COVID-19 and Gut Injury
Abstract
1. Introduction
2. Epidemiology
2.1. Sex Differences in COVID-19
2.2. Age Difference in COVID-19
3. Clinical Features of COVID-19
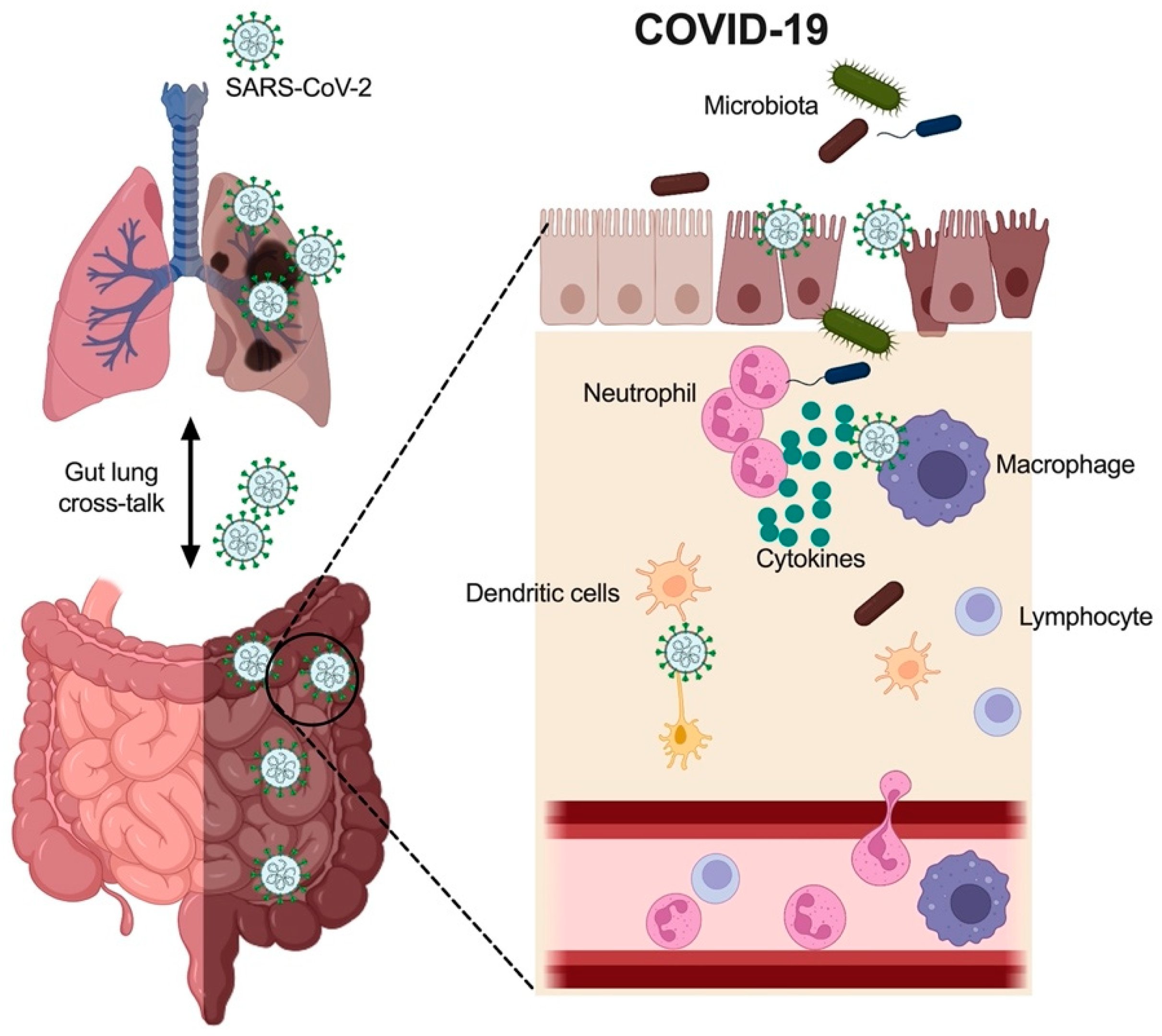
3.1. Immune Cells in COVID-19
3.2. Cytokines in the Gut with COVID-19
3.3. Gut Microbiota in COVID-19
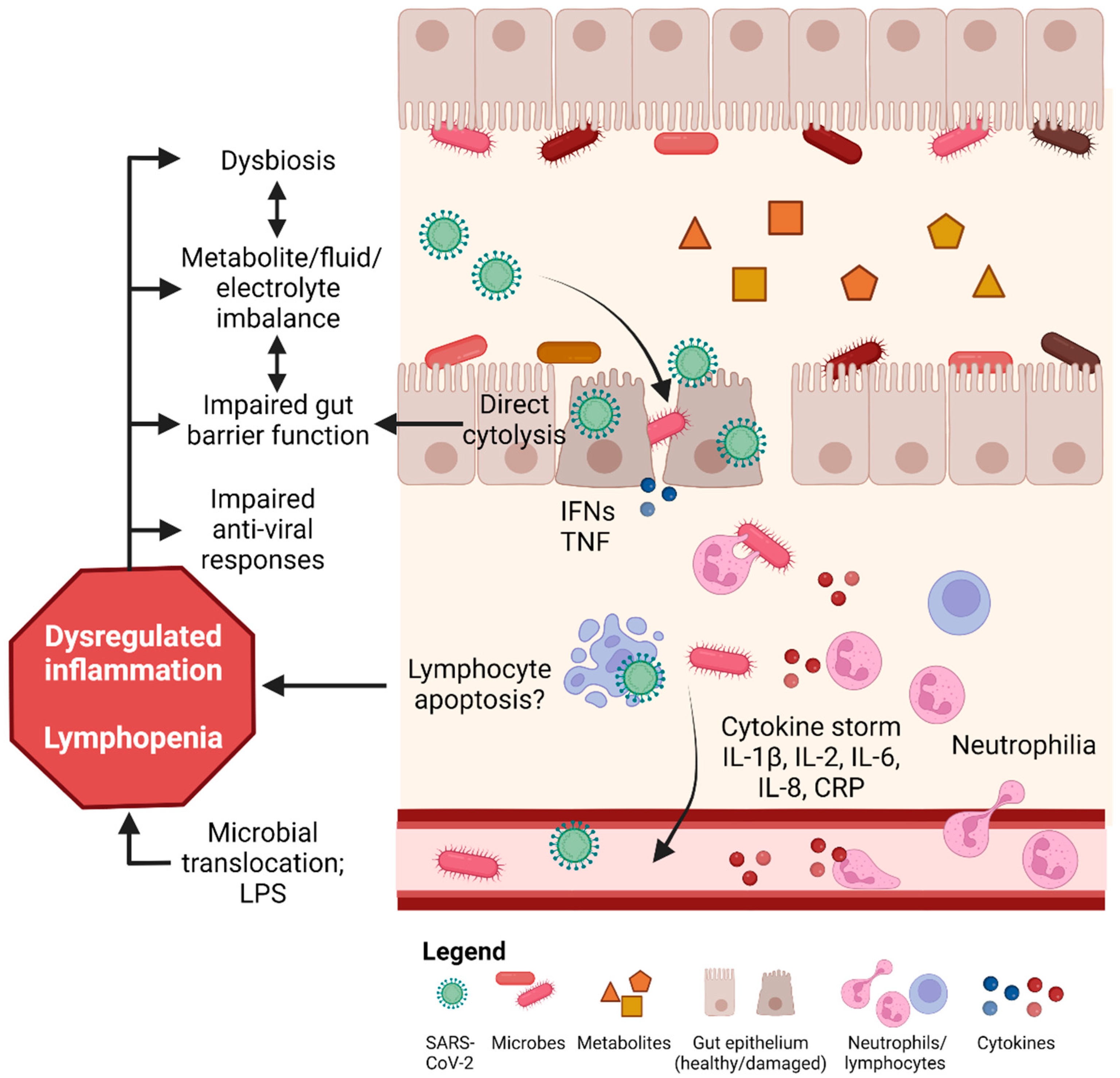
4. Mechanisms Underlying COVID-19 Induced GI Tract Injury
4.1. SARS-CoV-2 Infection via ACE2 Binding and Injury in the Gut
4.2. Dysbiosis and Metabolite Imbalance Induce Gut Inflammation and Injury
4.3. Other Mechanisms of COVID-19 Induced Gut Injury
4.4. Inflammatory Bowel Disease (IBD) and COVID-19
5. Treatment of COVID-19 and GI Injury
5.1. Current COVID-19 Treatment
5.2. Treatment for COVID-19 Induced Gut Injury
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, J.; Xu, X.; Jiang, L.; Dua, K.; Hansbro, P.M.; Liu, G. SARS-CoV-2 induces transcriptional signatures in human lung epithelial cells that promote lung fibrosis. Respir. Res. 2020, 21, 182. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.M.; Wang, J.; Liu, Y.; Zhang, X.; Wang, T.; Zhang, H.P.; Liang, Z.A.; Luo, F.M.; Li, W.M.; Liu, D.; et al. Combined and interactive effects of alcohol drinking and cigarette smoking on the risk of severe illness and poor clinical outcomes in patients with COVID-19: A multicentre retrospective cohort study. Public Health 2022, 205, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.; Xu, J.; Yang, D.; Shen, Y.; Wang, L.; Feng, Y.; Du, C.; Song, Y.; Wu, C.; Hu, X.; et al. COVID-19-associated gastrointestinal and liver injury: Clinical features and potential mechanisms. Signal Transduct. Target. Ther. 2020, 5, 256. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Coronavirus (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int (accessed on 10 October 2022).
