Genetic Determinants of 25-Hydroxyvitamin D Concentrations and Their Relevance to Public Health
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genome-Wide Association Studies on 25(OH)D
Variants with Evidence for Replicated Association with 25(OH)D
3.2. Heritability and the Genetic Contribution to the Prevalence of Deficiency
3.3. Genetic Differences in Response to Supplementation and the Need for Personalized Approaches
3.4. Mendelian Randomization to Establish Evidence for Causal Effects of 25(OH)D
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hunter, D.; De Lange, M.; Snieder, H.; MacGregor, A.J.; Swaminathan, R.; Thakker, R.V.; Spector, T.D. Genetic Contribution to Bone Metabolism, Calcium Excretion, and Vitamin D and Parathyroid Hormone Regulation. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2001, 16, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Orton, S.-M.; Morris, A.P.; Herrera, B.M.; Ramagopalan, S.V.; Lincoln, M.R.; Chao, M.J.; Vieth, R.; Sadovnick, A.D.; Ebers, G.C. Evidence for Genetic Regulation of Vitamin D Status in Twins with Multiple Sclerosis. Am. J. Clin. Nutr. 2008, 88, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Shea, M.K.; Benjamin, E.J.; Dupuis, J.; Massaro, J.M.; Jacques, P.F.; D’Agostino, R.B.; Ordovas, J.M.; O’Donnell, C.J.; Dawson-Hughes, B.; Vasan, R.S.; et al. Genetic and Non-Genetic Correlates of Vitamins K and D. Eur. J. Clin. Nutr. 2009, 63, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Wjst, M.; Altmüller, J.; Braig, C.; Bahnweg, M.; André, E. A Genome-Wide Linkage Scan for 25-OH-D(3) and 1,25-(OH)2-D3 Serum Levels in Asthma Families. J. Steroid Biochem. Mol. Biol. 2007, 103, 799–802. [Google Scholar] [CrossRef]
- Berry, D.; Hypponen, E. Determinants of Vitamin D Status: Focus on Genetic Variations. Curr. Opin. Nephrol. Hypertens. 2011, 20, 331–336. [Google Scholar] [CrossRef]
- Jablonski, N.G.; Chaplin, G. The Roles of Vitamin D and Cutaneous Vitamin D Production in Human Evolution and Health. Int. J. Paleopathol. 2018, 23, 54–59. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Ahn, J.; Yu, K.; Stolzenberg-Solomon, R.; Simon, K.C.; McCullough, M.L.; Gallicchio, L.; Jacobs, E.J.; Ascherio, A.; Helzlsouer, K.; Jacobs, K.B.; et al. Genome-Wide Association Study of Circulating Vitamin D Levels. Hum. Mol. Genet. 2010, 19, 2739–2745. [Google Scholar] [CrossRef]
- Engelman, C.D.; Meyers, K.J.; Ziegler, J.T.; Taylor, K.D.; Palmer, N.D.; Haffner, S.M.; Fingerlin, T.E.; Wagenknecht, L.E.; Rotter, J.I.; Bowden, D.W.; et al. Genome-Wide Association Study of Vitamin D Concentrations in Hispanic Americans: The IRAS Family Study. J. Steroid Biochem. Mol. Biol. 2010, 122, 186–192. [Google Scholar] [CrossRef]
- Wang, T.J.; Zhang, F.; Richards, J.B.; Kestenbaum, B.; Van Meurs, J.B.; Berry, D.; Kiel, D.P.; Streeten, E.A.; Ohlsson, C.; Koller, D.L.; et al. Common Genetic Determinants of Vitamin D Insufficiency: A Genome-Wide Association Study. Lancet 2010, 376, 180–188. [Google Scholar] [CrossRef]
