Abstract
Tocotrienol-rich fraction (TRF), a palm oil-derived vitamin E fraction, is reported to possess potent neuroprotective effects. However, the modulation of proteomes in differentiated human neuroblastoma SH-SY5Y cells (diff-neural cells) by TRF has not yet been reported. This study aims to investigate the proteomic changes implicated by TRF in human neural cells using a label-free liquid-chromatography-double mass spectrometry (LC-MS/MS) approach. Levodopa, a drug used in the treatment of Parkinson’s disease (PD), was used as a drug control. The human SH-SY5Y neuroblastoma cells were differentiated for six days and treated with TRF or levodopa for 24 h prior to quantitative proteomic analysis. A total of 81 and 57 proteins were differentially expressed in diff-neural cells following treatment with TRF or levodopa, respectively. Among these proteins, 32 similar proteins were detected in both TRF and levodopa-treated neural cells, with 30 of these proteins showing similar expression pattern. The pathway enrichment analysis revealed that most of the proteins regulated by TRF and levodopa are key players in the ubiquitin-proteasome, calcium signalling, protein processing in the endoplasmic reticulum, mitochondrial pathway and axonal transport system. In conclusion, TRF is an essential functional food that affects differential protein expression in human neuronal cells at the cellular and molecular levels.
1. Introduction
TRF, a palm oil extract, consists of 25% α-tocopherol (α-Toc) and 75% tocotrienols (T3). The tocotrienols that exist in TRF are a mixture of four naturally occurring isoforms i.e., alpha-T3 (α-T3), beta-T3 (β-T3), delta-T3 (δ-T3) and gamma-T3 (γ-T3). The T3 isoforms contain a chromanol ring with a hydroxyl (-OH) group that can donate a hydrogen (H) atom to counteract the deleterious effects of free radicals and a hydrophobic side-chain that can readily permeate cell membranes. In addition, the presence of three double bonds at positions 3′, 7′ and 11′ in the side-chain of the T3 may contribute to the superior antioxidant potency compared to the Toc isoforms [1]. Vitamin E accumulates in lipid-rich regions of cells such as organelles, cell membranes, mitochondrial membranes and lipoproteins. Studies have found α-Toc to be a potent antioxidant molecule that can break the propagation of the peroxyl chain in redox cycle reaction [2,3]. T3 and α-Toc in TRF may amplify the beneficial changes at cellular and molecular levels and result in disease prevention in humans.
Several studies have investigated the neuroprotective effects of TRF using toxin-induced damage in cell-based and animal models. TRF attenuated glutamate-induced injury in astrocytes by reducing mitochondrial damage and increasing the number of viable cells [4]. In a mouse model of Alzheimer disease (AD), TRF supplementation modulated the hippocampal gene expression affecting the biological process and pathways that delay the pathological changes [5]. In addition, TRF also regulated the metabolic pathways that improved exploratory activity, spatial learning and memory in mice [6]. In rats, TRF supplementation caused significant changes in L-arginine metabolism and age-associated biochemical changes in the entorhinal cortex and cerebellum, indicating the enhancement of memory and motor functions [7]. A recent clinical study also found that supplementation with TRF for six months or more significantly increased the activity of antioxidant enzymes, such as superoxide dismutase (SOD) and glutathione peroxidase, and the ratio of reduced glutathione (GSH)-to-oxidized glutathione (GSSG) in healthy individuals between 50–55 years old [8]. In another clinical study, stroke patients supplemented with TRF showed a remarkable reduction in the mean white matter lesion (WML) size at the end of two years, compared to placebo groups [9]. Although these studies have delineated the protective effects of TRF using various biochemical and clinical approaches, studies on the proteomic changes implicated by TRF at the molecular and cellular level in PD remain scarce.
A recent quantitative proteomic study reported that TRF supplemented AβPP/PS1 mice altered the expression of proteins, which improved cognitive functions and modulated amyloid pathology in the hippocampus, medial prefrontal cortex and striatum of these mice [10]. Given the lack of cell-based proteomics data on the effects of TRF in PD, we embarked on a comparative proteomic analysis to discover differential proteins altered by TRF in comparison with levodopa. We also investigated the common and exclusive pathways modulated by TRF and levodopa in diff-neural cells. Furthermore, we performed the comparative functional bioinformatics analyses on differentially expressed proteins between TRF and levodopa treatments on diff-neural cells. We evaluated the proteins targeted by TRF and/or levodopa that play vital roles in various cellular processes and signalling pathways that protect the neural cells. As levodopa is often prescribed to PD patients as a dopamine replacement agent, we have intricately scrutinized and compared the alteration in protein expression implicated by TRF and levodopa on PD pathway in neuronal cells using the online bioinformatic Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway database resource [11].
2. Materials and Methods
2.1. Reagents
The SH-SY5Y neuroblastoma cells (Cat# CRL-2266) were purchased from the American Tissue Culture Collection (ATCC), and cultured in Dulbecco Modified Eagle Medium (DMEM) (Biosera, Nuaille, France) supplemented with 4.5 g/L glucose (Biosera, Nuaille, France) and L-glutamine without sodium pyruvate (Biosera, Nuaille, France) and 10% fetal bovine serum (FBS) (Biosera, Nuaille, France). Penicillin-Streptomycin solution (P/S) and non-essential amino acid (NEAA) were obtained from Gibco, Carlsbad, CA, USA. Retinoic acid (RA), dimethylsulphoxide (DMSO), 3,4-Dihydroxy-L-phenylalanine (levodopa) were purchased from Sigma Aldrich, St. Louis, MO, USA. The tocotrienol-rich fraction (TRF) was a kind gift from Excelvite Sdn. Bhd, Ipoh, Malaysia. The EasyPrep Mini MS Sample Prep kit and Pierce BCA Protein Kit were obtained from Thermo Fisher Scientific, USA. For the PCR validation assay, the commercial primers i.e., All-in-One qPCR Primers for the human CALM1 (NM_006888.5), TUBB (NM_178014.3), ATP5F1A (NM_004046.5), UCHL1 (NM_0044181.4) and GAPDH (NM_002046.6) genes were purchased from Genecopoeia, MD, USA. The HiScript II One Step qRT-PCR SYBR Green Kit was purchased from Vazyme biotech Co., Ltd., Nanjing, China.
2.2. Cell Culture and Differentiation of SH-SY5Y Human Neuroblastoma Cells
The SH-SY5Y human neuroblastoma cells were cultured in culture medium consisting of DMEM, 10% heat-inactivated FBS (HI-FBS), 1% P/S and 1% NEAA at 37 °C in a humidified 5% CO2 incubator. Culture media was replaced every alternate day, and cells were sub-cultured when it reached 70% confluence. To differentiate these human neuroblastoma cells, 1 × 106 SH-SY5Y cells were seeded in complete culture medium in a T75 flask. After 24 h, the culture medium was replaced with fresh medium that consist of DMEM supplemented with 3% HI-FBS, 1% P/S, 1% NEAA and 10 μM retinoic acid. The cells were allowed to differentiate for six-days with the medium replaced once on day 4. After six days, the terminally differentiated SH-SY5Y neural cells (diff-neural cells) expresses the dopaminergic features at morphological, biochemical and genetic levels [12].
2.3. Treatment Protocol
On day 7, the culture medium was removed and replaced with serum-free medium containing TRF (0.1 μg/mL) or levodopa (0.1 μg/mL) and the diff-neural cells were incubated at 37 °C for 24 h in a humidified 5% CO2 incubator. The concentration (0.1 μg/mL) of TRF and levodopa were determined from preliminary cytotoxicity studies using a range of concentrations (0 to 20 μg/mL). After 24 h of exposure to either TRF or levodopa, the diff-neural cells were harvested and subjected to protein extraction. The untreated cells were simultaneously maintained in a serum-free culture medium and served as controls.
2.4. Protein Extraction, Reduction and Alkylation
Protein extraction and processing to yield MS-ready peptides was achieved using the EasyPep Mini MS Sample Prep kit. The treated (levodopa or TRF) and untreated diff-neural cells were centrifuged for 7 min at 120 g, and supernatants were discarded. Then, 100 μL of lysis buffer (provided with the kit) and 1 μL of Universal Nuclease (provided with the kit) were added to the pelleted cells and mixed vigorously until sample viscosity was reduced. The protein concentration was determined using Pierce BCA Protein Assay Kit. Then, lysis solution (provided with the kit) was used to prepare the final protein concentration of 100 μg for the protein reduction step.
A volume of 50 μL of Reduction Solution (provided with the kit) was added to the sample and gently mixed. This was followed by 50 μL of Alkylation solution (provided with the kit), again with gentle mixing. Following this, the samples were incubated at 95 °C using a heat block for 10 min to reduce and alkylate the samples. At the end of this incubation, the samples were cooled to room temperature.
2.5. Protein Digestion and Peptide Clean-up
In the protein digestion step, 50 μL of Trypsin/Lys-C Protease enzyme (provided with the kit) was added to the reduced and alkylated protein samples and incubated at 37 °C for 3 h with intermittent shaking. Then, 50 μL of digestion stop solution (provided with the kit) was added to the sample and mixed gently. The digested protein samples were transferred to individual Peptide Clean-up columns (provided with the kit) and centrifuged at 1500× g for 2 min. Then, 300 μL of Wash Solution A (provided with the kit) and Wash Solution B (provided with the kit) were sequentially added to the column and centrifuged (1500× g for 2 min). Next, the Peptide clean-up columns were transferred into clean 2 mL microcentrifuge tubes, and 300 μL of Elution buffer (provided with the kit) was added to these columns, and the tubes were centrifuged. Finally, the peptide samples were dried using a vacuum centrifuge for 4-h and stored at −80 °C prior to the LCMS/MS analysis. Prior to the LCMS/MS analysis, the dried samples were resuspended in 100 μL of 0.1% formic acid in water.
2.6. Liquid Chromatography Electrospray-Ionization Coupled with Tandem Mass Spectrometry Analysis
Proteomic analysis was conducted using LC-MS/MS according to the methods described previously [13]. Upon injection of 1 μL of 100 μg of extracted peptides suspended in 0.1% formic acid into the LC-MS/MS analyser, the samples underwent chromatographic separation in Agilent Large Capacity Chip, 300 Å, C18 (Agilent, Santa Clara, CA, USA) through a 75 µm × 150 mm analytical column. The samples were separated using a gradient mobile phase system with a total running time of 47 min and a constant flow rate of 4 μL/min from Agilent 1200 Series Capillary pump and 0.5 μL/min from Agilent 1200 Series Nano pump. The autosampler temperature was set at 4 °C. The quadrupole-time of flight (Q-TOF) polarity was set at positive with capillary and fragmenter voltage at 1900 V and 360 V, respectively, and 5 L/min of gas flow with a temperature of 325 °C. The collision energy was determined at 3.7 V (100 Da), and reference masses with positive polarity was set at 299.294457 and 1221.990637. The peptide spectrum was analysed in auto MS mode ranging from 110–3000 m/z for MS scan and 50–3000 m/z for MS/MS scan.
