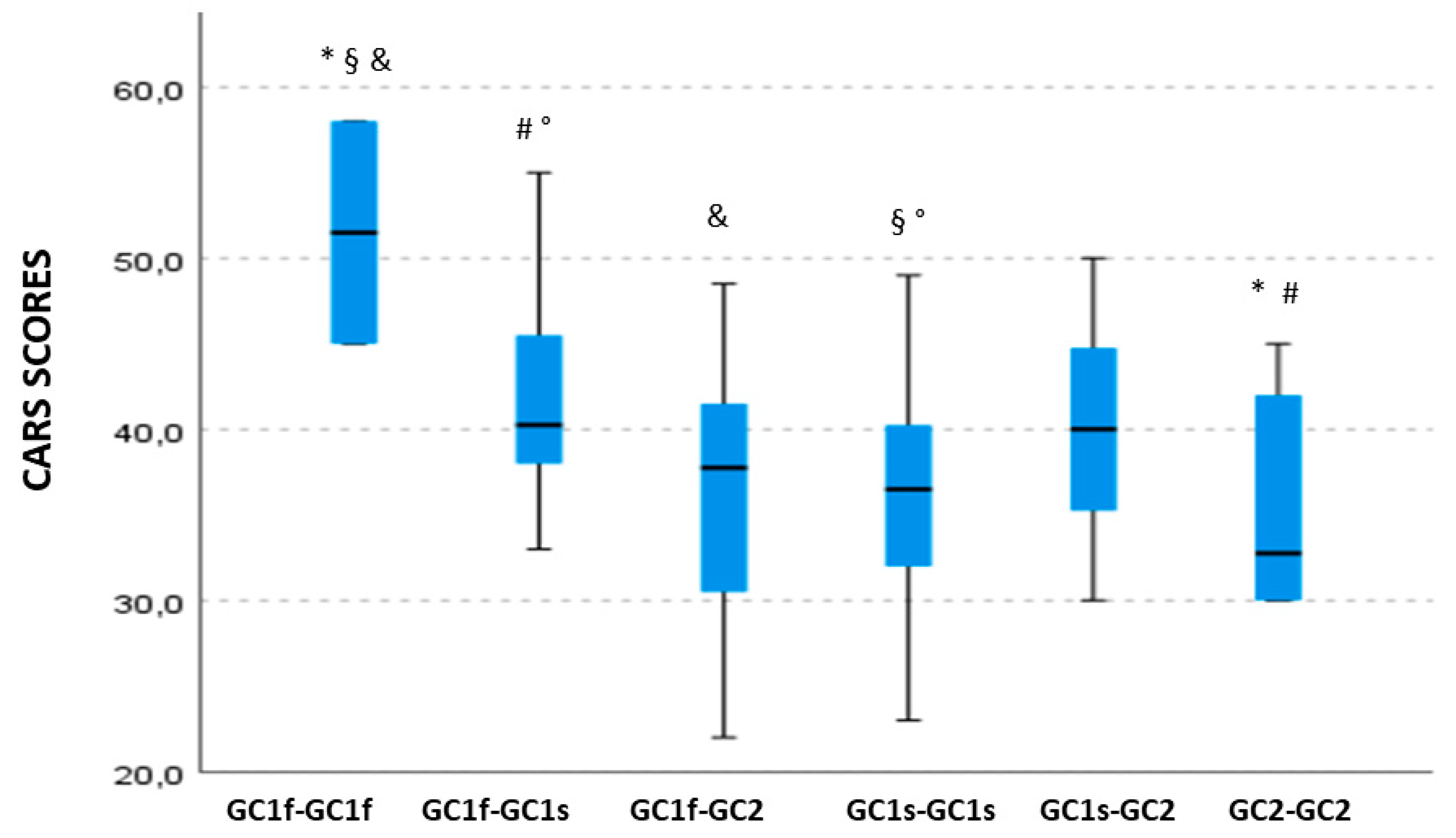
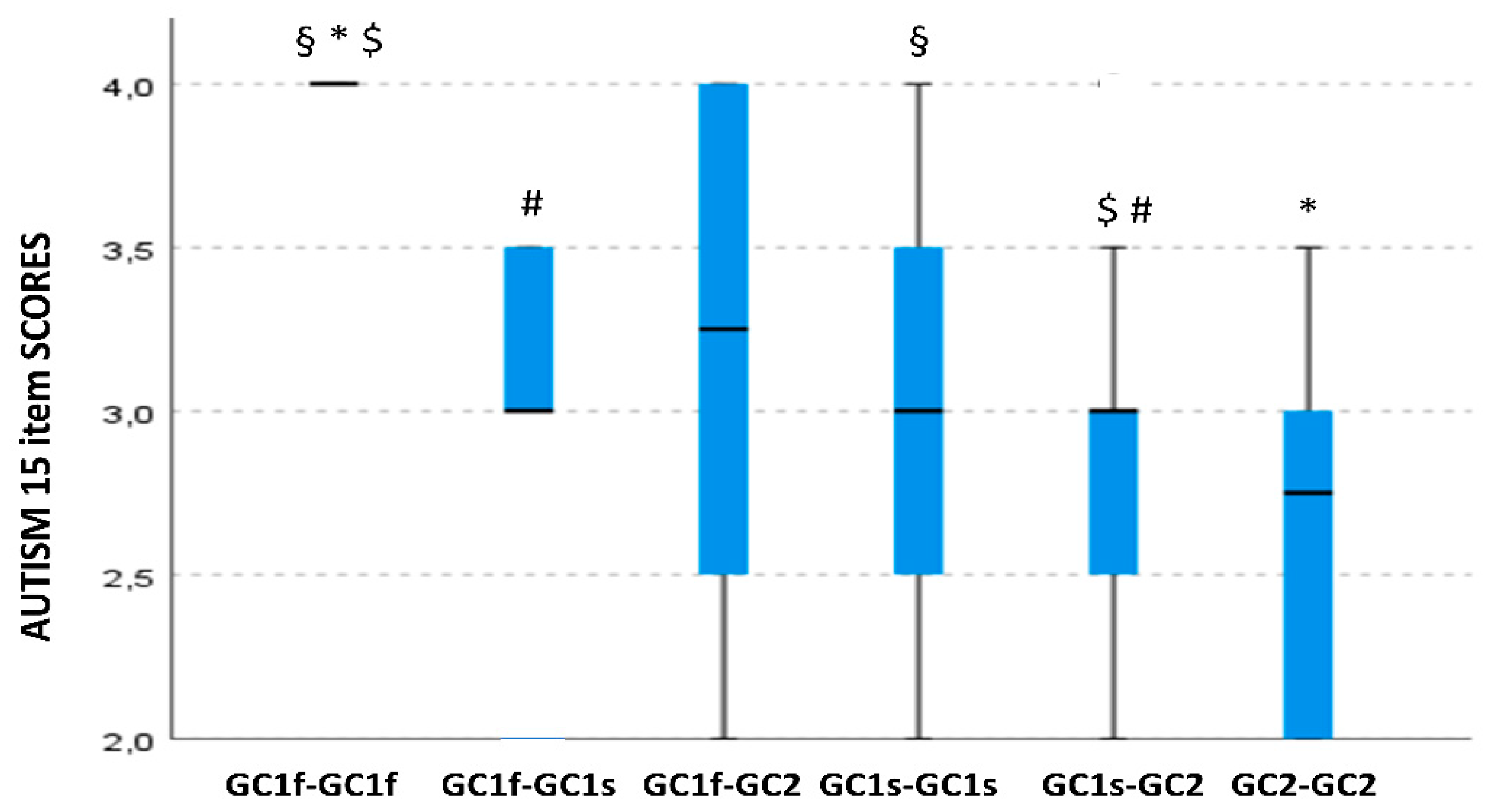
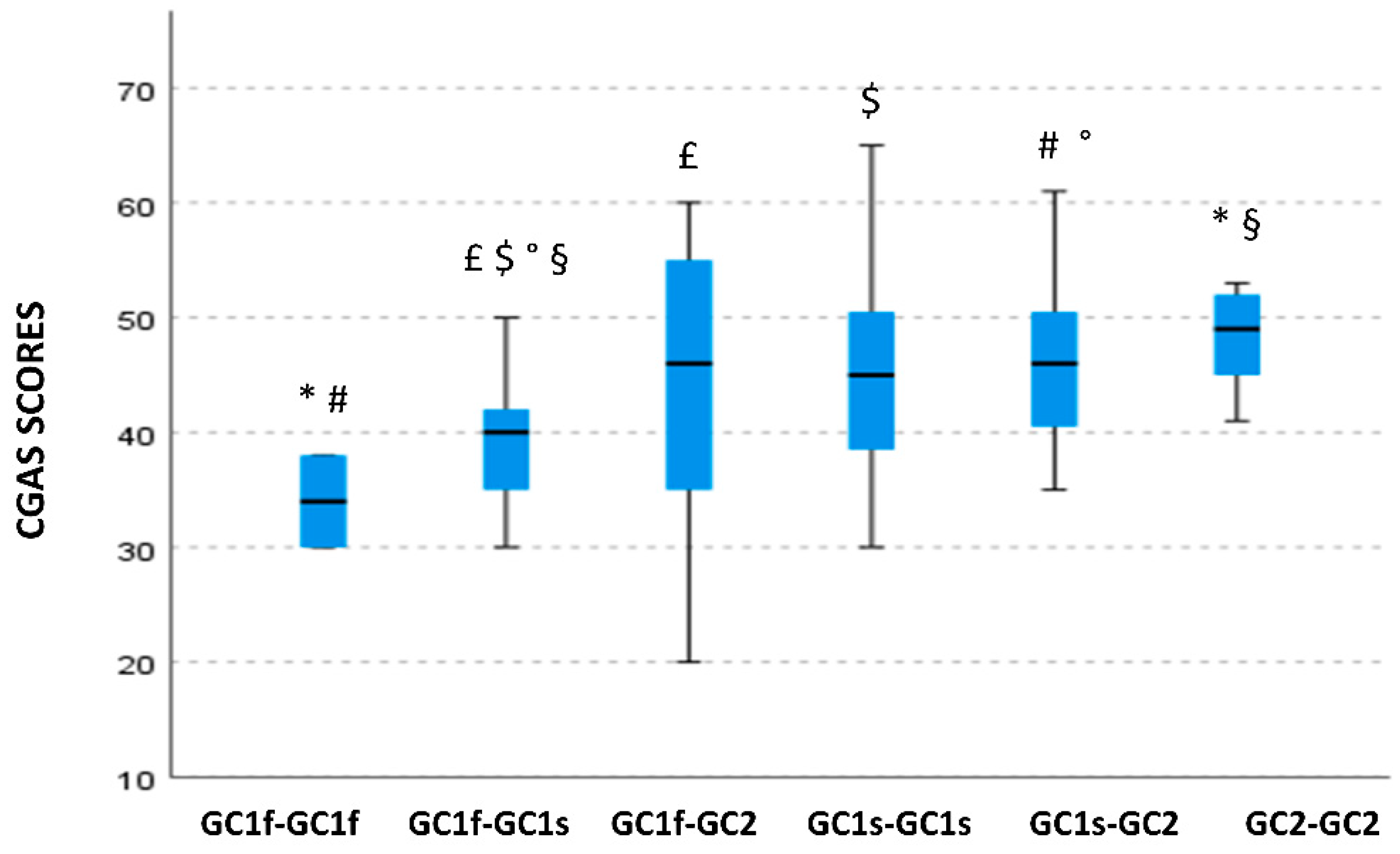
3.3. GC Isoform Genotype and Phenotype Correlation with Clinical Symptoms Severity, Cognitive, Behavioral, and Functioning Scales
3.4. GC Isoform Phenotype Correlation with CARS Total Scores
3.5. GC Isoform Phenotype Correlation with Item 15 of the CARS Scale Scores
3.6. GC Isoform Phenotype Correlation with CGAS Scores
3.7. Lack of Correlations between DBP rs2282679 and Clinical, Behavioral, and Functioning Scales
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, DSM-5, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Osservatorio Nazionale Autismo. Available online: https://osservatorionazionaleautismo.iss.it/ (accessed on 1 September 2022).
- Hodges, H.; Fealko, C.; Soares, N. Autism spectrum disorder: Definition, epidemiology, causes, and clinical evtion. Transl. Pediatr. 2020, 9, S55–S65. [Google Scholar] [CrossRef]
- Bölte, S.; Girdler, S.; Marschik, P.B. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell Mol. Life Sci. 2019, 76, 1275–1297. [Google Scholar] [CrossRef]
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J. Child Psychol. Psychiatry 2016, 57, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Alzghoul, L. Role of Vitamin D in Autism Spectrum Disorder. Curr. Pharm. Des. 2019, 25, 4357–4367. [Google Scholar] [CrossRef] [PubMed]
- Siracusano, M.; Riccioni, A.; Abate, R.; Benvenuto, A.; Curatolo, P.; Mazzone, L. Vitamin D Deficiency and Autism Spectrum Disorder. Curr. Pharm. Des. 2020, 26, 2460–2474. [Google Scholar] [CrossRef]
- Wang, Z.; Ding, R.; Wang, J. The Association between Vitamin D Status and Autism Spectrum Disorder (ASD): A Systematic Review and Meta-Analysis. Nutrients 2020, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Esposito, S. Vitamin D Deficiency During Pregnancy and Autism Spectrum Disorders Development. Front. Psychiatry 2020, 10, 987. [Google Scholar] [CrossRef]
- Holick, M.F. Ultraviolet B Radiation: The Vitamin D Connection. Adv. Exp. Med. Biol. 2017, 996, 137–154. [Google Scholar] [CrossRef]
- Uçar, N.; Grant, W.B.; Peraita-Costa, I.; Morales Suárez-Varela, M. How 25(OH)D Levels during Pregnancy Affect Prevalence of Autism in Children: Systematic Review. Nutrients 2020, 12, 2311. [Google Scholar] [CrossRef]
- Şengenç, E.; Kıykım, E.; Saltik, S. Vitamin D levels in children and adolescents with autism. J. Int. Med. Res. 2020, 48. [Google Scholar] [CrossRef]
- Petruzzelli, M.G.; Marzulli, L.; Margari, F.; De Giacomo, A.; Gabellone, A.; Giannico, O.V.; Margari, L. Vitamin D Deficiency in Autism Spectrum Disorder: A Cross-Sectional Study. Dis. Mark. 2020, 2020, 9292560. [Google Scholar] [CrossRef]