- Sultan, S.; Altayar, O.; Siddique, S.M.; Davitkov, P.; Feuerstein, J.D.; Lim, J.K.; Falck-Ytter, Y.; El-Serag, H.B.; AGA Institute. AGA Institute Rapid Review of the Gastrointestinal and Liver Manifestations of COVID-19, Meta-Analysis of International Data, and Recommendations for the Consultative Management of Patients with COVID-19. Gastroenterology 2020, 159, 320–334.e27. [Google Scholar] [CrossRef]
- Zhang, J.; Garrett, S.; Sun, J. Gastrointestinal symptoms, pathophysiology, and treatment in COVID-19. Genes Dis. 2021, 8, 385–400. [Google Scholar] [CrossRef]
- Abate, B.B.; Kassie, A.M.; Kassaw, M.W.; Aragie, T.G.; Masresha, S.A. Sex difference in coronavirus disease (COVID-19): A systematic review and meta-analysis. BMJ Open 2020, 10, e040129. [Google Scholar] [CrossRef]
- Qi, S.; Ngwa, C.; Morales Scheihing, D.A.; Al Mamun, A.; Ahnstedt, H.W.; Finger, C.E.; Colpo, G.D.; Sharmeen, R.; Kim, Y.; Choi, H.A.; et al. Sex differences in the immune response to acute COVID-19 respiratory tract infection. Biol. Sex Differ. 2021, 12, 66. [Google Scholar] [CrossRef]
- Strandberg, T.E.; Pentti, J.; Kivimaki, M. Sex Difference in Serious Infections: Not Only COVID-19. Epidemiology 2021, 32, e26–e27. [Google Scholar] [CrossRef]
- Team, C.-F. Variation in the COVID-19 infection-fatality ratio by age, time, and geography during the pre-vaccine era: A systematic analysis. Lancet 2022, 399, 1469–1488. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y.; et al. SARS-CoV-2 Infection in Children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef]
- Xiong, X.L.; Wong, K.K.; Chi, S.Q.; Zhou, A.F.; Tang, J.Q.; Zhou, L.S.; Chung, P.H.; Chua, G.; Tung, K.; Wong, I.; et al. Comparative study of the clinical characteristics and epidemiological trend of 244 COVID-19 infected children with or without GI symptoms. Gut 2021, 70, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Abrams, J.Y.; Godfred-Cato, S.E.; Oster, M.E.; Chow, E.J.; Koumans, E.H.; Bryant, B.; Leung, J.W.; Belay, E.D. Multisystem Inflammatory Syndrome in Children Associated with Severe Acute Respiratory Syndrome Coronavirus 2: A Systematic Review. J. Pediatr. 2020, 226, 45–54.e41. [Google Scholar] [CrossRef]
- Guimarães, D.; Pissarra, R.; Reis-Melo, A.; Guimarães, H. Multisystem inflammatory syndrome in children (MISC): A systematic review. Int. J. Clin. Pract. 2021, 75, e14450. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, M.; Nandi, A.; Banerjee, P.; Gupta, P.; Sarkar, S.D.; Basu, S.; Pal, P. A comparative study of IL-6, CRP and NT-proBNP levels in post-COVID multisystem inflammatory syndrome in children (MISC) and Kawasaki disease patients. Int. J. Rheum. Dis. 2022, 25, 27–31. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Khanmohammadi, S.; Rezaei, N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021, 93, 2735–2739. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Donovan, C.; Liu, G.; Shen, S.; Marshall, J.E.; Kim, R.Y.; Alemao, C.A.; Budden, K.F.; Choi, J.P.; Kohonen-Corish, M.; El-Omar, E.M.; et al. The role of the microbiome and the NLRP3 inflammasome in the gut and lung. J. Leukoc. Biol. 2020, 108, 925–935. [Google Scholar] [CrossRef]
- Shen, S.; Wong, C.H. Bugging inflammation: Role of the gut microbiota. Clin. Transl. Immunol. 2016, 5, e72. [Google Scholar] [CrossRef]
- Hung, Y.P.; Lee, C.C.; Lee, J.C.; Tsai, P.J.; Ko, W.C. Gut Dysbiosis during COVID-19 and Potential Effect of Probiotics. Microorganisms 2021, 9, 1605. [Google Scholar] [CrossRef]
- Poletti, M.; Treveil, A.; Csabai, L.; Gul, L.; Modos, D.; Madgwick, M.; Olbei, M.; Bohar, B.; Valdeolivas, A.; Turei, D.; et al. Mapping the epithelial-immune cell interactome upon infection in the gut and the upper airways. NPJ Syst. Biol. Appl. 2022, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Pang, I.K.; Iwasaki, A. Control of antiviral immunity by pattern recognition and the microbiome. Immunol. Rev. 2012, 245, 209–226. [Google Scholar] [CrossRef]
- Manik, M.; Singh, R.K. Role of toll-like receptors in modulation of cytokine storm signaling in SARS-CoV-2-induced COVID-19. J. Med. Virol. 2022, 94, 869–877. [Google Scholar] [CrossRef]
- Nainu, F.; Shiratsuchi, A.; Nakanishi, Y. Induction of Apoptosis and Subsequent Phagocytosis of Virus-Infected Cells as an Antiviral Mechanism. Front. Immunol. 2017, 8, 1220. [Google Scholar] [CrossRef] [PubMed]
- Shokri-Afra, H.; Alikhani, A.; Moradipoodeh, B.; Noorbakhsh, F.; Fakheri, H.; Moradi-Sardareh, H. Elevated fecal and serum calprotectin in COVID-19 are not consistent with gastrointestinal symptoms. Sci. Rep. 2021, 11, 22001. [Google Scholar] [CrossRef]
- Shi, H.; Zuo, Y.; Yalavarthi, S.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Woodward, W.; Lezak, S.P.; Lugogo, N.L.; et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J. Leukoc. Biol. 2021, 109, 67–72. [Google Scholar] [CrossRef]