2.7. Data Computation and Protein Identification
The mass spectra data were analysed with Agilent MassHunter (Agilent, Santa Clara, CA, USA) data acquisition software and PEAKS X 7.0 software [14] was used to identify proteins against Homo sapiens sequences obtained from Uniprot, Swissprot and TrEMBL databases. Trypsin was selected as the digestive enzyme, while the maximum missed cleavage by the enzymatic digestion was set at three sites for the protein sequence with carbamidomethylation set as a fixed modification. The protein identification was filtered using a target-decoy approach with a global false discovery rate (FDR) of 1% determined by the in-built FDR tool within PEAKSX+ software. The unique peptide ≥1 was used to filter out inaccurate proteins. The filtration parameter of protein abundance score of −10lgP ≥ 20 and peptide −10lgP ≥15: the identified proteins are relatively high in confidence as it targets very few decoy-matches above that threshold.
2.8. Differential Protein Expression Analysis
The raw mass spectra data obtained from MS/MS analysis were visualized using PEAKS X+ and the differentially expressed protein between two treatment groups were evaluated and corroborated with Uniprot search engine by species selection, i.e., Homo sapiens. The protein data were filtered for three valid values from three biological replicates, and proteins that were only presented in one or two biological replicates were eliminated. The biological replicates from all samples were clustered under the same matrix, and the outliers were removed, and the missing data was assigned with a random number derived from a normal distribution. The TRF-treated protein sets were compared to the protein sets of untreated controls using a two-tailed, Student t-test to identify significant proteins altered by TRF in diff-neural cells. The proteins were regarded as significantly regulated if the difference between the two groups showed a p-value of less than 0.05 (p < 0.05). Similar computation was performed on levodopa-treated cells against untreated control cells. The significantly upregulated and down-regulated proteins in response to TRF or levodopa treatment with average mass, p-value and log2 fold expression are listed in Table 1 (TRF) or Table 2 (Levodopa).

Table 1.
Differentially expressed total protein extract from TRF-treated diff-neural cells compared to untreated control.

Table 2.
Differentially expressed total protein extract from levodopa-treated diff-neural cells compared to untreated control.
2.9. Determination of Common and Unique Proteins between TRF and Levodopa Treatment
A Venn diagram was generated to identify unique and overlapping proteins between the differentially expressed proteins (p < 0.05) in TRF-treated vs. untreated control or levodopa-treated vs. untreated control using an online software (http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 5 April 2021)). The unique proteins regulated by TRF or levodopa represent distinct pathways regulated by these compounds in diff-neural cells. The overlapping proteins indicate the mutual biological pathways mediated by TRF and levodopa in neuronal cells.
2.10. Gene Ontology (GO) Term and Pathway Enrichment Analysis
The functional GO annotations of the significantly expressed proteins were analysed using the PANTHER version 16 (http://pantherdb.org (accessed on 24 September 2021)), and the proteins were classified based on their molecular function, biological process and cellular components. The protein classification was analysed by uploading the differentially expressed proteins regulated by TRF or levodopa in the PANTHER GO term interface. Only the top 10 GO terms that showed the highest fold enrichments for each classification was analysed. All results shown expressed adjusted p-value < 0.05 as determined by the Fischer’s Exact test and False Discovery Rate. We also identified the functional protein class of the 32 overlapping proteins between TRF and levodopa using the PANTHER database. The Pathway enrichment analysis was performed using the KEGG (Kyoto Encyclopaedia of Genes and Genomes) https://www.genome.jp/kegg/pathway.html (accessed on 20 October 2021) database for the differentially expressed proteins regulated by TRF or levodopa. We highlighted the top 5 enriched pathways implicated by TRF or levodopa treatment on diff-neural cells. The differentially expressed proteins regulated by TRF or levodopa enriching the KEGG-Parkinson’s disease pathway. Besides applying the bioinformatics platform to understand the biological roles of the expressed proteins, cross-referencing with published reports was also conducted to validate these proteins’ functions.
2.11. Validation of mRNA by One Step Quantitative PCR (qPCR) Assay
Four candidate genes were chosen from each PD pathway as curated by KEGG to validate the proteomic data using qPCR technology. The four genes are responsible for coding proteins that play a pivotal role in the PD pathway, i.e., CALM1 protein in the calcium signalling pathway, ATP5F1A in the mitochondrial pathway, TUBB in the axonal transport system and UCHL1 in the ubiquitin pathway. Purified RNA was processed using the HiScript II One Step qPCR SYBR Green Kit according to the manufacturer’s instruction. The total reaction volume was 20 μL consisting of 100 pg of RNA, 10 μL of 2x One Step SYBR Green Mix, 1 μL of One Step SYBR Green Enzyme Mix, 0.4 μL of 50x ROX Reference Dye and 2 μL of forward and reverse primers (10 μM) of CALM1, TUBB, ATP5F1A, UCHL1 or GAPDH genes. The assay was conducted in One Step qRT-PCR protocol commencing with reverse transcription at 50 °C for 3 min, followed by pre-denaturation at 95 °C for 30 s. After this step, the samples went through 40 cycles involving denaturation at 95 °C for 10 s, followed by 30 s at 60 °C of extension. A dissociation curve analysis was performed at the end of the cycle to confirm amplification specificity. All samples were run in triplicates. The signal was detected using ABI StepOne Plus (ThermoFisher Scientific, Waltham, MA, USA). The CALM1, TUBB, ATP5F1A and UCHL1 messenger RNA (mRNA) levels were normalised to the mRNA of the reference gene GAPDH. The non-template control (sample without RNA template) was run parallel in triplicate for each primer to assess for non-specific amplification.
2.12. Statistical Analysis
Statistical analysis comparing the quantitative proteomic data from TRF-treated vs. untreated control cells and levodopa-treated vs. untreated controls in diff-neural cells were performed using a two-tailed Student’s t-test. All statistical analyses were performed with SPSS v20. Differentially expressed proteins that displayed the difference in Log2 Fold change with p < 0.05 were regarded as statistically significant. For the qPCR validation assay, the treatment groups (TRF, levodopa) were compared with the untreated group using the Bonferroni multiple-comparison post-hoc test. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. The Effects of TRF and Levodopa Treatments on Total Protein Extract from Diff-Neural Cells
We conducted a shotgun proteomic investigation using label-free tandem liquid mass spectrometry (LC-MS/MS) on TRF or levodopa’s treated diff-neural cells. The pre-processing of raw tandem mass spectrometry data was conducted using PEAKS X+ to explore the alterations in diff-neural cells’ proteome profile in response to TRF or levodopa treatment. A total of 528 statistically significant proteins were identified from a total of 3828 proteins in three biological replicates of untreated control cells. As from TRF treatment on diff-neural cells, a sum of 4019 proteins were detected in all three biological replicates that constituted 707 common protein data sets. A total of 163 proteins from TRF treated diff-neural cells matched with protein sets from untreated control cells. Among these 163 proteins, 81 proteins showed a significant differential regulation, with 53 upregulated and 28 down-regulated (Table 1).
Meanwhile, the levodopa treated diff-neural cells yielded a total of 3603 proteins, with 682 proteins distinct all three biological replicates. When we compared the common protein sets between levodopa treated and untreated control cells, we found a total of 166 proteins that matched both the data sets with 57 proteins expressing a significant difference in expression with p < 0.05. Among the 57 significantly expressed proteins, 32 proteins demonstrated a significant upregulation, while 25 were down-regulated (Table 2).
3.2. Venn Diagram Analysis
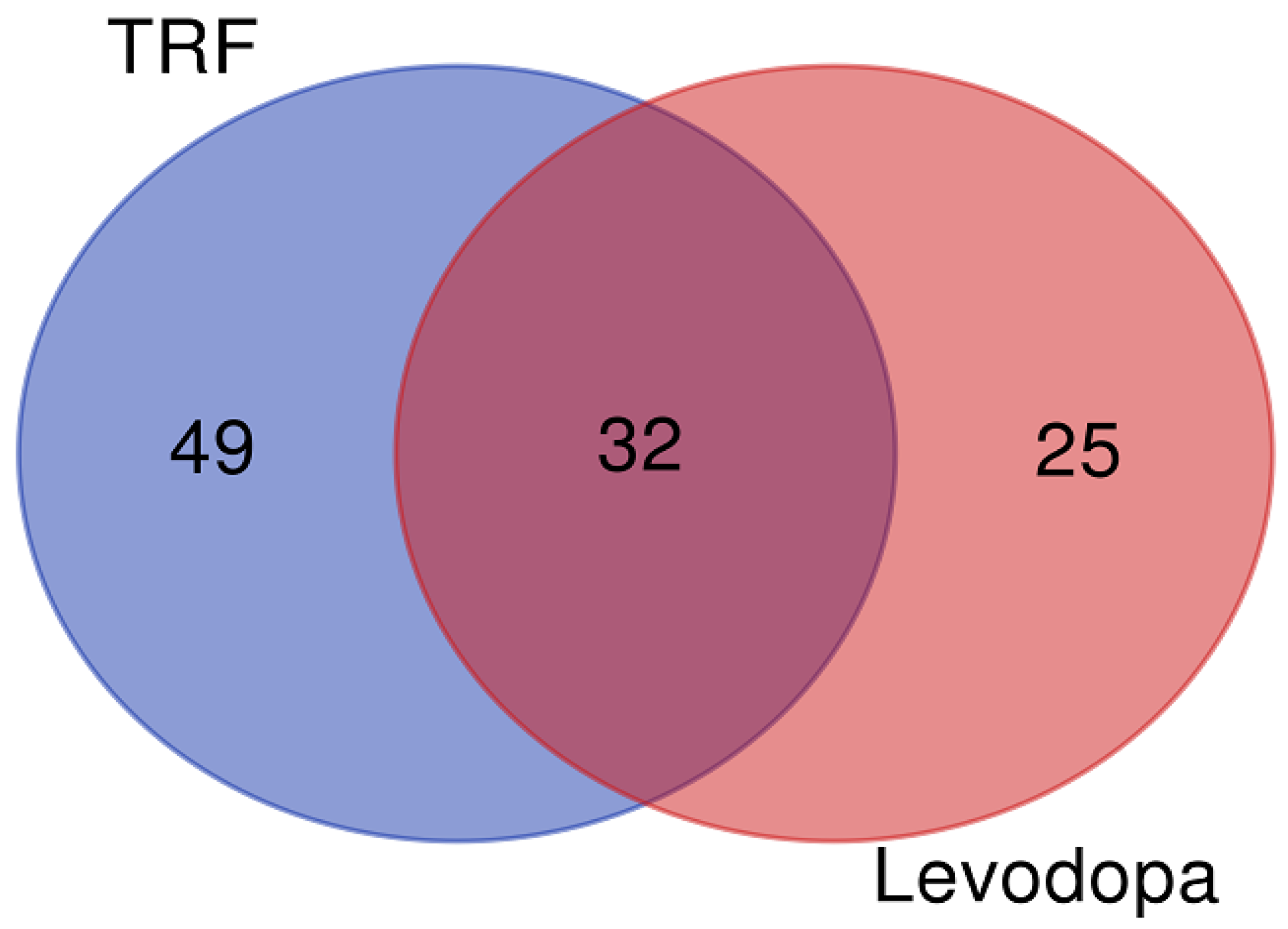
The differentially expressed proteins regulated by TRF and levodopa treatments were further stratified using a Venn diagram to identify the unique and overlapping proteins. The overlapping regions illustrate the proteins’ involvement in common biological and molecular processes in TRF and levodopa treated cells. The Venn diagram revealed 49 and 25 unique proteins regulated by TRF and levodopa, respectively. Importantly, a total of 32 differentially expressed proteins were common to both TRF and levodopa treatments (Figure 1). The list of unique and overlapping proteins expressed in response to TRF or levodopa treatment in diff-neural cells are shown in Table 3.