- Wang, T.; Shan, L.; Du, L.; Feng, J.; Xu, Z.; Staal, W.G.; Jia, F. Serum concentration of 25-hydroxyvitamin D in autism spectrum disorder: A systematic review and meta-analysis. Eur. Child Adolesc. Psychiatry 2016, 25, 341–350. [Google Scholar] [CrossRef]
- Kittana, M.; Ahmadani, A.; Stojanovska, L.; Attlee, A. The Role of Vitamin D Supplementation in Children with Autism Spectrum Disorder: A Narrative Review. Nutrients 2021, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, H.; Liu, C.; Zhang, Y.; Wang, W.; Zou, Z.; Yang, L.; He, X.; Wu, J.; Ma, J.; et al. Research Progress on the Role of Vitamin D in Autism Spectrum Disorder. Front. Behav. Neurosci. 2022, 16, 859151. [Google Scholar] [CrossRef]
- Uitterlinden, A.G.; Fang, Y.; Van Meurs, J.B.; Pols, H.A.; Van Leeuwen, J.P. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004, 338, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Schuster, I. Cytochromes P450 are essential players in the vitamin D signaling system. Biochim. Biophys. Acta 2011, 1814, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Chun, R.F.; Shieh, A.; Gottlieb, C.; Yacoubian, V.; Wang, J.; Hewison, M.; Adams, J.S. Vitamin D Binding Protein and the Biological Activity of Vitamin D. Front. Endocrinol. 2019, 10, 718. [Google Scholar] [CrossRef]
- Bouillon, R.; Pauwels, S. The Vitamin D-Binding Protein. In Vitamin D, 4th ed.; Feldman, D., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 97–115. [Google Scholar] [CrossRef]
- Braun, A.; Bichlmaier, R.; Cleve, H. Molecular analysis of the gene for the human vitamin-D-binding protein (group-specific component): Allelic differences of the common genetic GC types. Hum. Genet. 1992, 89, 401–406. [Google Scholar] [CrossRef]
- Newton, D.A.; Baatz, J.E.; Kindy, M.S.; Gattoni-Celli, S.; Shary, J.R.; Hollis, B.W.; Wagner, C.L. Vitamin D binding protein polymorphisms significantly impact vitamin D status in children. Pediatr. Res. 2019, 86, 662–669. [Google Scholar] [CrossRef]
- Powe, C.E.; Evans, M.K.; Wenger, J.; Zonderman, A.B.; Berg, A.H.; Nalls, M.; Tamez, H.; Zhang, D.; Bhan, I.; Karumanchi, S.A.; et al. Vitamin D-binding protein and vitamin D status of black Americans and white Americans. N. Engl. J. Med. 2013, 369, 1991–2000. [Google Scholar] [CrossRef]
- Slater, N.A.; Rager, M.L.; Havrda, D.E.; Harralson, A.F. Genetic Variation in CYP2R1 and GC Genes Associated with Vitamin D Deficiency Status. J. Pharm. Pract. Res. 2017, 30, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Nissen, J.; Vogel, U.; Ravn-Haren, G.; Andersen, E.W.; Madsen, K.H.; Nexø, B.A.; Andersen, R.; Mejborn, H.; Bjerrum, P.J.; Rasmussen, L.B.; et al. Common variants in CYP2R1 and GC genes are both determinants of serum 25-hydroxyvitamin D concentrations after UVB irradiation and after consumption of vitamin D3–fortified bread and milk during winter in Denmark. Am. J. Clin. Nutr. 2015, 101, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Enlund-Cerullo, M.; Koljonen, L.; Holmlund-Suila, E.; Hauta-Alus, H.; Rosendahl, J.; Valkama, S.; Helve, O.; Hytinantti, T.; Viljakainen, H.; Andersson, S.; et al. Genetic Variation of the Vitamin D Binding Protein Affects Vitamin D Status and Response to Supplementation in Infants. J. Clin. Endocrinol. Metab. 2019, 104, 5483–5498. [Google Scholar] [CrossRef] [PubMed]