- Saithong, S.; Worasilchai, N.; Saisorn, W.; Udompornpitak, K.; Bhunyakarnjanarat, T.; Chindamporn, A.; Tovichayathamrong, P.; Torvorapanit, P.; Chiewchengchol, D.; Chancharoenthana, W.; et al. Neutrophil Extracellular Traps in Severe SARS-CoV-2 Infection: A Possible Impact of LPS and (1→3)-beta-D-glucan in Blood from Gut Translocation. Cells 2022, 11, 1103. [Google Scholar] [CrossRef]
- Martin-Gayo, E.; Yu, X.G. Role of Dendritic Cells in Natural Immune Control of HIV-1 Infection. Front. Immunol. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Greene, T.T.; Zuniga, E.I. Type I Interferon Induction and Exhaustion during Viral Infection: Plasmacytoid Dendritic Cells and Emerging COVID-19 Findings. Viruses 2021, 13, 1839. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Barragan, L.; Zust, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 2007, 109, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Kindler, E.; Thiel, V.; Weber, F. Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response. Adv. Virus. Res. 2016, 96, 219–243. [Google Scholar] [CrossRef] [PubMed]
- Winheim, E.; Rinke, L.; Lutz, K.; Reischer, A.; Leutbecher, A.; Wolfram, L.; Rausch, L.; Kranich, J.; Wratil, P.R.; Huber, J.E.; et al. Impaired function and delayed regeneration of dendritic cells in COVID-19. PLoS Pathog. 2021, 17, e1009742. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Kundu, R.; Narean, J.S.; Wang, L.; Fenn, J.; Pillay, T.; Fernandez, N.D.; Conibear, E.; Koycheva, A.; Davies, M.; Tolosa-Wright, M.; et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022, 13, 80. [Google Scholar] [CrossRef]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef]
- Zhuang, Z.; Lai, X.; Sun, J.; Chen, Z.; Zhang, Z.; Dai, J.; Liu, D.; Li, Y.; Li, F.; Wang, Y.; et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J. Exp. Med. 2021, 218, e20202187. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.Q.; Wang, Q.; Miao, H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct. Target. Ther. 2020, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, C.; Zhang, S.; Duan, C.; Shang, H.; Bai, T.; Hou, X. Diarrhea and altered inflammatory cytokine pattern in severe coronavirus disease 2019: Impact on disease course and in-hospital mortality. J. Gastroenterol. Hepatol. 2021, 36, 421–429. [Google Scholar] [CrossRef]
- Liu, G.; Jarnicki, A.G.; Paudel, K.R.; Lu, W.; Wadhwa, R.; Philp, A.M.; van Eeckhoutte, H.; Marshall, J.E.; Malyla, V.; Katsifis, A.; et al. Adverse roles of mast cell chymase-1 in chronic obstructive pulmonary disease. Eur. Respir. J. 2022, 60. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W. Interferons. In Viruses and Man: A History of Interactions; Springer: Berlin/Heidelberg, Germany, 2014; pp. 101–119. [Google Scholar] [CrossRef]
- Raftery, N.; Stevenson, N.J. Advances in anti-viral immune defence: Revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci. 2017, 74, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.C.; Young, H.A. Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014, 25, 369–376. [Google Scholar] [CrossRef]
- Welsh, R.M.; Bahl, K.; Marshall, H.D.; Urban, S.L. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog. 2012, 8, e1002352. [Google Scholar] [CrossRef]
- García-Sastre, A. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 2002, 4, 647–655. [Google Scholar] [CrossRef]
- Schreiber, G. The Role of Type I Interferons in the Pathogenesis and Treatment of COVID-19. Front. Immunol. 2020, 11, 595739. [Google Scholar] [CrossRef]
- Broggi, A.; Ghosh, S.; Sposito, B.; Spreafico, R.; Balzarini, F.; lo Cascio, A.; Clementi, N.; de Santis, M.; Mancini, N.; Granucci, F.; et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 2020, 369, 706–712. [Google Scholar] [CrossRef]
- Lee, J.S.; Shin, E.C. The type I interferon response in COVID-19: Implications for treatment. Nat. Rev. Immunol. 2020, 20, 585–586. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Webster, R.G. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 2002, 76, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Van der Ploeg, K.; Kirosingh, A.S.; Mori, D.A.M.; Chakraborty, S.; Hu, Z.; Sievers, B.L.; Jacobson, K.B.; Bonilla, H.; Parsonnet, J.; Andrews, J.R.; et al. TNF-alpha(+) CD4(+) T cells dominate the SARS-CoV-2 specific T cell response in COVID-19 outpatients and are associated with durable antibodies. Cell Rep. 2022, 3, 100640. [Google Scholar] [CrossRef]
- Alexander, J.L.; Kennedy, N.A.; Ibraheim, H.; Anandabaskaran, S.; Saifuddin, A.; Castro Seoane, R.; Liu, Z.; Nice, R.; Bewshea, C.; D’Mello, A.; et al. COVID-19 vaccine-induced antibody responses in immunosuppressed patients with inflammatory bowel disease (VIP): A multicentre, prospective, case-control study. Lancet Gastroenterol. Hepatol. 2022, 7, 342–352. [Google Scholar] [CrossRef]