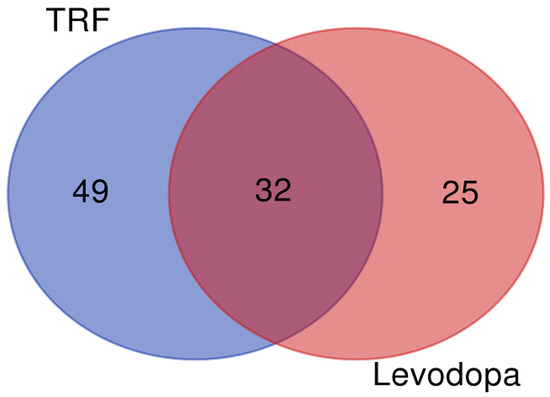
Figure 1.
The Venn diagram. The union of intersection of differentially expressed proteins (p < 0.05) in diff-neural cell in response to TRF and levodopa treatments.

Table 3.
List of unique and common proteins between the levodopa and TRF treatment on diff-neural cell.
3.3. Functional Gene Ontology (GO) Enrichment Analysis
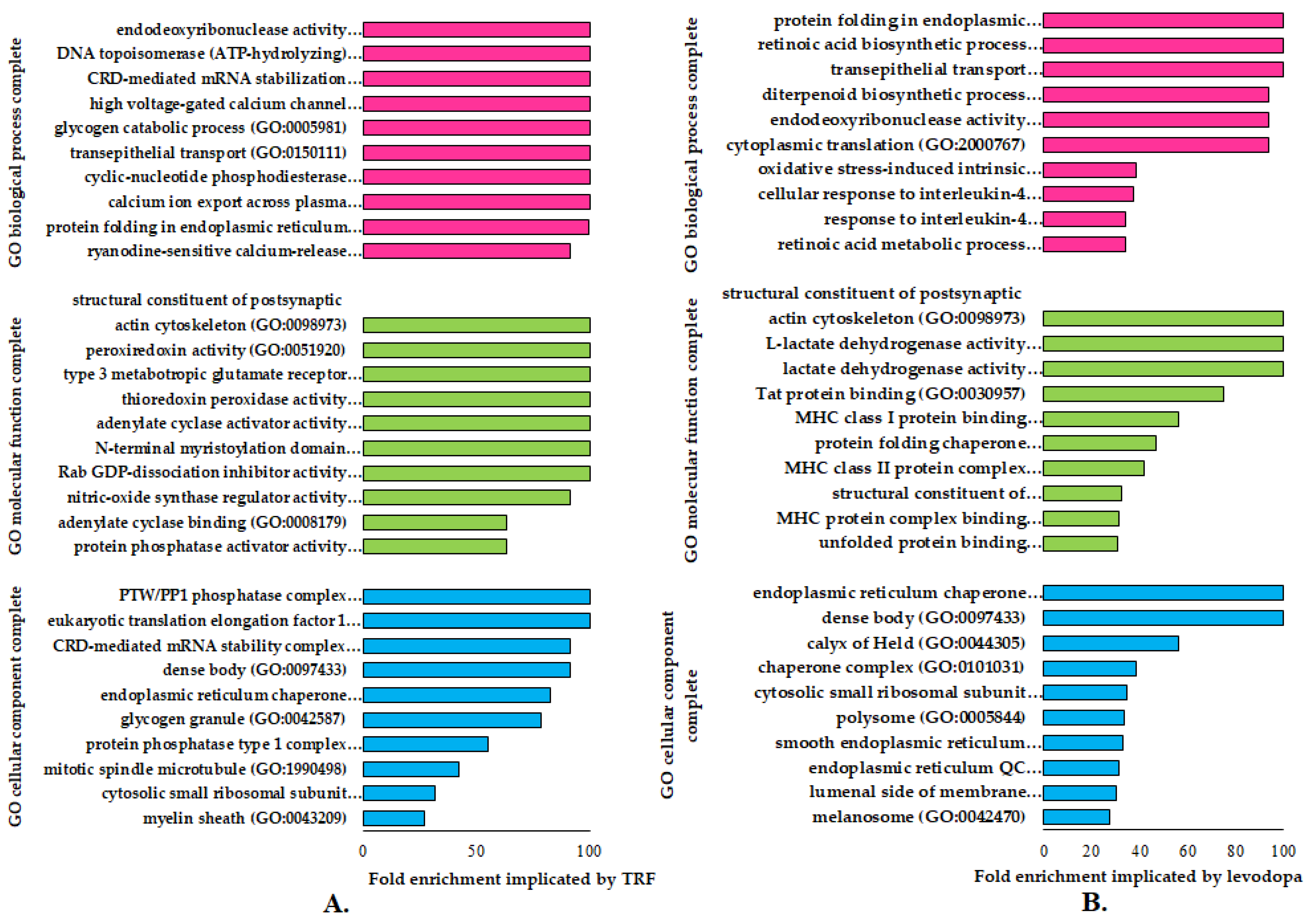
The GO enrichment analysis identifies specific genes or proteins’ biological and functional roles. To further understand the alterations in cellular and molecular processes regulated by TRF or levodopa treatments in diff-neural cells, the differentially expressed proteins (p < 0.05) were mapped to their enriched functional annotation GO terms from the PANTHER bioinformatics database. The functional annotations of differentially expressed proteins modulated by TRF or levodopa compared against untreated controls were classified into three classes-molecular function, biological processes and cellular components, as shown in Figure 2A,B.
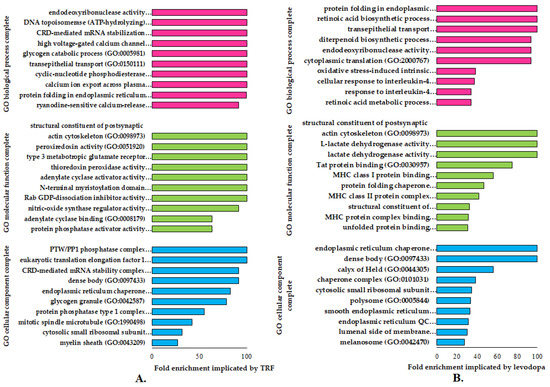
Figure 2.
The functional gene ontology (GO) terms of molecular function, biological process and cellular component using PANTHER (version 16) database on differentially regulated proteins (p < 0.05) in (A) TRF and (B) levodopa treatments on diff-neural cells. The horizontal coordinate (x-axis) represents the logarithmic transformation at the base of 10 of the raw p-value, and the vertical (Y-axis) displays the GO term classifications.
A total of nine GO terms biological processes were significantly overexpressed (>100) in TRF implicated diff-neural cells. Examples of these GO term biological processes are endodeoxyribonuclease activity, DNA topoisomerase (ATP hydrolysing) activity and CRD-mediated mRNA stabilization. As for the molecular function, TRF implicated differentially expressed proteins modulated the GO term classes of structural constituents of the postsynaptic actin cytoskeleton, peroxiredoxin activity and type 3 metabotropic glutamate receptor biding. These molecular and biological processes were found to be highly prevalent at cellular component regions, particularly PTW/PP1 phosphatase complex, eukaryotic translation elongation factor 1 complex and CRD-mediated mRNA stability complex, as displayed in Figure 2A.
A total of three GO terms biological processes exhibited a significant fold change overexpression (>100) in levodopa implicated diff-neural cells. Examples of these GO term biological processes are protein folding in endoplasmic reticulum, retinoic acid biosynthetic process and transepithelial process. As for the molecular function, levodopa implicated differentially expressed proteins modulated the GO term classes of structural constituents of the postsynaptic actin cytoskeleton, L-lactate dehydrogenase activity and lactate dehydrogenase activity. These molecular and biological processes were found to be highly prevalent at cellular component regions, particularly endoplasmic reticulum chaperone complex and unfolded protein binding, as displayed in Figure 2B.