- Guerini, F.R.; Bolognesi, E.; Chiappedi, M.; Mensi, M.M.; Fumagalli, O.; Rogantini, C.; Zanzottera, M.; Ghezzo, A.; Zanette, M.; Agliardi, C.; et al. Vitamin D Receptor Polymorphisms Associated with Autism Spectrum Disorder. Autism Res. 2020, 13, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Amer, Y.S.; Alenezi, S.; Bashiri, F.A.; Alawami, A.H.; Alhazmi, A.S.; Aladamawi, S.A.; Alnemary, F.; Alqahtani, Y.; Buraik, M.W.; AlSuwailem, S.S.; et al. AGREEing on Clinical Practice Guidelines for Autism Spectrum Disorders in Children: A Systematic Review and Quality Assessment. Children 2022, 9, 1050. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, F.; Feldman, I.; Lavelle, T.A.; Skokauskas, N. The cost-effectiveness of treatments for attention deficit-hyperactivity disorder and autism spectrum disorder in children and adolescents: A systematic review. Eur. Child Adolesc. Psychiatry 2022, 31, 1655–1670. [Google Scholar] [CrossRef]
- Roid, G.H.; Miller, L.J.; Pomplun, M.; Koch, C. Leiter international performance, Leiter-3 scale. In Los Angeles: Western Psychological Services Italian Edition, 3rd ed.; Cornoldi, C., Giofrè, D., Belacchi, C., Eds.; Giunti Organizzazioni Speciali: Florence, Italy, 2013. [Google Scholar]
- Wechsler, D. Wechsler Intelligence Scale for Children, 4th ed.; Harcourt Assessment: San Antonio, TX, USA, 2003. [Google Scholar]
- Raven, J.C.; Court, J.H.; Raven, J. Manual for Raven’s progressive matrices and vocabulary scales. Section 2: Coloured progressive matrices. In London: H. K. Lewis Italian Edition; Belacchi, C., Scalisi, T.G., Cannoni, E., Cornoldi, C., Eds.; Giunti Organizzazioni Speciali: Florence, Italy, 1984. [Google Scholar]
- Lord, C.; Rutter, M.; DiLavore, P.C.; Risi, S.; Gotham, K.; Bishop, S. Autism diagnostic observation schedule—Second edition (ADOS-2). In Los Angeles: Western Psychological Services Italian Edition; Colombi, C., Tancredi, R., Persico, A., Faggioli, A., Eds.; Hogrefe: Florence, Italy, 2012. [Google Scholar]
- Lord, C.; Rutter, M.; le Couteur, A. Autism diagnostic interview-revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994, 24, 659–685. [Google Scholar] [CrossRef] [PubMed]
- Schopler, E.; Reichler, R.J.; Renner, B.R. The Childhood Autism Rating Scale (CARS), for Diagnostic Screening and Classification in Autism; Irvington: New York, NY, USA, 1986. [Google Scholar]