- Haberman, R.H.; Um, S.; Axelrad, J.E.; Blank, R.B.; Uddin, Z.; Catron, S.; Neimann, A.L.; Mulligan, M.J.; Herat, R.S.; Hong, S.J.; et al. Methotrexate and TNF inhibitors affect long-term immunogenicity to COVID-19 vaccination in patients with immune-mediated inflammatory disease. Lancet Rheumatol. 2022, 4, e384–e387. [Google Scholar] [CrossRef]
- Venerito, V.; Stefanizzi, P.; Fornaro, M.; Cacciapaglia, F.; Tafuri, S.; Perniola, S.; Iannone, F.; Lopalco, G. Immunogenicity of BNT162b2 mRNA SARS-CoV-2 vaccine in patients with psoriatic arthritis on TNF inhibitors. RMD Open 2022, 8, e001847. [Google Scholar] [CrossRef]
- Ruder, B.; Atreya, R.; Becker, C. Tumour Necrosis Factor Alpha in Intestinal Homeostasis and Gut Related Diseases. Int. J. Mol. Sci. 2019, 20, 1887. [Google Scholar] [CrossRef]
- Naito, Y.; Takagi, T.; Handa, O.; Ishikawa, T.; Nakagawa, S.; Yamaguchi, T.; Yoshida, N.; Minami, M.; Kita, M.; Imanishi, J.; et al. Enhanced intestinal inflammation induced by dextran sulfate sodium in tumor necrosis factor-alpha deficient mice. J. Gastroenterol. Hepatol. 2003, 18, 560–569. [Google Scholar] [CrossRef]
- Noti, M.; Corazza, N.; Mueller, C.; Berger, B.; Brunner, T. TNF suppresses acute intestinal inflammation by inducing local glucocorticoid synthesis. J. Exp. Med. 2010, 207, 1057–1066. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, Y.; Zuo, T.; Yeoh, Y.K.; Liu, Q.; Zhang, L.; Zhan, H.; Lu, W.; Xu, W.; Lui, G.C.Y.; et al. Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients with COVID-19. Gastroenterology 2022, 162, 548–561.e4. [Google Scholar] [CrossRef]
- Vestad, B.; Ueland, T.; Lerum, T.V.; Dahl, T.B.; Holm, K.; Barratt-Due, A.; Kasine, T.; Dyrhol-Riise, A.M.; Stiksrud, B.; Tonby, K.; et al. Respiratory dysfunction three months after severe COVID-19 is associated with gut microbiota alterations. J. Intern. Med. 2022, 291, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Schult, D.; Reitmeier, S.; Koyumdzhieva, P.; Lahmer, T.; Middelhoff, M.; Erber, J.; Schneider, J.; Kager, J.; Frolova, M.; Horstmann, J.; et al. Gut bacterial dysbiosis and instability is associated with the onset of complications and mortality in COVID-19. Gut Microbes 2022, 14, 2031840. [Google Scholar] [CrossRef]
- Debourdeau, A.; Vitton, V.; Barthet, M.A.; Gonzalez, J.M. If pneumatic dilation is not enough efficient for post fundoplication dysphagia, is Per Oral Endoscopic Myotomy a good answer to manage it? Gut 2022, 71, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; del Chierico, F.; Macari, G.; Pane, S.; Ristori, M.V.; Guarrasi, V.; Gardini, S.; Pascucci, G.R.; Cotugno, N.; Perno, C.F.; et al. The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection. Front. Cell. Infect. Microbiol. 2022, 12, 908492. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W. Diarrhoea may be underestimated: A missing link in 2019 novel coronavirus. Gut 2020, 69, 1141–1143. [Google Scholar] [CrossRef]
- Hikmet, F.; Mear, L.; Edvinsson, A.; Micke, P.; Uhlen, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef]
- Senapati, S.; Banerjee, P.; Bhagavatula, S.; Kushwaha, P.P.; Kumar, S. Contributions of human ACE2 and TMPRSS2 in determining host–pathogen interaction of COVID-19. J. Genet. 2021, 100, 12. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.M.; Raoult, D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020, 53, 425–435. [Google Scholar] [CrossRef]
- Perlot, T.; Penninger, J.M. ACE2—From the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013, 15, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Singer, D.; Camargo, S.M.; Ramadan, T.; Schafer, M.; Mariotta, L.; Herzog, B.; Huggel, K.; Wolfer, D.; Werner, S.; Penninger, J.M.; et al. Defective intestinal amino acid absorption in Ace2 null mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G686–G695. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.; Angus, P.W.; Burrell, L.M.; Herath, C.; Gibson, P.R.; Lubel, J.S. Review article: The pathophysiological roles of the renin-angiotensin system in the gastrointestinal tract. Aliment. Pharmacol. Ther. 2012, 35, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.H. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Song, Z.G.; Liu, C.; Tan, S.; Lin, S.; Zhu, J.; Dai, F.H.; Gao, J.; She, J.L.; Mei, Z.; et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022, 20, 24. [Google Scholar] [CrossRef]
- Nagpal, R.; Newman, T.M.; Wang, S.; Jain, S.; Lovato, J.F.; Yadav, H. Obesity-Linked Gut Microbiome Dysbiosis Associated with Derangements in Gut Permeability and Intestinal Cellular Homeostasis Independent of Diet. J. Diabetes Res. 2018, 2018, 3462092. [Google Scholar] [CrossRef]
- Lauritano, E.C.; Valenza, V.; Sparano, L.; Scarpellini, E.; Gabrielli, M.; Cazzato, A.; Ferraro, P.M.; Gasbarrini, A. Small intestinal bacterial overgrowth and intestinal permeability. Scand. J. Gastroenterol. 2010, 45, 1131–1132. [Google Scholar] [CrossRef]