3.4. Common Protein Expression between TRF and Levodopa Treatment in Diff-Neural Cells
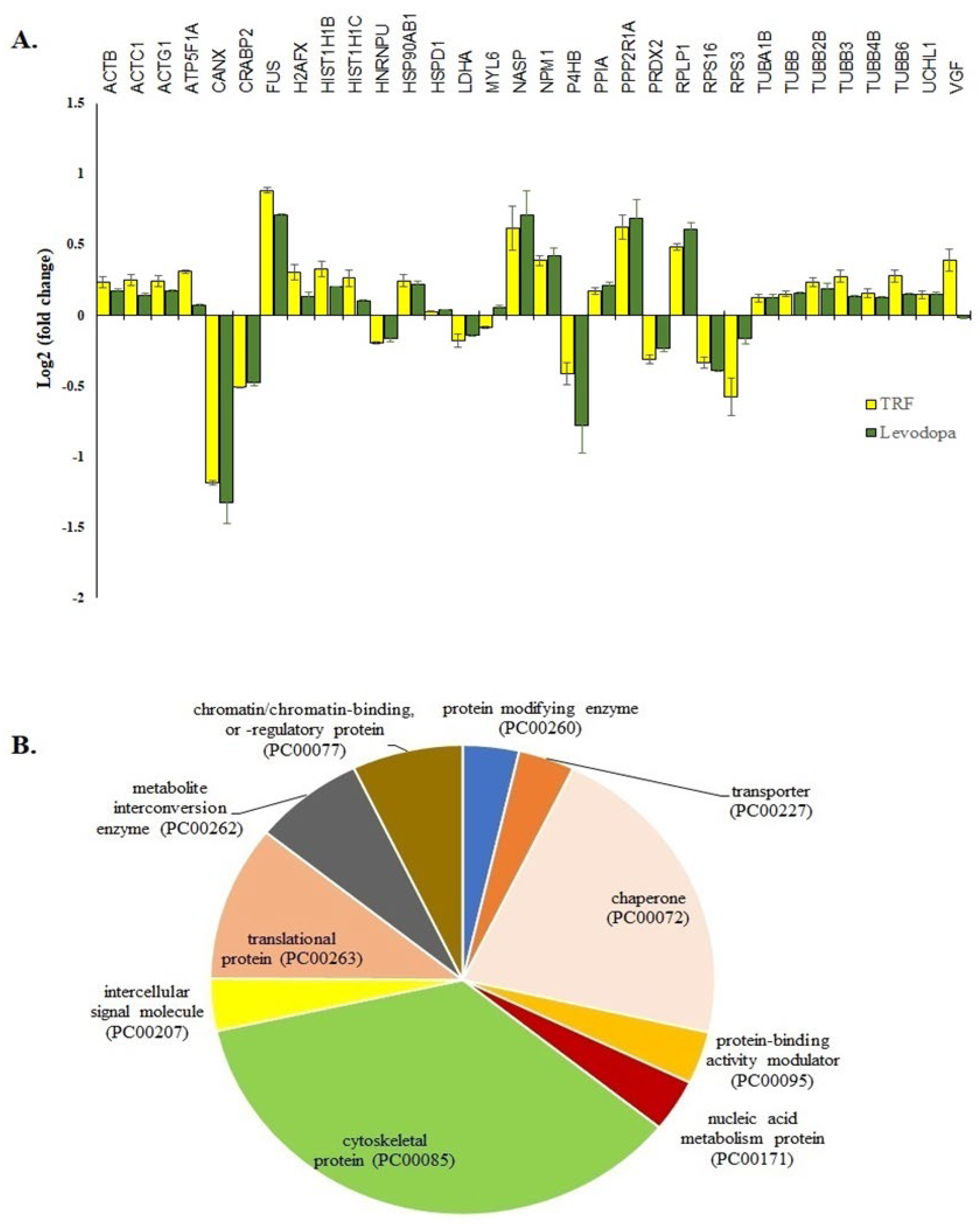
The treatment of TRF or levodopa on diff-neural cells yielded 32 significant common proteins (p < 0.05). The fold expression patterns of these 32 common proteins between TRF vs. untreated and levodopa vs. untreated diff-neural cells are illustrated in Figure 3A. Among the 32 proteins, 30 proteins regulated by TRF displayed similar expression pattern with the levodopa in diff-neural cells. In contrary, two out of the 32 proteins displayed an opposite expression pattern between levodopa and TRF: MYL6 and VGF. The similar protein expression patterns regulated by TRF and levodopa treatments indicate the enrichment of common neuroprotective pathways in diff-neural cells.
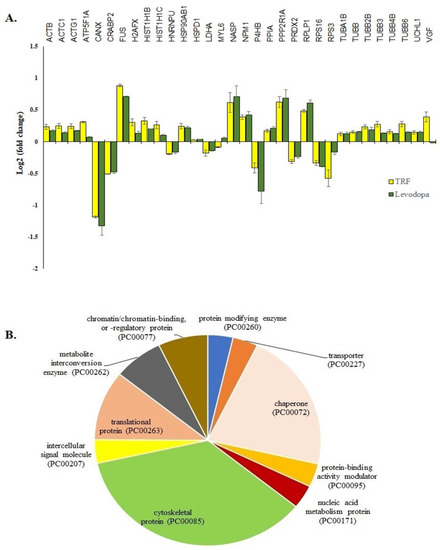
Figure 3.
The 32 common proteins expressed in TRF and levodopa treated diff-neural cells. (A) Histogram shows the different fold change expressions of the 32 common proteins (p < 0.05) regulated by treatment of levodopa or TRF on diff-neural cells. The horizontal coordinate (x-axis) represents the types of protein, and the vertical coordinate (Y-axis) denotes the difference in fold change (logarithmic transformation at the base of 2). (B) Pie chart shows the classification of 32 proteins into 10 functional categories using PANTHER online database.
Using the PANTHER online database, 28 out of the 32 proteins grouped into 10 functional categories; cytoskeletal protein (TUBB, MYL6, TUBB2B, ACTB, TUBB4B, ACTG1, ACTC1, TUBB6, TUBA1B, TUBB3), chaperone (PPIA, P4HB, NPM1, NASP, CANX, HSP90AB1), chromatin/ chromatin-binding or regulatory protein (HIST1H1B, H2AFX), intercellular signal molecule (VGF), metabolite interconversion enzyme (PRDX2, LDHA), nucleic acid metabolism protein (FUS), protein modifying enzyme (UCHL1), protein-binding activity modulator (PPP2R1A), translational protein (RPS3, RPS16, RPLP1) and transporter (ATP5F1A) as shown in Figure 3B. The four proteins that did not belong to any protein classes are CRABP2, HNRNPU, H2AFX, HIST1H1C).
3.5. Pathway Enrichment Analysis of the Differentially Expressed Proteins
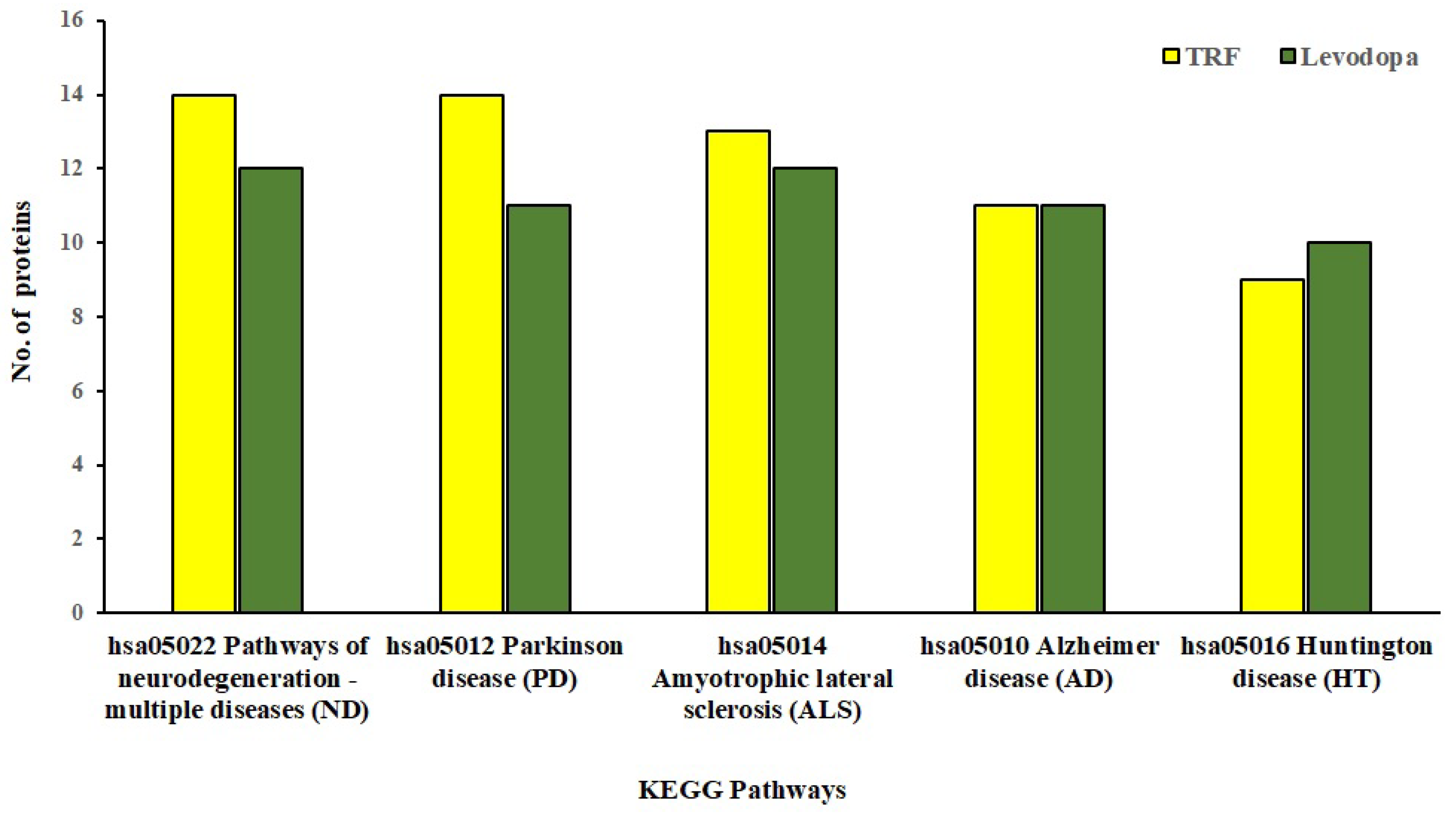
The KEGG database provides mechanistic insights into biological pathways enriched by differentially expressed protein set following treatment conditions. The pathway construction is based on the molecular interaction network of how genes/proteins interact and regulate. When 81 and 57 differentially expressed proteins implicated by TRF and levodopa, respectively, were uploaded in the KEGG mapper, we identified the top five enriched neurodegenerative disease pathways as shown in Figure 4. These five significantly enriched pathways modulated by TRF or levodopa in diff-neural cells are pathways associated with neurodegenerative disease-multiple disease (hsa05022), PD (hsa05012), Amyotrophic lateral sclerosis (hsa05014), AD (hsa05010) and Huntington’s disease (hsa05016). The identity of the differentially expressed proteins enriching the respective pathways modulated by TRF or levodopa are listed in Table 4.
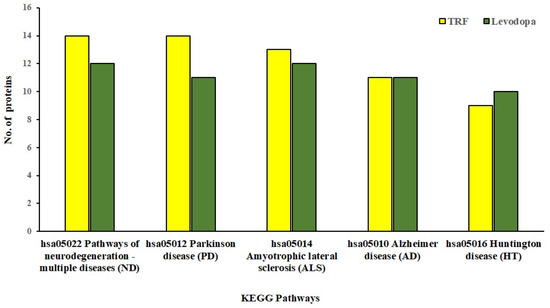
Figure 4.
KEGG Pathway enrichment analysis of significantly regulated proteins (p < 0.05) by TRF and levodopa on diff-neural cells. Bar chart shows the TRF and levodopa regulated top 5 enriched neurodegenerative pathways.

Table 4.
List of TRF and levodopa regulated proteins in the top 5 enriched neurodegenerative pathways.