- Shaffer, D.; Gould, M.S.; Brasic, J.; Ambrosini, P.; Fisher, P.; Bird, H.; Aluwahlia, S. A children’s global assessment scale (CGAS). Arch. Gen. Psychiatry 1983, 40, 1228–1231. [Google Scholar] [CrossRef]
- Theodoratou, E.; Palmer, T.; Zgaga, L.; Farrington, S.M.; McKeigue, P.; Din, F.V.; Tenesa, A.; Davey-Smith, G.; Dunlop, M.G.; Campbell, H. Instrumental variable estimation of the causal effect of plasma 25-hydroxy-vitamin D on colorectal cancer risk: A mendelian randomization analysis. PLoS ONE 2012, 7, e37662. [Google Scholar] [CrossRef]
- Rozmus, D.; Płomiński, J.; Augustyn, K.; Cieślińska, A. rs7041 and rs4588 Polymorphisms in Vitamin D Binding Protein Gene (VDBP) and the Risk of Diseases. Int. J. Mol. Sci. 2022, 23, 933. [Google Scholar] [CrossRef]
- Constans, J.; Lefevre-Witier, P.; Richard, P.; Jaeger, G. Gc (vitamin D binding protein) subtype polymorphism and variants distribution among Saharan, Middle East, and African populations. Am. J. Phys. Anthropol. 1980, 52, 435–441. [Google Scholar] [CrossRef]
- Bouillon, R.; van Baelen, H.; de Moor, P. Comparative study of the affinity of the serum vitamin D-binding protein. J. Steroid Biochem. 1980, 13, 1029–1034. [Google Scholar] [CrossRef]
- Boutin, B.; Galbraith, R.M.; Arnaud, P. Comparative affinity of the major genetic variants of human group-specific component (vitamin D-binding protein) for 25-(OH) vitamin D. J. Steroid Biochem. 1989, 32, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Almesri, N.; Das, N.S.; Ali, M.E.; Gumaa, K.; Giha, H.A. Independent associations of polymorphisms in vitamin D binding protein (GC) and vitamin D receptor (VDR) genes with obesity and plasma 25OHD3 levels demonstrate sex dimorphism. Appl. Physiol. Nutr. Metab. 2016, 41, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, J.; Constans, J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP). Hum. Genet. 1993, 92, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Lauridsen, A.L.; Vestergaard, P.; Hermann, A.P.; Brot, C.; Heickendorff, L.; Mosekilde, L.; Nexo, E. Plasma concentrations of 25-hydroxy-vitamin D and 1,25-dihydroxy-vitamin D are related to the phenotype of Gc (vitamin D-binding protein): A cross-sectional study on 595 early postmenopausal women. Calcif. Tissue Int. 2005, 77, 15–22. [Google Scholar] [CrossRef]
- Taes, Y.E.; Goemaere, S.; Huang, G.; Van Pottelbergh, I.; De Bacquer, D.; Verhasselt, B.; Van den Broeke, C.; Delanghe, J.R.; Kaufman, J.M. Vitamin D binding protein, bone status and body composition in community-dwelling elderly men. Bone 2006, 38, 701–707. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Hansen, R.L.; Hartiala, J.; Allayee, H.; Sconberg, J.L.; Schmidt, L.C.; Volk, H.E.; Tassone, F. Selected vitamin D metabolic gene variants and risk for autism spectrum disorder in the CHARGE Study. Early Hum. Dev. 2015, 91, 483–489. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, K.; Feng, C.; Chen, Y.; Li, S.; Iqbal, J.; Liao, L.; Zhao, Y.; Zhai, J. iTRAQ-Based Proteomic Analysis Reveals Protein Profile in Plasma from Children with Autism. Proteomics Clin. Appl. 2018, 12, e1700085. [Google Scholar] [CrossRef]