- Stevens, B.R.; Goel, R.; Seungbum, K.; Richards, E.M.; Holbert, R.C.; Pepine, C.J.; Raizada, M.K. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut 2018, 67, 1555–1557. [Google Scholar] [CrossRef]
- Gill, P.A.; van Zelm, M.C.; Muir, J.G.; Gibson, P.R. Review article: Short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharmacol. Ther. 2018, 48, 15–34. [Google Scholar] [CrossRef]
- Rizvi, A.; Patel, Z.; Liu, Y.; Satapathy, S.K.; Sultan, K.; Trindade, A.J. Gastrointestinal Sequelae 3 and 6 Months After Hospitalization for Coronavirus Disease 2019. Clin. Gastroenterol. Hepatol. 2021, 19, 2438–2440. [Google Scholar] [CrossRef]
- Wolfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Muller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Jiang, H.; Chen, Y.; Gu, S.; Xia, J.; Zhang, H.; Lu, Y.; Yan, R.; Li, L. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal. Chim. Acta 2021, 1152, 338267. [Google Scholar] [CrossRef] [PubMed]
- Di Filippo, L.; de Lorenzo, R.; D’Amico, M.; Sofia, V.; Roveri, L.; Mele, R.; Saibene, A.; Rovere-Querini, P.; Conte, C. COVID-19 is associated with clinically significant weight loss and risk of malnutrition, independent of hospitalisation: A post-hoc analysis of a prospective cohort study. Clin. Nutr. 2021, 40, 2420–2426. [Google Scholar] [CrossRef]
- Prasad, R.; Patton, M.J.; Floyd, J.L.; Fortmann, S.; DuPont, M.; Harbour, A.; Wright, J.; Lamendella, R.; Stevens, B.R.; Oudit, G.Y.; et al. Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. Int. J. Mol. Sci. 2022, 23, 9141. [Google Scholar] [CrossRef] [PubMed]
- Lunjani, N.; Albrich, W.C.; Suh, N.; Barda, B.; Finnegan, L.A.; Dam, S.A.; Walter, J.; Sadlier, C.; Horgan, M.; O’Toole, P.W.; et al. Higher levels of bacterial DNA in serum associate with severe and fatal COVID-19. Allergy 2022, 77, 1312–1314. [Google Scholar] [CrossRef]
- Wang, X.; Wei, J.; Zhu, R.; Chen, L.; Ding, F.; Zhou, R.; Ge, L.; Xiao, J.; Zhao, Q. Contribution of CD4+ T cell-mediated inflammation to diarrhea in patients with COVID-19. Int. J. Infect. Dis. 2022, 120, 1–11. [Google Scholar] [CrossRef]
- Santinelli, L.; Laghi, L.; Innocenti, G.P.; Pinacchio, C.; Vassalini, P.; Celani, L.; Lazzaro, A.; Borrazzo, C.; Marazzato, M.; Tarsitani, L.; et al. Oral Bacteriotherapy Reduces the Occurrence of Chronic Fatigue in COVID-19 Patients. Front. Nutr. 2021, 8, 756177. [Google Scholar] [CrossRef]
- Sirohiya, P.; Elavarasi, A.; Sagiraju, H.K.R.; Baruah, M.; Gupta, N.; Garg, R.K.; Paul, S.S.; Ratre, B.K.; Singh, R.; Kumar, B.; et al. Silent Hypoxia in Coronavirus disease-2019: Is it more dangerous? -A retrospective cohort study. Lung India 2022, 39, 247–253. [Google Scholar] [CrossRef]
- Adriana, D.N.; Sugihartono, T.; Nusi, I.A.; Setiawan, P.B.; Purbayu, H.; Maimunah, U.; Kholili, U.; Widodo, B.; Thamrin, H.; Vidyani, A.; et al. Role of fecal calprotectin as a hypoxic intestinal damage biomarker in COVID-19 patients. Gut Pathog. 2022, 14, 34. [Google Scholar] [CrossRef]
- Wang, F.; Zou, J.; Xu, H.; Huang, W.; Zhang, X.; Wei, Z.; Li, X.; Liu, Y.; Zou, J.; Liu, F.; et al. Effects of Chronic Intermittent Hypoxia and Chronic Sleep Fragmentation on Gut Microbiome, Serum Metabolome, Liver and Adipose Tissue Morphology. Front. Endocrinol. 2022, 13, 820939. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.S.; Liang, C.H.; Liu, Y.L.; Wei, W.; Deng, X.R.; Shi, X.Y.; Wang, L.M.; Zhang, L.J.; Yuan, H.J. Intermittent hypoxia is involved in gut microbial dysbiosis in type 2 diabetes mellitus and obstructive sleep apnea-hypopnea syndrome. World J. Gastroenterol. 2022, 28, 2320–2333. [Google Scholar] [CrossRef] [PubMed]
- Pourfridoni, M.; Abbasnia, S.M.; Shafaei, F.; Razaviyan, J.; Heidari-Soureshjani, R. Fluid and Electrolyte Disturbances in COVID-19 and Their Complications. Biomed. Res. Int. 2021, 2021, 6667047. [Google Scholar] [CrossRef] [PubMed]
- Corrales-Medina, V.F.; Musher, D.M.; Shachkina, S.; Chirinos, J.A. Acute pneumonia and the cardiovascular system. Lancet 2013, 381, 496–505. [Google Scholar] [CrossRef]
- Regenhardt, R.W.; Takase, H.; Lo, E.H.; Lin, D.J. Translating concepts of neural repair after stroke: Structural and functional targets for recovery. Restor. Neurol. Neurosci. 2020, 38, 67–92. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Li, F.; Peng, J.; Qin, D.; Yuan, M.; Liu, G. Risk Factors and Predictive Model of Diarrhea Among Patients with Severe Stroke. World Neurosurg. 2020, 136, 213–219. [Google Scholar] [CrossRef]
- Liu, G.; Mateer, S.W.; Hsu, A.; Goggins, B.J.; Tay, H.; Mathe, A.; Fan, K.; Neal, R.; Bruce, J.; Burns, G.; et al. Platelet activating factor receptor regulates colitis-induced pulmonary inflammation through the NLRP3 inflammasome. Mucosal. Immunol. 2019, 12, 862–873. [Google Scholar] [CrossRef]