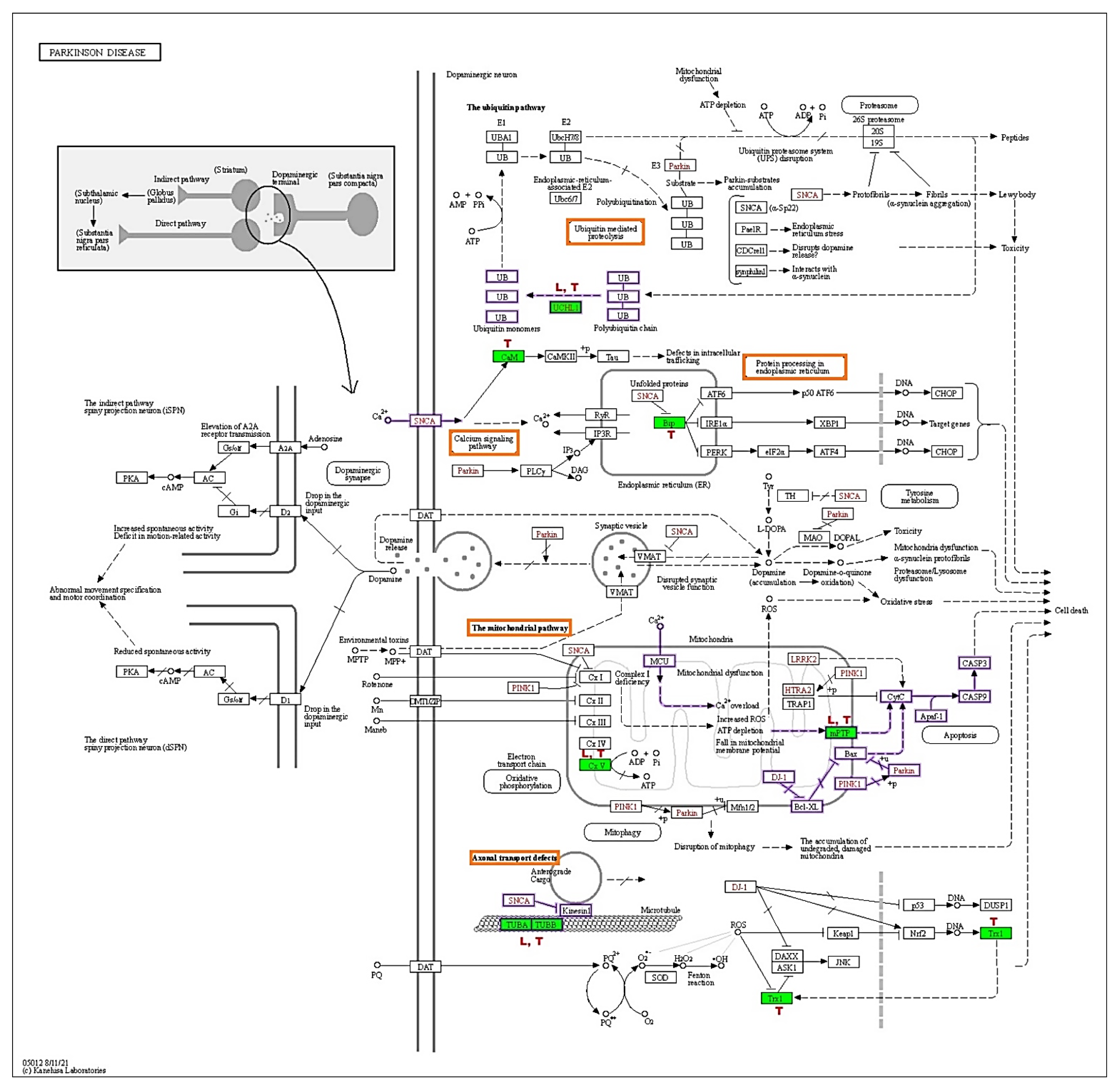
3.6. TRF and Levodopa Implicated Differential Protein Expression in PD Pathways
The KEGG-PD pathway depicts the consolidation of multiple pathways that lead to dopaminergic neuronal demise. The four vital pathways that cause neurodegeneration are ubiquitin, calcium, mitochondrial, and axonal transport defects (red box). Diff-neural cells treated with TRF and levodopa resulted in upregulation of UCHL1 protein in the ubiquitin pathway. Moreover, TRF upregulated the CALM1, CALM2 and CALM3 [CaM] proteins in the calcium signalling pathway and downregulated the HSPA5 [Bip] in protein processing the endoplasmic reticulum. TRF and levodopa upregulated the ATP5F1A [CxV] in the mitochondrial pathway. While SLC25A6 [mPTP] was upregulated by TRF and VDAC1 and VDAC3 [mPTP] were significantly suppressed by levodopa. Notably, both TRF and levodopa upregulated the TUBA1A, TUBA1B, TUBB, TUBB2B, TUBB3, TUBB4B, TUBB6 [TUBA, TUBB] that mediate the Axonal transport system. The effects of TRF (T) and levodopa (L) in regulating differential protein expression in KEGG-PD pathways are illustrated in Figure 5.
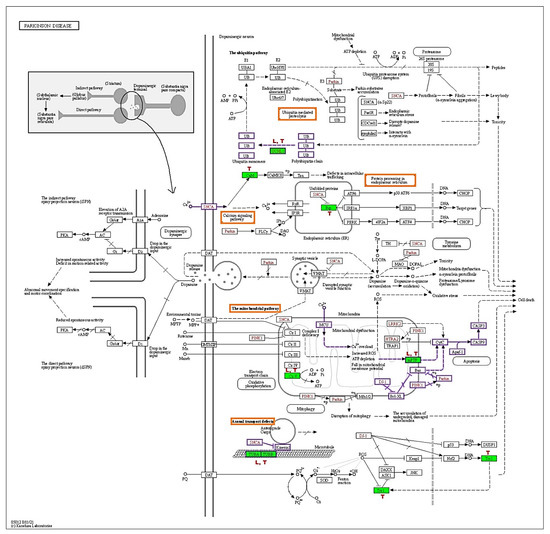
Figure 5.
KEGG Parkinson disease pathway (hsa05012). The red boxes indicate the pathways involved by TRF and levodopa, green shaded boxes are proteins that were differentially regulated by (T) TRF and (L) levodopa in differ-neural cells. The proteins behind the KEGG symbol are as follows: UCHL1:UCHL1, CaM: CALM1, CALM2, CALM3, Bip: HSPA5, mPTP: SLC25A6, VDAC1, VDAC3, CxV: ATP5F1A, TUBA/TUBB: TUBA1B, TUBB, TUBB2B, TUBB3, TUBB4B, TUBB6 (Adapted from KEGG-Parkinson’s disease pathway- hsa05012 with permission from Ref. [11]. 2019. Kanehisa, M.).
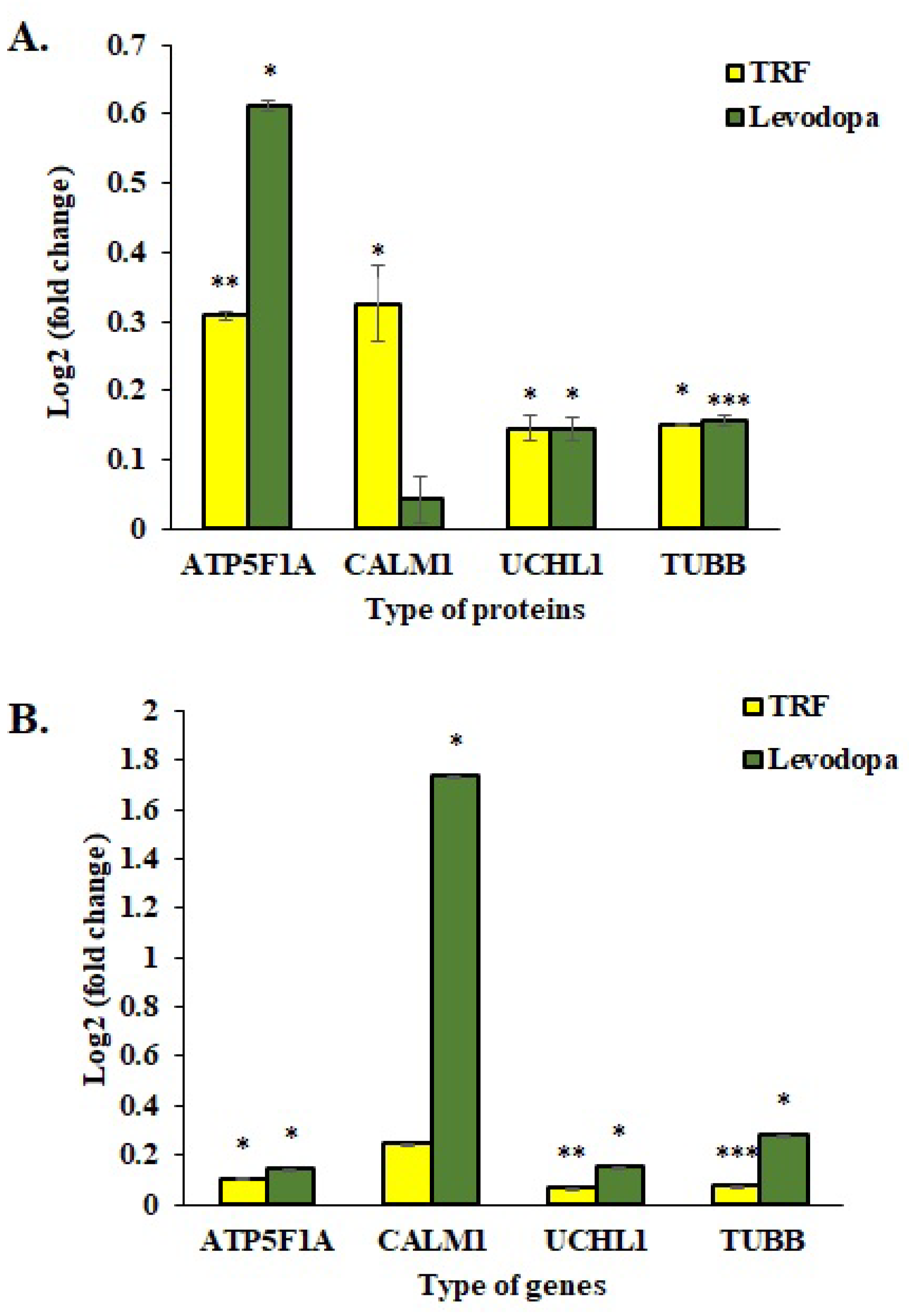
3.7. Validation of Protein Expression Using qPCR Assay
The mRNA expressions of four selected genes were investigated using the One Step qPCR assay to validate the proteomic data obtained using the LC-MS/MS assay. As shown in Figure 6, the qPCR data correlated with proteomic findings with all four genes displayed similar expression patterns as respective protein expression. The ATP5F1A, UCHL1 and TUBB proteins were significantly upregulated in TRF and levodopa treated diff-neural cells compared to untreated control cells. Although CALM1 protein expression was upregulated in both TRF and levodopa treated neural cells, but the significant difference with at least p < 0.05 was only demonstrated by TRF (Figure 6A).
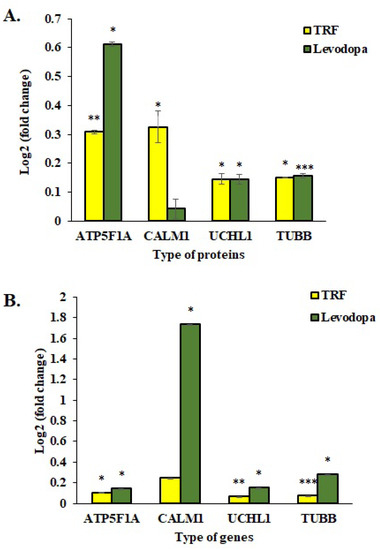
Figure 6.