- Gong, Z.L.; Luo, C.M.; Wang, L.; Shen, L.; Wei, F.; Tong, R.J.; Liu, Y. Serum 25-hydroxyvitamin D levels in Chinese children with autism spectrum disorders. Neuroreport 2014, 25, 23–27. [Google Scholar] [CrossRef]
- Mostafa, G.A.; Al-Ayadhi, L.Y. Reduced serum concentrations of 25-hydroxy vitamin D in children with autism: Relation to autoimmunity. J. Neuroinflamm. 2012, 9, 201. [Google Scholar] [CrossRef] [PubMed]
- Delanghe, J.R.; Speeckaert, R.; Speeckaert, M.M. Behind the scenes of vitamin D binding protein: More than vitamin D binding. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 773–786. [Google Scholar] [CrossRef]
- Lee, D.H.; Kang, H.; Kim, J.H.; Jung, M.H.; Cho, M.C. Cerebrospinal fluid vitamin D-binding protein as a new biomarker for the diagnosis of meningitis. Neurol. Sci. 2019, 40, 1597–1605. [Google Scholar] [CrossRef]
- Swamy, N.; Dutta, A.; Ray, R. Roles of the structure and orientation of ligands and ligand mimics inside the ligand-binding pocket of the vitamin D-binding protein. Biochemistry 1997, 36, 7432–7436. [Google Scholar] [CrossRef] [PubMed]
- Gomme, P.T.; Bertolini, J. Therapeutic potential of vitamin D-binding protein. Trends Biotechnol. 2004, 22, 340–345. [Google Scholar] [CrossRef]
- Schellenberg, D.; Paré, P.D.; Weir, T.D.; Spinelli, J.J.; Walker, B.A.; Sandford, A.J. Vitamin D binding protein variants and the risk of COPD. Am. J. Respir. Crit. Care Med. 1998, 157, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ran, R.; Li, X.; Liu, G.; Xie, X.; Li, J. Genetic Variants Associated with Chronic Obstructive Pulmonary Disease Risk: Cumulative Epidemiological Evidence from Meta-Analyses and Genome-Wide Association Studies. Can. Respir. J. 2022, 2022, 3982335. [Google Scholar] [CrossRef]
- Oriano, M.; Aliberti, S.; Guerini, F.R.; Agliardi, C.; Di Francesco, C.; Gelmini, A.; Terranova, L.; Zanzottera, M.; Marchisio, P.; Clerici, M.; et al. The Isoform GC1f of the Vitamin D Binding Protein Is Associated with Bronchiectasis Severity. Biomedicines 2021, 9, 1573. [Google Scholar] [CrossRef]
- Eloranta, J.J.; Wenger, C.; Mwinyi, J.; Hiller, C.; Gubler, C.; Vavricka, S.R.; Fried, M.; Kullak-Ublick, G.A.; Swiss IBD Cohort Study Group. Association of a common vitamin D-binding protein polymorphism with inflammatory bowel disease. Pharmacogenet Genomics 2011, 21, 559–564. [Google Scholar] [CrossRef]