- Cortes, G.M.; Marcialis, M.A.; Bardanzellu, F.; Corrias, A.; Fanos, V.; Mussap, M. Inflammatory Bowel Disease and COVID-19: How Microbiomics and Metabolomics Depict Two Sides of the Same Coin. Front. Microbiol. 2022, 13, 856165. [Google Scholar] [CrossRef]
- Kawade, Y.; Sakai, M.; Okamori, M.; Morita, M.; Mizushima, K.; Ueda, T.; Takagi, T.; Naito, Y.; Itoh, Y.; Shimada, T. Administration of live, but not inactivated, Faecalibacterium prausnitzii has a preventive effect on dextran sodium sulfate-induced colitis in mice. Mol. Med. Rep. 2019, 20, 25–32. [Google Scholar] [CrossRef]
- Tripathi, K.; Godoy Brewer, G.; Thu Nguyen, M.; Singh, Y.; Saleh Ismail, M.; Sauk, J.S.; Parian, A.M.; Limketkai, B.N. COVID-19 and Outcomes in Patients with Inflammatory Bowel Disease: Systematic Review and Meta-Analysis. Inflamm. Bowel Dis. 2022, 28, 1265–1279. [Google Scholar] [CrossRef]
- Kokkotis, G.; Kitsou, K.; Xynogalas, I.; Spoulou, V.; Magiorkinis, G.; Trontzas, I.; Trontzas, P.; Poulakou, G.; Syrigos, K.; Bamias, G. Systematic review with meta-analysis: COVID-19 outcomes in patients receiving anti-TNF treatments. Aliment. Pharmacol. Ther. 2022, 55, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Hobartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef]
- Consortium, W.H.O.S.T.; Pan, H.; Peto, R.; Henao-Restrepo, A.M.; Preziosi, M.P.; Sathiyamoorthy, V.; Abdool Karim, Q.; Alejandria, M.M.; Hernandez Garcia, C.; Kieny, M.P.; et al. Repurposed Antiviral Drugs for COVID-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Kabinger, F.; Stiller, C.; Schmitzova, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Hobartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.A., 2nd; Eron, J.J., Jr.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2022, 14, eabl7430. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Stebbing, J.; Krishnan, V.; de Bono, S.; Ottaviani, S.; Casalini, G.; Richardson, P.J.; Monteil, V.; Lauschke, V.M.; Mirazimi, A.; Youhanna, S.; et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 2020, 12, e12697. [Google Scholar] [CrossRef]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with COVID-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 2020, 370, 1110–1115. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Group, R.C.; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef] [PubMed]
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and Inputs from Lactic Acid Bacteria and Their Bacteriocins as Alternatives to Antibiotic Growth Promoters During Food-Animal Production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef]
- Preidis, G.A.; Saulnier, D.M.; Blutt, S.E.; Mistretta, T.A.; Riehle, K.P.; Major, A.M.; Venable, S.F.; Barrish, J.P.; Finegold, M.J.; Petrosino, J.F.; et al. Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Wischmeyer, P.E.; Tang, H.; Ren, Y.; Bohannon, L.; Ramirez, Z.E.; Andermann, T.M.; Messina, J.A.; Sung, J.A.; Jensen, D.; Jung, S.-H. Daily Lactobacillus Probiotic versus Placebo in COVID-19-Exposed Household Contacts (PROTECT-EHC): A Randomized Clinical Trial. medRxiv 2022. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Hazan, S.; Stollman, N.; Bozkurt, H.S.; Dave, S.; Papoutsis, A.J.; Daniels, J.; Barrows, B.D.; Quigley, E.M.; Borody, T.J. Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022, 9, e000871. [Google Scholar] [CrossRef]
- Xu, J.; Chu, M.; Zhong, F.; Tan, X.; Tang, G.; Mai, J.; Lai, N.; Guan, C.; Liang, Y.; Liao, G. Digestive symptoms of COVID-19 and expression of ACE2 in digestive tract organs. Cell Death Discov. 2020, 6, 76. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Penninger, J.M. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ. J. 2013, 77, 301–308. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkruys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef]
- Zabetakis, I.; Lordan, R.; Norton, C.; Tsoupras, A. COVID-19: The Inflammation Link and the Role of Nutrition in Potential Mitigation. Nutrients 2020, 12, 1466. [Google Scholar] [CrossRef] [PubMed]
- Hajipour, A.; Afsharfar, M.; Jonoush, M.; Ahmadzadeh, M.; Gholamalizadeh, M.; Hassanpour Ardekanizadeh, N.; Doaei, S.; Mohammadi-Nasrabadi, F. The effects of dietary fiber on common complications in critically ill patients; with a special focus on viral infections; a systematic reveiw. Immun. Inflamm. Dis. 2022, 10, e613. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, F.; Akbarzadeh-Khiavi, M.; Lotfi, Z.; Rahbarnia, L.; Safary, A.; Zarredar, H.; Baghbanzadeh, A.; Naghili, B.; Baradaran, B. Micronutrient therapy and effective immune response: A promising approach for management of COVID-19. Infection 2021, 49, 1133–1147. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.O. Therapeutic supplementation with zinc in the management of COVID-19-related diarrhea and ageusia/dysgeusia: Mechanisms and clues for a personalized dosage regimen. Nutr. Rev. 2022, 80, 1086–1093. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, G.; Zhao, L.; Wang, W. Nutritional Modulation of Gut Microbiota Alleviates Severe Gastrointestinal Symptoms in a Patient with Post-Acute COVID-19 Syndrome. mBio 2022, 13, e0380121. [Google Scholar] [CrossRef]