Validation study of mRNA expressions of four selected genes, ATP5F1A, CALM1, UCHL1 and TUBB. (A) The log2 fold change protein expression following TRF and levodopa treatment on diff-neural cells. (B) The log2 fold change values of selected mRNA expression following TRF and levodopa treatment on diff-neural cells. For calculation of p value, the normalized data set from TRF and levodopa treated group were compared against normalized data obtained from untreated control group. The values are expressed as mean ± SEM (n = 3), Significant difference * p < 0.05, ** p < 0.01, *** p < 0.001 compared to untreated control group.
In the qPCR assay, TRF upregulated the mRNA expression of ATP5F1A, UCHL1 and TUBB with p < 0.05. However, the CALM1 upregulation by TRF was not statistically significant. Levodopa showed a remarkable overexpression of ATP5F1A, CALM1, UCHL1 and TUBB with p < 0.05 (Figure 6B).
4. Discussion
The present study was aimed to provide insights into TRF modulated proteome footprints in diff-neural cells using an in vitro model of human dopaminergic neural cells. We compared these findings with proteomes regulated by levodopa, a standard drug used in the treatment of PD. The diff-neural cells were established by culturing cells in retinoic acid and low-serum culture medium for 6 days. The established diff-neural cells exhibit neuronal characteristics such as suppression of proliferation rate, generation of long and branched neurites and secretion of neuronal marker proteins [15]. Our previous study has shown that diff-neural cells express characteristics of dopaminergic cells at morphology, biochemical and genetic levels. This study demonstrated that differentiated human neural cells are the most appropriate platform for understanding the effects of potential drugs targeting the various cellular and molecular signalling processes [12]. In the present quantitative proteomic study, we discovered that TRF and levodopa treatment on diff-neural cells significantly regulated 81 and 57 proteins, respectively. TRF treated diff-neural cells upregulated 53 proteins and suppressed 28 proteins (Table 1). While levodopa treated diff-neural cells upregulated 32 proteins and down-regulated 25 proteins (Table 2).
In addition, TRF and levodopa treated diff-neural cells significantly upregulated the chaperone proteins, including HSP90AB1, HSPD1 and PPIA (Figure 3A,B). The heat shock proteins (HSP) HSP90AB1(HS90B) and HSPD1(Hsp60) are evolutionarily well-conserved molecular chaperones that assist in protein folding and maintain protein stability. As the accumulation of misfolded proteins can cause deleterious effects to cells, the HSP90AB1 interacts with more than 20 co-chaperones that facilitate recognising unfolded client proteins and uses ATP energy to transform these proteins to their biologically active conformations via hydrolysis [16]. Whereas HSPD1, a nuclear-encoded mitochondrial chaperonin, is readily expressed in the event of oxidative stress, heat shock and DNA damage. In mitochondria, HSPD1 coupled with Hsp10 forms a folding apparatus for the correct folding of mitochondrial proteins [17]. Previous studies have documented that levodopa administration in rats significantly upregulated HSPD1in the substantia nigra region. The over-expression of HSPD1 was in line with dose-dependent increased activity of mitochondrial complex I as well as nitrosative stress in rats [18]. On the other hand, we also showed that TRF and levodopa stimulated the expression of peptidylprolyl isomerase A (PPIA) in diff-neural cells. The PPIA exists in the highest amount in the brain cells and functions as a molecular chaperone and plays a crucial role in the maturation, folding and assembly of neuron-specific membrane proteins. A report from Pasetto et al. [19] has further validated that the intracellular expression of PPIA is beneficial as this enzyme functions as foldase to correct proteins to achieve its normal conformation.
The dopaminergic neurodegeneration at substantia nigra pars compacta (SNpc) and dopamine depletion at striatum have been documented to be the consequences of the impairment of multiple neurophysiological pathways. Genetic mutations, oxidative stress, mitochondrial defects, failure in the clearance of toxic proteins, aggregation of misfolded proteins (Lewy bodies) are some of the reported mechanisms involved in the pathogenesis of PD [20]. In our investigation, we explored the role of TRF in modulating the crucial pathways that may impede the neurodegenerative process in diff-neural cells. On that note, we applied the online bioinformatics database—KEGG, to understand the effects of differential protein expression implicated by TRF on various pathways leading to PD. Here, we report that TRF treatment on diff-neural cells resulted in differential expression of proteins in the ubiquitin pathway, calcium signalling pathway, mitochondrial pathway and axonal transport system (Figure 5).
The ubiquitin pathway is the fundamental enzymatic cascade of reaction that functions in the clearance of impaired and misfolded proteins in nucleus and cytoplasm of eukaryotic cells. Mutation in any component enzymes or proteins of the ubiquitin pathway is linked to the blockade in protein homeostasis leading to proteinopathy and neurodegeneration [21]. The ubiquitin carboxyterminal hydrolases 1 (UCHL1), also known as PARK 5, has been suggested to be involved in monoubiquitin processing via the short C terminal peptide cleavage and ubiquitin recovery from proteasomal degradation protein remnants. These processes maintain the optimal ubiquitin pool to remove damaged proteins in the ubiquitin proteosome pathway (UPP), as depletion in ubiquitin pools has been noted in UCHL1 knockout neurons [22]. A recent study has proposed the different roles of UCHL1 in normal and diseased conditions. According to this study, the suppression of UCHL1 in primary neurons and hippocampal tissues from mice resulting in overexpression and accumulation of α-synuclein in these neurons. On the contrary, inhibition of UCHL1 activity in α-synuclein overexpressing neurons strikingly reduced the α-synuclein level and promoted its clearance at neuron synapses [23]. Our finding shows the UCHL1 protein expression was significantly upregulated following treatment with TRF and levodopa compared to untreated controls. We postulate that the increase in UCHL1 protein expression indicates TRF and levodopa’s neuroprotective actions of in boosting up the monomeric ubiquitin levels and maintaining the normal synaptic structure of the neurons. Our claim is in line with a previous study that showed the correlation between the increased expression of UCHL1 in neurons and restoration of the normal synaptic remodelling through upregulation of free monomeric ubiquitin levels [24]. Therefore, TRF is a safe and effective disease-modifying therapy to be considered in treating neurodegenerative diseases.
Calcium (Ca2+) via its signalling pathway in neurons, plays a central role in regulating a plethora of cellular activities, including synaptic plasticity and transmission, differentiation of neurons, gene expression and regulation of complex protein networks [25]. Neurons effectively maintain a steep Ca2+ gradient between intra- and extracellular compartments, notably low intracellular Ca2+ concentration compared to extracellular space. The alteration in intracellular Ca2+ concentration is triggered by the opening of Ca2+ channels such as voltage-dependent Ca2+ channel, N-Methyl-D-Aspartate (NMDA) receptor and transient receptor potential (TRP). At resting state, the Ca2+ level is restored by efflux of Ca2+ through plasma membrane channels (Ca2+-ATPase, sodium calcium exchanger), binding to Ca2+ sensor proteins (calmodulin, calcineurin, calretinin) and uptake by organelles (endoplasmic reticulum and mitochondria) [26]. Calmodulin (CALM) is an important calcium-sensing protein that modulates the presynaptic release of neurotransmitter thru activation of calmodulin-dependent kinase-IIα (CAMKIIα) and CAMKIIβ. In addition, CALM was also reported to control the function of pre-and postsynaptic Ca2+ channels [27], synaptic plasticity [28] and modify gene expression [29]. In the present study, TRF treatment of diff-neural cells caused significant upregulation of CALM proteins, namely CALM1, CALM2 and CALM3 proteins (Figure 5, CaM=CALM). The CALM knockdown studies using lentiviral-delivered short-hairpin RNAs have reported no significant changes in neuronal survivability and synapse formation. However, neurons with low levels of the CALM protein, exhibited a spontaneous depression of neuronal network activity [27]. Since CALM proteins are copiously found in the human brain and overexpression studies are relatively scarce, it is difficult to interpret the physiological overexpression of these proteins in our study. Hence, future studies should be directed to understanding the effects of CALM overexpression in modulating the calcium signalling pathway and activity of CAMKII proteins.
Another critical protein that TRF differentially regulated in the calcium signalling pathway is the endoplasmic reticulum chaperone Bip (HSPA5/GRP78/Bip). The HSPA5 protein was significantly down-regulated by TRF compared to untreated controls. No significant changes of this protein were noted in levodopa treated diff-neural cells (Figure 5, Bip=HSPA5). The HSPA5 is a critical protein in the unfolded protein response (UPR) mechanism, an adaptive response that localizes in the endoplasmic reticulum (ER). The UPR is mediated by three ER local proteins: IRE1, PERK and AT6 [30]. At relaxed conditions, HSPA5 binds with these three localized proteins and remain inactive. During ER stress, in which is triggered by disruption in Ca2+ homeostasis and redox state and cellular energy supply, HSPA5 dissociates with IRE1, PERK and ATF6. It stimulates the UPR signalling pathway to exert their roles in proteolytic processing of unfolded proteins to prevent aggregation of these proteins [31]. A substantial body of evidence has reported that UPR is activated in post mortem human brain samples of PD patients, indicating ER stress that leads to neurodegeneration [32]. Nevertheless, compelling data from a recent study suggested that HSPA5 mRNA level was significantly upregulated in caudate nucleus, cingulate gyrus, prefrontal and parietal cortex regions of post mortem tissue obtained from PD patients [33]. The suppression of HSPA5 protein has been identified as one of the therapeutic target models in PD. In this regard, Hu and their team have revealed that luteolin, a naturally occurring flavonoid compound, has immense potency in attenuating the 6-OHDA induced augmentation of HSPA5 and down-regulating the ubiquitin proteosome activity. The suppression of HSPA5 protein and UPS signal is one of the neuroprotective approaches in treating neurodegenerative diseases, i.e., PD and AD [34]. Therefore, we suggest that the down-regulation of HSPA5 in diff-neural cells treated with TRF may imply the presence of antioxidant effects that resulted in the absence of ER stress, which kept the HSPA5 at considerably lower concentration in neural cells compared to untreated control cells.