- Lasky-Su, J.; Lange, N.; Brehm, J.M.; Damask, A.; Soto-Quiros, M.; Avila, L.; Celedon, J.C.; Canino, G.; Cloutier, M.M.; Hollis, B.W.; et al. Genome-Wide Association Analysis of Circulating Vitamin D Levels in Children with Asthma. Hum. Genet. 2012, 131, 1495–1505. [Google Scholar] [CrossRef]
- Anderson, D.; Holt, B.J.; Pennell, C.E.; Holt, P.G.; Hart, P.H.; Blackwell, J.M. Genome-Wide Association Study of Vitamin D Levels in Children: Replication in the Western Australian Pregnancy Cohort (Raine) Study. Genes Immun. 2014, 15, 578–583. [Google Scholar] [CrossRef]
- Sapkota, B.R.; Hopkins, R.; Bjonnes, A.; Ralhan, S.; Wander, G.S.; Mehra, N.K.; Singh, J.R.; Blackett, P.R.; Saxena, R.; Sanghera, D.K. Genome-Wide Association Study of 25(OH) Vitamin D Concentrations in Punjabi Sikhs: Results of the Asian Indian Diabetic Heart Study. J. Steroid Biochem. Mol. Biol. 2016, 158, 149–156. [Google Scholar] [CrossRef]
- Manousaki, D.; Dudding, T.; Haworth, S.; Hsu, Y.-H.; Liu, C.-T.; Medina-Gomez, C.; Voortman, T.; van der Velde, N.; Melhus, H.; Robinson-Cohen, C.; et al. Low-Frequency Synonymous Coding Variation in CYP2R1 Has Large Effects on Vitamin D Levels and Risk of Multiple Sclerosis. Am. J. Hum. Genet. 2017, 101, 227–238. [Google Scholar] [CrossRef]
- Hong, J.; Hatchell, K.E.; Bradfield, J.P.; Bjonnes, A.; Chesi, A.; Lai, C.-Q.; Langefeld, C.D.; Lu, L.; Lu, Y.; Lutsey, P.L.; et al. Transethnic Evaluation Identifies Low-Frequency Loci Associated with 25-Hydroxyvitamin D Concentrations. J. Clin. Endocrinol. Metab. 2018, 103, 1380–1392. [Google Scholar] [CrossRef]
- Jiang, X.; O’Reilly, P.F.; Aschard, H.; Hsu, Y.-H.; Richards, J.B.; Dupuis, J.; Ingelsson, E.; Karasik, D.; Pilz, S.; Berry, D.; et al. Genome-Wide Association Study in 79,366 European-Ancestry Individuals Informs the Genetic Architecture of 25-Hydroxyvitamin D Levels. Nat. Commun. 2018, 9, 260. [Google Scholar] [CrossRef]
- O’Brien, K.M.; Shi, M.; Weinberg, C.R.; Sandler, D.P.; Harmon, Q.E.; Taylor, J.A. Genome-Wide Association Study of Serum 25-Hydroxyvitamin D in US Women. Front. Genet. 2018, 9, 67. [Google Scholar] [CrossRef]
- Kampe, A.; Enlund-Cerullo, M.; Valkama, S.; Holmlund-Suila, E.; Rosendahl, J.; Hauta-Alus, H.; Pekkinen, M.; Andersson, S.; Makitie, O. Genetic Variation in GC and CYP2R1 Affects 25-Hydroxyvitamin D Concentration and Skeletal Parameters: A Genome-Wide Association Study in 24-Month-Old Finnish Children. PLoS Genet. 2019, 15, e1008530. [Google Scholar] [CrossRef]
- Manousaki, D.; Mitchell, R.; Dudding, T.; Haworth, S.; Harroud, A.; Forgetta, V.; Shah, R.L.; Luan, J.; Langenberg, C.; Timpson, N.J.; et al. Genome-Wide Association Study for Vitamin D Levels Reveals 69 Independent Loci. Am. J. Hum. Genet. 2020, 106, 327–337. [Google Scholar] [CrossRef]