- Li, F.; Jiang, L.; Willis-Owen, S.A.; Zhang, Y.; Gao, J. Vitamin D binding protein variants associate with asthma susceptibility in the Chinese Han population. BMC Med. Genet. 2011, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Carta, A.; Manca, M.A.; Scoppola, C.; Simula, E.R.; Noli, M.; Ruberto, S.; Conti, M.; Zarbo, I.R.; Antonucci, R.; Sechi, L.A.; et al. Antihuman Endogenous Retrovirus Immune Response and Adaptive Dysfunction in Autism. Biomedicines 2022, 10, 1365. [Google Scholar] [CrossRef]
- Perry, V.H.; Holmes, C. Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 2014, 10, 217–224. [Google Scholar] [CrossRef]
- Cunningham, C. Microglia and neurodegeneration: The role of systemic inflammation. Glia 2013, 61, 71–90. [Google Scholar] [CrossRef] [PubMed]
- Sotgiu, S.; Manca, S.; Gagliano, A.; Minutolo, A.; Melis, M.C.; Pisuttu, G.; Scoppola, C.; Bolognesi, E.; Clerici, M.; Guerini, F.R.; et al. Immune regulation of neurodevelopment at the mother-foetus interface: The case of autism. Clin. Transl. Immunol. 2020, 9, e1211. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.T.; Chana, G.; Pardo, C.A.; Achim, C.; Semendeferi, K.; Buckwalter, J.; Courchesne, E.; Everall, I.P. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol. Psychiatry 2010, 68, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Edmonson, C.A.; Ziats, M.N.; Rennert, O.M. A Non-inflammatory Role for Microglia in Autism Spectrum Disorders. Front. Neurol. 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.; Melnyk, S.; Pavliv, O.; Bai, S.; Nick, T.G.; Frye, R.E.; James, S.J. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Transl. Psychiatry 2012, 2, e134. [Google Scholar] [CrossRef]
- Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Pessah, I.; van de Water, J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav. Immun. 2011, 25, 40–45. [Google Scholar] [CrossRef]
- Chez, M.G.; Dowling, T.; Patel, P.B.; Khanna, P.; Kominsky, M. Elevation of tumor necrosis factor-alpha in cerebrospinal fluid of autistic children. Pediatr. Neurol. 2007, 36, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Napolioni, V.; Ober-Reynolds, B.; Szelinger, S.; Corneveaux, J.J.; Pawlowski, T.; Ober-Reynolds, S.; Kirwan, J.; Persico, A.M.; Melmed, R.D.; Craig, D.W.; et al. Plasma cytokine profiling in sibling pairs discordant for autism spectrum disorder. J. Neuroinflamm. 2013, 10, 38. [Google Scholar] [CrossRef]
- Morgan, J.T.; Chana, G.; Abramson, I.; Semendeferi, K.; Courchesne, E.; Everall, I.P. Abnormal microglial-neuronal spatial organization in the dorsolateral prefrontal cortex in autism. Brain Res. 2012, 1456, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sugihara, G.; Ouchi, Y.; Nakamura, K.; Futatsubashi, M.; Takebayashi, K.; Yoshihara, Y.; Omata, K.; Matsumoto, K.; Tsuchiya, K.J.; et al. Microglial activation in young adults with autism spectrum disorder. JAMA Psychiatry 2013, 70, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Berk, M. The many roads to mitochondrial dysfunction in neuroimmune and neuropsychiatric disorders. BMC Med. 2015, 13, 68. [Google Scholar] [CrossRef]