The mitochondria are the principal site for energy metabolism via the Krebs cycle, oxidation phosphorylation and fatty acid β-oxidation pathways. Dysfunction in any of these pathways could compromise the energy synthesis in the form of ATP and cause degeneration of neurons in PD [35]. The exposure of diff-neural cells to TRF yielded a significant upregulation of two significant proteins involved in energy synthesis, which are SLC25A6/ANT3 (ADP/ATP translocase 3) and ATP5F1A (ATP synthase F1 subunit α) (Figure 5, mPTP=SLC25A6, CxV=ATP5F1A). Intriguingly, our data also revealed that diff-neural cells exposed to L-DOPA, displayed a significant upregulation of these two proteins compared to untreated controls. The SLC25 proteins are the largest transporters with 26 members (SLC25A1-46) found abundantly in brain regions such as the hippocampus, thalamus and brainstem, are essential proteins for energy production in neurons [36]. Particularly, the SLC25A6, which is an inherent carrier protein located at the mitochondrial inner membrane and functions as mitochondrial carriers that allow the electrogenic exchange of two native substrates, namely ADP and ATP between mitochondria matrix and intermembrane gap [37]. A study on cancer cells revealed that the SLC25A6 antiport plays a critical role by increasing the metabolic demands for DNA and protein synthesis [38]. Previous studies have delineated that dysfunction in SLC carriers caused fatal neurological disorders. Mutation and decreased SLC30A10 protein levels were discovered in the brain autopsy of Alzheimer’s Disease and PD patients [39]. Besides, an allelic variant in the SLC6A3 gene has been associated with modulation in dopamine metabolism in PD [40]. However, no studies have reported the effect of SLC25A6 protein dysfunction in neuronal cells.
On the other hand, ATP5F1A is found in mitochondria’s cristae and inner membrane and functions in synthesis of ATP from the precursor molecules, ADP and Pi. According to Song et al. [41] the ATP5F1A overexpression may enhance genes related to immune response, angiogenesis and collagen catabolic processes. This study also demonstrated that ATP5F1A might interfere with alternative splicing of genes associated with glucose homeostasis. Notably, a recent study has documented that a lower level of ATP5F1A was detected in circulating small vascular vesicles from PD patients [42]. Along this line, using whole exome sequencing technology, Jonckheere et al. [43] have discovered a heterogenous mutation in ATP5A1 gene in two siblings with severe neonatal encephalopathy. Of note, gene expression profiling of substantia nigra autopsy from PD patients has shown that the ATP5F1A gene was significantly suppressed compared to the control group.
Tubulin is a heterodimer protein that contains a pair of polypeptide chains known as a monomer. It appears as a hollow fibre that polymerizes into long filaments made of α/β tubulin heterodimers that form microtubules [44]. Microtubules are a pertinent component of the cell cytoskeleton that mediate internal trafficking, cell mitosis and repositioning of neurons [45]. A total of 13 protofilaments of microtubules exist but only certain protofilaments can be detected in specialized cells or organisms [46]. We detected six protofilaments of microtubules specific to neural cells, namely TUBA1B, TUBB, TUBB2B, TUBB3, TUBB4B and TUBB6 in our studies. All of these tubulins were significantly upregulated in TRF-treated and levodopa-treated diff-neural cells compared with untreated controls (Figure 3). Microtubule defects have been known to cause neurodegeneration in PD and AD and this impairment is known as axonal transport defects as curated in KEGG-PD pathway [47] (Figure 6). The axonal transport machinery consists of four elements-microtubules, kinesin, dynein and neurofilaments. The microtubules extend along with the axon and transport cargoes of proteins, lipids, RNAs and organelles between soma and distal axonal terminals. Kinesin and dynein are motor proteins that aid in moving the cargoes along the microtubules. Neurofilaments are abundant in axons and play critical roles in preserving axonal stability, transport and shape. Genetic mutation in any component of the axonal transport system is linked to neurodegenerative diseases [48]. Increased expression of TUBB3 in neurons is an indication that differentiating neurons have achieved morphologically and functionally established neuron states [49]. Consistent with this finding, Guo et al. [50] have investigated the roles of TUBB1, TUBB2 and TUBB3 in differentiating neuroblastoma cells. According to this report, TUBB1 and TUBB3 exist in cell bodies and neurites, whereas TUBB2 occurs predominantly in neurites. The gene knock down experiment revealed TUBB1 has a profound effect on cell viability, TUBB2 for neurite outgrowth and TUBB3 for neuroprotection against reactive oxygen species and free radicals. Hence, our study’s increased expression of tubulins in TRF or levodopa treated diff-neural cells may have contributed to enhanced microtubule reorganisation and stabilization that occur in matured differentiated neuronal cells. These functions are important to preserve the physiology and morphology of neurons.
5. Conclusions
The label-free mass spectrometry quantification revealed that TRF exhibited a significant role at the molecular level in protecting neuronal cells. We also showed that the TRF regulated key proteins overlaps with levodopa treated neural cells, which indicate the manifestation of common neuroprotective pathways between TRF and levodopa. The KEGG pathway enrichment analysis showed that the Parkinson’s disease (hsa05012) pathway was enriched with the most populated proteins found to be regulated by TRF and levodopa treatment. Upon further investigation into the KEGG-Parkinson’s disease pathway, we discovered that TRF significantly altered the expression of proteins that play a crucial role in the ubiquitin-proteasome pathway, calcium signalling pathway, protein processing in the endoplasmic reticulum, mitochondrial pathway and axonal transport system when compared with levodopa.
Author Contributions
Conceptualization, A.K.R. and K.B.M.; methodology, A.K.R. and K.B.M.; software, K.B.M. and P.R.; validation, K.B.M.; formal analysis, K.B.M.; investigation, K.B.M.; resources, A.K.R.; data curation, K.B.M.; writing—original draft preparation, K.B.M.; writing—review and editing, A.K.R., S.D.S., P.R. and S.B.; visualization, K.B.M.; supervision, A.K.R. and S.D.S.; project administration, A.K.R. and S.D.S.; funding acquisition, A.K.R. and K.R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Jeffrey Cheah School of Medicine and Health Sciences Internal grant scheme.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to Bryan See Ka Leong (ExcelVite Sdn. Bhd. Malaysia) for generously providing the palm TRF. We thank the Monash Liquid Chromatography Mass Spectrometry (LCMS) platform for the instrumentation and technical support.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AD | Alzheimer’s disease |
| ATP5F1A | ATP synthase F1 subunit α |
| CALM | calmodulin |
| CAMKII | calmodulin-dependent kinase-IIα |
| diff-neural cells | differentiated human SH-SY5Y neuroblastoma cells |
| DMSO | dimethyl sulfoxide |
| ER | endoplasmic reticulum |
| HSPA5 | heat shock protein family A member 5 |
| HSPD1 | heat shock protein family D member 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PD | Parkinson’s disease |
| PPIA | peptidylprolyl isomerase A |
| RA | retinoic acid |
| SLC25 | ADP/ATP translocase 3 |
| SOD | superoxide dismutase |
| T3 | tocotrienol |
| Toc | tocopherol |
| TUBA | tubulin α |
| TUBB | tubulin β |
References
- Ahsan, H.; Ahad, A.; Siddiqui, W.A. A Review of Characterization of Tocotrienols from Plant Oils and Foods. J. Chem. Biol. 2015, 8, 45–59. [Google Scholar] [CrossRef]
- Niki, E. Role of Vitamin e as a Lipid-Soluble Peroxyl Radical Scavenger: In Vitro and in Vivo Evidence. Free Radic. Biol. Med. 2014, 66, 3–12. [Google Scholar] [CrossRef]
- Wang, X.; Quinn, P.J. Vitamin E and Its Function in Membranes. Prog. Lipid Res. 1999, 38, 309–336. [Google Scholar] [CrossRef]
- Selvaraju, T.R.; Khaza’ai, H.; Vidyadaran, S.; Mutalib, M.S.A.; Vasudevan, R. The Neuroprotective Effects of Tocotrienol Rich Fraction and Alpha Tocopherol against Glutamate Injury in Astrocytes. Bosn. J. Basic Med. Sci. 2014, 14, 195–204. [Google Scholar] [CrossRef]
- Nasri, W.N.W.; Makpol, S.; Mazlan, M.; Tooyama, I.; Ngah, W.Z.W.; Damanhuri, H.A. Tocotrienol Rich Fraction Supplementation Modulate Brain Hippocampal Gene Expression in APPswe/PS1dE9 Alzheimer’s Disease Mouse Model. J. Alzheimers Dis. 2019, 70, S239–S254. [Google Scholar] [CrossRef]
- Durani, L.W.; Hamezah, H.S.; Ibrahim, N.F.; Yanagisawa, D.; Nasaruddin, M.L.; Mori, M.; Azizan, K.A.; Damanhuri, H.A.; Makpol, S.; Ngah, W.Z.W.; et al. Tocotrienol-Rich Fraction of Palm Oil Improves Behavioral Impairments and Regulates Metabolic Pathways in AβPP/PS1 Mice. J. Alzheimers Dis. 2018, 64, 249–267. [Google Scholar] [CrossRef]
- Mazlan, M.; Hamezah, H.S.; Taridi, N.M.; Jing, Y.; Liu, P.; Zhang, H.; Ngah, W.Z.W.; Damanhuri, H.A. Effects of Aging and Tocotrienol-Rich Fraction Supplementation on Brain Arginine Metabolism in Rats. Oxid. Med. Cell. Longev. 2017, 2017, 6019796. [Google Scholar] [CrossRef]
- Azman, N.H.E.N.; Goon, J.A.; Ghani, S.M.A.; Hamid, Z.; Ngah, W.Z.W. Comparing Palm Oil, Tocotrienol-Rich Fraction and α-Tocopherol Supplementation on the Antioxidant Levels of Older Adults. Antioxidants 2018, 7, 74. [Google Scholar] [CrossRef]