- Silveira, D.; Prieto-Garcia, J.M.; Boylan, F.; Estrada, O.; Fonseca-Bazzo, Y.M.; Jamal, C.M.; Magalhães, P.O.; Pereira, E.O.; Tomczyk, M.; Heinrich, M. COVID-19: Is There Evidence for the Use of Herbal Medicines as Adjuvant Symptomatic Therapy? Front. Pharmacol. 2020, 11, 581840. [Google Scholar] [CrossRef]
- Yonker, L.M.; Swank, Z.; Gilboa, T.; Senussi, Y.; Kenyon, V.; Papadakis, L.; Boribong, B.P.; Carroll, R.W.; Walt, D.R.; Fasano, A. Zonulin Antagonist, Larazotide (AT1001), As an Adjuvant Treatment for Multisystem Inflammatory Syndrome in Children: A Case Series. Crit. Care Explor. 2022, 10, e0641. [Google Scholar] [CrossRef]
- Miyakawa, K.; Machida, M.; Kawasaki, T.; Nishi, M.; Akutsu, H.; Ryo, A. Reduced Replication Efficacy of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant in “Mini-gut” Organoids. Gastroenterology 2022, 163, 514–516. [Google Scholar] [CrossRef]
- Ahmadi Badi, S.; Malek, A.; Paolini, A.; Rouhollahi Masoumi, M.; Seyedi, S.A.; Amanzadeh, A.; Masotti, A.; Khatami, S.; Siadat, S.D. Downregulation of ACE, AGTR1, and ACE2 genes mediating SARS-CoV-2 pathogenesis by gut microbiota members and their postbiotics on Caco-2 cells. Microb. Pathog. 2022, 173, 105798. [Google Scholar] [CrossRef]
| Sponsor | Targets | Clinical Trial Identifier | Status |
|---|---|---|---|
| I.M. Sechenov First Moscow State Medical University, Moscow, Russia | Lactobacillus rhamnosus; Bifidobacterium bifidum; B. longum subsp. Infantis; Bifidobacterium longum | NCT04854941 | Completed |
| Centre de recherche du Centre hospitalier universitaire de Sherbrooke, Sherbrooke, Canada | 2 strains (unknown) | NCT05080244 | Recruiting |
| King’s College Hospital NHS Trust | Bifidobacterium; Lactobacillus | NCT04877704 | Not yet recruiting |
| Medi Pharma Vision | Probiotics (unknown) | NCT05474144 | Completed |
| Bioithas SL, London, UK | Probiotics (unknown) | NCT04390477 | Completed |
| Centre hospitalier de l’Université de Montréal (CHUM), Montréal, Canada | Probiotics (unknown) | NCT04458519 | Completed |
| Nordic Biotic Sp. Z o.o., Warsaw, Poland | Probiotics (unknown) | NCT04907877 | Recruiting |
| Université de Sherbrooke, Sherbrooke, Canada | Probiotics (unknown) | NCT05195151 | Not yet recruiting |
| Biosearch S.A., Granada, Spain | Lactobacillus Coryniformis K8 | NCT04366180 | Recruiting |
| ProbiSearch SL, Madrid, Spain | Lactobacillus salivarius + Vitamin D + Zinc | NCT04937556 | Completed |
| Vanderbilt University Medical Center, Nashville, USA | Magnesium Citrate + probiotics (unknown) | NCT04941703 | Recruiting |
| King Edward Medical University, Punjab, Pakistan | Streptococcus salivarius K12 | NCT05043376 | Completed |
| Universidade de Passo Fundo, Passo Fundo, Brazil | Streptococcus salivarius K12; Lactobacillus brevis CD2 | NCT05175833 | Completed |
| Indonesia University, Jakarta, Indonesia | Probiotics (unknown) + Vitamin D | NCT04979065 | Recruiting |
| Biosearch S.A. Granada, Spain | Lactobacillus | NCT04756466 | Active, not recruiting |
| Chinese University of Hong Kong, Hong Kong, China | Microbiome immunity formula (unknown) | NCT04950803 | Recruiting |
| Rutgers, The State University of New Jersey, New Brunswick, USA | A combination of live microbials (unknown) | NCT04847349 | Completed |
| Medical University of Graz, Graz, Austria | Omni-Biotic Pro Vi 5 (unknown probiotics) | NCT04813718 | Active, not recruiting |
| AB Biotek, Barcelona, Spain | Saccharomyces cerevisiae | NCT04798677 | Completed |
| Medical College of Wisconsin, Milwaukee, USA | Lactobacillus Plantarum | NCT05227170 | Recruiting |
| Universidad Complutense de Madrid, Madrid, Spain | Ligilactobacillus salivarius | NCT04922918 | Recruiting |
| AB Biotics, SA, Mexico city, Mexico | Lactobacillus plantarum, Lactobacillus plantarum, Lactobacillus plantarum, Pediococcus acidilactici | NCT04517422 | Completed |
| Duke University, Durham, USA | Lactobacillus rhamnosus | NCT04399252 | Completed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, S.; Gong, M.; Wang, G.; Dua, K.; Xu, J.; Xu, X.; Liu, G. COVID-19 and Gut Injury. Nutrients 2022, 14, 4409. https://doi.org/10.3390/nu14204409
Shen S, Gong M, Wang G, Dua K, Xu J, Xu X, Liu G. COVID-19 and Gut Injury. Nutrients. 2022; 14(20):4409. https://doi.org/10.3390/nu14204409
Chicago/Turabian StyleShen, Sj, Muxue Gong, Gang Wang, Kamal Dua, Jincheng Xu, Xiaoyue Xu, and Gang Liu. 2022. "COVID-19 and Gut Injury" Nutrients 14, no. 20: 4409. https://doi.org/10.3390/nu14204409
APA StyleShen, S., Gong, M., Wang, G., Dua, K., Xu, J., Xu, X., & Liu, G. (2022). COVID-19 and Gut Injury. Nutrients, 14(20), 4409. https://doi.org/10.3390/nu14204409