- Morris, G.; Berk, M.; Galecki, P.; Walder, K.; Maes, M. The Neuro-Immune Pathophysiology of Central and Peripheral Fatigue in Systemic Immune-Inflammatory and Neuro-Immune Diseases. Mol. Neurobiol. 2016, 53, 1195–1219. [Google Scholar] [CrossRef] [PubMed]
- Saresella, M.; Marventano, I.; Guerini, F.R.; Mancuso, R.; Ceresa, L.; Zanzottera, M.; Rusconi, B.; Maggioni, E.; Tinelli, C.; Clerici, M. An autistic endophenotype results in complex immune dysfunction in healthy siblings of autistic children. Biol. Psychiatry 2009, 66, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Saresella, M.; Piancone, F.; Marventano, I.; Zoppis, M.; Hernis, A.; Zanette, M.; Trabattoni, D.; Chiappedi, M.; Ghezzo, A.; Canevini, M.P.; et al. Multiple inflammasome complexes are activated in autistic spectrum disorders. Brain Behav. Immun. 2016, 57, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, P.; Corbett, B.A.; Kantor, A.; Schulman, H.; Van de Water, J.; Amaral, D.G. In search of cellular immunophenotypes in the blood of children with autism. PLoS ONE 2011, 6, e19299. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, H.; Uto, Y.; Sasaki, H.; Okamura, N.; Murakami, A.; Kubo, S.; Kirk, K.L.; Hori, H. Gc protein (vitamin D-binding protein): Gc genotyping and GcMAF precursor activity. Anticancer Res. 2005, 25, 3689–3695. [Google Scholar] [PubMed]
- Nabeshima, Y.; Abe, C.; Kawauchi, T.; Hiroi, T.; Uto, Y.; Nabeshima, Y.I. Simple method for large-scale production of macrophage activating factor GcMAF. Sci. Rep. 2020, 10, 19122. [Google Scholar] [CrossRef]
- Eric Matamoros, M. GcMAF: A polemic or a highly promising molecule? World Sci.News 2017, 65, 20–36. [Google Scholar]
- Bishop, E.L.; Ismailova, A.; Dimeloe, S.; Hewison, M.; White, J.H. Vitamin D and Immune Regulation: Antibacterial, Antiviral, Anti-Inflammatory. JBMR Plus 2020, 5, e10405. [Google Scholar] [CrossRef] [PubMed]
- Aggeletopoulou, I.; Thomopoulos, K.; Mouzaki, A.; Triantos, C. Vitamin D-VDR Novel Anti-Inflammatory Molecules-New Insights into Their Effects on Liver Diseases. Int. J. Mol. Sci. 2022, 23, 8465. [Google Scholar] [CrossRef] [PubMed]
- Wöbke, T.K.; Sorg, B.L.; Steinhilber, D. Vitamin D in inflammatory diseases. Front. Physiol. 2014, 5, 244. [Google Scholar] [CrossRef] [PubMed]
| Allele Frequency | Continental ASD N (%) | Sardinian ASD N (%) | p Value | ASD N (%) | HC N (%) | p Value |
|---|---|---|---|---|---|---|
| rs2282679 | ||||||
| T | 265 (75.7) | 198 (73.8) | 463 (74.9) | 1223 (73.6) | ||
| G | 85 (24.3) | 70 (26.2) | 0.6 | 155 (25.1) | 439 (26.4) | 0.5 |
| rs7041 | ||||||
| A | 157 (44.8) | 111 (41.4) | 268 (43.4) | 680 (40.9) | ||