- Gopalan, Y.; Shuaib, I.L.; Magosso, E.; Ansari, M.A.; Abu Bakar, M.R.; Wong, J.W.; Khan, N.A.K.; Liong, W.C.; Sundram, K.; Ng, B.H.; et al. Clinical Investigation of the Protective Effects of Palm Vitamin e Tocotrienols on Brain White Matter. Stroke 2014, 45, 1422–1428. [Google Scholar] [CrossRef]
- Hamezah, H.S.; Durani, L.W.; Yanagisawa, D.; Ibrahim, N.F.; Aizat, W.M.; Makpol, S.; Ngah, W.Z.W.; Damanhuri, H.A.; Tooyama, I. Modulation of Proteome Profile in AβPP/PS1 Mice Hippocampus, Medial Prefrontal Cortex, and Striatum by Palm Oil Derived Tocotrienol-Rich Fraction. J. Alzheimers Dis. 2019, 72, 229–246. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Magalingam, K.B.; Radhakrishnan, A.K.; Somanath, S.D.; Md, S.; Haleagrahara, N. Influence of Serum Concentration in Retinoic Acid and Phorbol Ester Induced Differentiation of SH-SY5Y Human Neuroblastoma Cell Line. Mol. Biol. Rep. 2020, 47, 8775–8788. [Google Scholar] [CrossRef]
- Magalingam, K.B.; Somanath, S.D.; Haleagrahara, N.; Selvaduray, K.R.; Radhakrishnan, A.K. Unravelling the Neuroprotective Mechanisms of Carotenes in Differentiated Human Neural Cells: Biochemical and Proteomic Approaches. Food Chem. Mol. Sci. 2022, 4, 100088. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful Software for Peptide de Novo Sequencing by Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Lopes, F.M.; da Motta, L.L.; De Bastiani, M.A.; Pfaffenseller, B.; Aguiar, B.W.; de Souza, L.F.; Zanatta, G.; Vargas, D.M.; Schönhofen, P.; Londero, G.F.; et al. RA Differentiation Enhances Dopaminergic Features, Changes Redox Parameters, and Increases Dopamine Transporter Dependency in 6-Hydroxydopamine-Induced Neurotoxicity in SH-SY5Y Cells. Neurotox. Res. 2017, 31, 545–559. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the Hub of Protein Homeostasis: Emerging Mechanistic Insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef]
- Bross, P.; Fernandez-Guerra, P. Disease-Associated Mutations in the HSPD1 Gene Encoding the Large Subunit of the Mitochondrial HSP60/HSP10 Chaperonin Complex. Front. Mol. Biosci. 2016, 3, 49. [Google Scholar] [CrossRef]
- Calabrese, V.; Mancuso, C.; Ravagna, A.; Perluigi, M.; Cini, C.; de Marco, C.; Butterfield, D.A.; Stella, A.M.G. In Vivo Induction of Heat Shock Proteins in the Substantia Nigra Following L-DOPA Administration Is Associated with Increased Activity of Mitochondrial Complex I and Nitrosative Stress in Rats: Regulation by Glutathione Redox State. J. Neurochem. 2007, 101, 709–717. [Google Scholar] [CrossRef]
- Pasetto, L.; Pozzi, S.; Castelnovo, M.; Basso, M.; Estevez, A.G.; Fumagalli, S.; De Simoni, M.G.; Castellaneta, V.; Bigini, P.; Restelli, E.; et al. Targeting Extracellular Cyclophilin a Reduces Neuroinflammation and Extends Survival in a Mouse Model of Amyotrophic Lateral Sclerosis. J. Neurosci. 2017, 37, 1413–1427. [Google Scholar] [CrossRef]
- Maiti, P.; Manna, J.; Dunbar, G.L.; Maiti, P.; Dunbar, G.L. Current Understanding of the Molecular Mechanisms in Parkinson’s Disease: Targets for Potential Treatments. Transl. Neurodegener. 2017, 6, 1–35. [Google Scholar]
- Dantuma, N.P.; Bott, L.C. The Ubiquitin-Proteasome System in Neurodegenerative Diseases: Precipitating Factor, yet Part of the Solution. Front. Mol. Neurosci. 2014, 7, 70. [Google Scholar] [CrossRef]
- Coulombe, J.; Gamage, P.; Gray, M.T.; Zhang, M.; Tang, M.Y.; Woulfe, J.; Saffrey, M.J.; Gray, D.A. Loss of UCHL1 Promotes Age-Related Degenerative Changes in the Enteric Nervous System. Front. Aging Neurosci. 2014, 6, 129. [Google Scholar] [CrossRef]
- Cartier, A.E.; Ubhi, K.; Spencer, B.; Vazquez-Roque, R.A.; Kosberg, K.A.; Fourgeaud, L.; Kanayson, P.; Patrick, C.; Rockenstein, E.; Patrick, G.N.; et al. Differential Effects of UCHL1 Modulation on Alpha-Synuclein in PD-Like Models of Alpha-Synucleinopathy. PLoS ONE 2012, 7, e34713. [Google Scholar] [CrossRef]
- Cartier, A.E.; Djakovic, S.N.; Salehi, A.; Wilson, S.M.; Masliah, E.; Patrick, G.N. Regulation of Synaptic Structure by Ubiquitin C-Terminal Hydrolase L1. J. Neurosci. 2009, 29, 7857–7868. [Google Scholar] [CrossRef]
- Kawamoto, E.M.; Vivar, C.; Camandola, S. Physiology and Pathology of Calcium Signaling in the Brain. Front. Pharmacol. 2012, 3, 61. [Google Scholar] [CrossRef]
- Pivovarova, N.B.; Andrews, S.B. Calcium-Dependent Mitochondrial Function and Dysfunction in Neurons. FEBS J. 2010, 277, 3622–3636. [Google Scholar] [CrossRef]
- Pang, Z.P.; Cao, P.; Xu, W.; Südhof, T.C. Calmodulin Controls Synaptic Strength via Presynaptic Activation of Calmodulin Kinase II. J. Neurosci. 2010, 30, 4132–4142. [Google Scholar] [CrossRef]
- Xia, Z.; Storm, D.R. The Role of Calmodulin as a Signal Integrator for Synaptic Plasticity. Nat. Rev. Neurosci. 2005, 6, 267–276. [Google Scholar] [CrossRef]
- Greer, P.L.; Greenberg, M.E. From Synapse to Nucleus: Calcium-Dependent Gene Transcription in the Control of Synapse Development and Function. Neuron 2008, 59, 846–860. [Google Scholar] [CrossRef]
- Bahar, E.; Kim, H.; Yoon, H. ER Stress-Mediated Signaling: Action Potential and Ca2+ as Key Players. Int. J. Mol. Sci. 2016, 17, 1558. [Google Scholar] [CrossRef]
- Yadav, R.K.; Chae, S.-W.; Kim, H.-R.; Chae, H.J. Endoplasmic Reticulum Stress and Cancer. J. Cancer Prev. 2014, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Hoozemans, J.J.M.; van Haastert, E.S.; Eikelenboom, P.; de Vos, R.A.I.; Rozemuller, J.M.; Scheper, W. Activation of the Unfolded Protein Response in Parkinson’s Disease. Biochem. Biophys. Res. Commun. 2007, 354, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.H.; Mamula, D.; Tingstam, B.; Pereira, M.; He, Y.; Svenningsson, P. GRP78 Level Is Altered in the Brain, but Not in Plasma or Cerebrospinal Fluid in Parkinson’s Disease Patients. Front. Neurosci. 2019, 13, 697. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.W.; Yen, J.H.; Shen, Y.T.; Wu, K.Y.; Wu, M.J. Luteolin Modulates 6-Hydroxydopamine-Induced Transcriptional Changes of Stress Response Pathways in PC12 Cells. PLoS ONE 2014, 9, e97880. [Google Scholar] [CrossRef] [PubMed]
- Prasuhn, J.; Davis, R.L.; Kumar, K.R. Targeting Mitochondrial Impairment in Parkinson’s Disease: Challenges and Opportunities. Front. Cell Dev. Biol. 2021, 8, 1704. [Google Scholar] [CrossRef] [PubMed]
- Aykaç, A.; Şehirli, A.Ö. The Role of the SLC Transporters Protein in the Neurodegenerative Disorders. Clin. Psychopharmacol. Neurosci. 2020, 18, 174. [Google Scholar] [CrossRef]
- Brenner, C.; Subramaniam, K.; Pertuiset, C.; Pervaiz, S. Adenine Nucleotide Translocase Family: Four Isoforms for Apoptosis Modulation in Cancer. Oncogene 2010, 30, 883–895. [Google Scholar] [CrossRef]
- Gottlieb, E.; Tomlinson, I.P.M. Mitochondrial Tumour Suppressors: A Genetic and Biochemical Update. Nat. Rev. Cancer 2005, 5, 857–866. [Google Scholar] [CrossRef]
- Bröer, A.; Wagner, C.; Lang, F.; Bröer, S. Neutral Amino Acid Transporter ASCT2 Displays Substrate-Induced Na+ Exchange and a Substrate-Gated Anion Conductance. Biochem. J. 2000, 346, 705–710. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Chen, J. Identification of Critical Genes and MiRNAs Associated with the Development of Parkinson’s Disease. J. Mol. Neurosci. 2018, 65, 527–535. [Google Scholar] [CrossRef]
- Song, B.Y.; Wang, M.F.; Wei, M.Y.; Chen, B.D.; Deng, B.G. ATP5A1 Participates in Transcriptional and Posttranscriptional Regulation of Cancer-Associated Genes by Modulating Their Expression and Alternative Splicing Profiles in HeLa Cells. Technol. Cancer Res. Treat. 2021, 20, 15330338211039. [Google Scholar]
- Picca, A.; Guerra, F.; Calvani, R.; Marini, F.; Biancolillo, A.; Landi, G.; Beli, R.; Landi, F.; Bernabei, R.; Bentivoglio, A.R.; et al. Mitochondrial Signatures in Circulating Extracellular Vesicles of Older Adults with Parkinson’s Disease: Results from the Exosomes in Parkinson’s Disease (EXPAND) Study. J. Clin. Med. 2020, 9, 504. [Google Scholar] [CrossRef]
- Jonckheere, A.I.; Renkema, G.H.; Bras, M.; Van Den Heuvel, L.P.; Hoischen, A.; Gilissen, C.; Nabuurs, S.B.; Huynen, M.A.; De Vries, M.C.; Smeitink, J.A.M.; et al. A Complex V ATP5A1 Defect Causes Fatal Neonatal Mitochondrial Encephalopathy. Brain 2013, 136, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Binarová, P.; Tuszynski, J. Tubulin: Structure, Functions and Roles in Disease. Cells 2019, 8, 1294. [Google Scholar] [CrossRef] [PubMed]
- Bodakuntla, S.; Janke, C.; Magiera, M.M. Tubulin Polyglutamylation, a Regulator of Microtubule Functions, Can Cause Neurodegeneration. Neurosci. Lett. 2021, 746, 135656. [Google Scholar] [CrossRef]
- Chaaban, S.; Brouhard, G.J. A Microtubule Bestiary: Structural Diversity in Tubulin Polymers. Mol. Biol. Cell 2017, 28, 2924–2931. [Google Scholar] [CrossRef]
- Cartelli, D.; Casagrande, F.; Busceti, C.L.; Bucci, D.; Molinaro, G.; Traficante, A.; Passarella, D.; Giavini, E.; Pezzoli, G.; Battaglia, G.; et al. Microtubule Alterations Occur Early in Experimental Parkinsonism and The Microtubule Stabilizer Epothilone D is Neuroprotective. Sci. Reports 2013, 3, 1–10. [Google Scholar] [CrossRef]
- Guo, W.; Dittlau, K.S.; Van Den Bosch, L. Axonal Transport Defects and Neurodegeneration: Molecular Mechanisms and Therapeutic Implications. Semin. Cell Dev. Biol. 2020, 99, 133–150. [Google Scholar] [CrossRef]
- Marei, H.E.S.; El-Gamal, A.; Althani, A.; Afifi, N.; Abd-Elmaksoud, A.; Farag, A.; Cenciarelli, C.; Thomas, C.; Anwarul, H. Cholinergic and Dopaminergic Neuronal Differentiation of Human Adipose Tissue Derived Mesenchymal Stem Cells. J. Cell. Physiol. 2018, 233, 936–945. [Google Scholar] [CrossRef]
- Guo, J.; Walss-Bass, C.; Ludueña, R.F. The β Isotypes of Tubulin in Neuronal Differentiation. Cytoskeleton 2010, 67, 431–441. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).