| C | 193 (55.2) | 157 (58.2) | 0.4 | 350 (56.6) | 982 (59.1) | 0.3 |
| rs4588 | ||||||
| T | 83 (23.7) | 70 (26.1) | 153 (26.1) | 439 (26.4) | ||
| G | 267 (76.3) | 198 (73.9) | 0.5 | 465 (75.3) | 1223 (73.6) | 0.42 |
| Total | 350 | 268 | 618 | 1662 | ||
| Genotype frequency | ||||||
| rs2282679 | ||||||
| TT | 101 (57.7) | 72 (57.7) | 173 (56.0) | 447 (53.8) | ||
| GT | 63 (36.0) | 54 (40.3) | 117 (37.9) | 329 (39.6) | ||
| GG | 11 (6.3) | 8 (6.0) | 0.7 | 19 (6.1) | 55 (6.6) | 0.8 |
| rs7041 | ||||||
| AA | 37 (21.1) | 23 (17.2) | 60 (19.4) | 134 (16.1) | ||
| AC | 83 (47.4) | 65 (48.5) | 148 (47.9) | 412 (49.6) | ||
| CC | 55 (31.4) | 46 (34.3) | 0.6 | 101 (32.7) | 285 (34.3) | 0.4 |
| rs4588 | ||||||
| TT | 10 (5.7) | 8 (6.0) | 18 (5.8) | 54 (6.5) | ||
| GT | 63 (36.0) | 54 (40.3) | 117 (37.9) | 331 (39.8) | ||
| GG | 102 (58.3) | 72 (53.7) | 0.7 | 174 (56.3) | 446 (53.7) | 0.7 |
| Total | 175 | 134 | 309 | 831 |
| Isoform | Genotype (rs7041/rs4588) | ASD N (%) | HC N (%) | p | OR, 95% IC |
|---|---|---|---|---|---|
| GC1s | C/G | 350 (56.6) | 1002 (60.3) | ||
| GC1f | A/G | 115 (18.6) | 241 (14.5) | 0.01, pc = 0.02 | 1.367, (1.07–1.74) |
| GC2 | A/T | 153 (24.8) | 439 (26.4) | ||
| 0.04 | |||||
| Isoform phenotype | |||||
| GC1f-GC1f | A/G-A/G | 8 (2.6) | 16 (1.9) | ||
| GC1f-GC1s | A/G-C/G | 65(21.0) | 145 (17.4) | ||
| GC1s-GC1s | C/G-C/G | 101 (32.7) | 285 (34.3) | ||
| GC1f-GC2 | A/G-A/T | 34(11.0) | 64 (7.7) | ||
| GC1s-GC2 | C/G-A/T | 83(26.9) | 267 (32.1) | ||
| GC2-GC2 | A/T-A/T | 18 (5.8) | 54 (6.5) | ||
| 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolognesi, E.; Guerini, F.R.; Sotgiu, S.; Chiappedi, M.; Carta, A.; Mensi, M.M.; Agliardi, C.; Zanzottera, M.; Clerici, M. GC1f Vitamin D Binding Protein Isoform as a Marker of Severity in Autism Spectrum Disorders. Nutrients 2022, 14, 5153. https://doi.org/10.3390/nu14235153
Bolognesi E, Guerini FR, Sotgiu S, Chiappedi M, Carta A, Mensi MM, Agliardi C, Zanzottera M, Clerici M. GC1f Vitamin D Binding Protein Isoform as a Marker of Severity in Autism Spectrum Disorders. Nutrients. 2022; 14(23):5153. https://doi.org/10.3390/nu14235153
Chicago/Turabian StyleBolognesi, Elisabetta, Franca Rosa Guerini, Stefano Sotgiu, Matteo Chiappedi, Alessandra Carta, Martina Maria Mensi, Cristina Agliardi, Milena Zanzottera, and Mario Clerici. 2022. "GC1f Vitamin D Binding Protein Isoform as a Marker of Severity in Autism Spectrum Disorders" Nutrients 14, no. 23: 5153. https://doi.org/10.3390/nu14235153
APA StyleBolognesi, E., Guerini, F. R., Sotgiu, S., Chiappedi, M., Carta, A., Mensi, M. M., Agliardi, C., Zanzottera, M., & Clerici, M. (2022). GC1f Vitamin D Binding Protein Isoform as a Marker of Severity in Autism Spectrum Disorders. Nutrients, 14(23), 5153. https://doi.org/10.3390/nu14235153