- Revez, J.A.; Lin, T.; Qiao, Z.; Xue, A.; Holtz, Y.; Zhu, Z.; Zeng, J.; Wang, H.; Sidorenko, J.; Kemper, K.E.; et al. Genome-Wide Association Study Identifies 143 Loci Associated with 25 Hydroxyvitamin D Concentration. Nat. Commun. 2020, 11, 1647. [Google Scholar] [CrossRef]
- Traglia, M.; Windham, G.C.; Pearl, M.; Poon, V.; Eyles, D.; Jones, K.L.; Lyall, K.; Kharrazi, M.; Croen, L.A.; Weiss, L.A. Genetic Contributions to Maternal and Neonatal Vitamin D Levels. Genetics 2020, 214, 1091–1102. [Google Scholar] [CrossRef]
- Zheng, J.-S.; Luan, J.; Sofianopoulou, E.; Sharp, S.J.; Day, F.R.; Imamura, F.; Gundersen, T.E.; Lotta, L.A.; Sluijs, I.; Stewart, I.D.; et al. The Association between Circulating 25-Hydroxyvitamin D Metabolites and Type 2 Diabetes in European Populations: A Meta-Analysis and Mendelian Randomisation Analysis. PLoS Med. 2020, 17, e1003394. [Google Scholar] [CrossRef]
- Kim, Y.A.; Yoon, J.W.; Lee, Y.; Choi, H.J.; Yun, J.W.; Bae, E.; Kwon, S.-H.; Ahn, S.E.; Do, A.-R.; Jin, H.; et al. Unveiling Genetic Variants Underlying Vitamin D Deficiency in Multiple Korean Cohorts by a Genome-Wide Association Study. Endocrinol. Metab. 2021, 36, 1189–1200. [Google Scholar] [CrossRef]
- Ong, J.-S.; Dixon-Suen, S.C.; Han, X.; An, J.; Fitzgerald, R.; Buas, M.; Gammon, M.D.; Corley, D.A.; Shaheen, N.J.; Hardie, L.J.; et al. A Comprehensive Re-Assessment of the Association between Vitamin D and Cancer Susceptibility Using Mendelian Randomization. Nat. Commun. 2021, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.D.; Lu, L.; Register, T.C.; Lenchik, L.; Carr, J.J.; Hicks, P.J.; Smith, S.C.; Xu, J.; Dimitrov, L.; Keaton, J.; et al. Genome-Wide Association Study of Vitamin D Concentrations and Bone Mineral Density in the African American-Diabetes Heart Study. PLoS ONE 2021, 16, e0251423. [Google Scholar] [CrossRef] [PubMed]
- Sallinen, R.J.; Dethlefsen, O.; Ruotsalainen, S.; Mills, R.D.; Miettinen, T.A.; Jääskeläinen, T.E.; Lundqvist, A.; Kyllönen, E.; Kröger, H.; Karppinen, J.I.; et al. Genetic Risk Score for Serum 25-Hydroxyvitamin D Concentration Helps to Guide Personalized Vitamin D Supplementation in Healthy Finnish Adults. J. Nutr. 2021, 151, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Sampathkumar, A.; Tan, K.M.; Chen, L.; Chong, M.F.F.; Yap, F.; Godfrey, K.M.; Chong, Y.S.; Gluckman, P.D.; Ramasamy, A.; Karnani, N. Genetic Link Determining the Maternal-Fetal Circulation of Vitamin D. Front. Genet. 2021, 12, 721488. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Ge, J.; Xu, W.; Ma, H.; Chen, L.; Xia, M.; Pan, B.; Lin, H.; Wang, S.; Gao, X. Type 2 Diabetes Is Causally Associated With Reduced Serum Osteocalcin: A Genomewide Association and Mendelian Randomization Study. J. Bone Miner. Res. 2021, 36, 1694–1707. [Google Scholar] [CrossRef]
- Backman, J.D.; Li, A.H.; Marcketta, A.; Sun, D.; Mbatchou, J.; Kessler, M.D.; Benner, C.; Liu, D.; Locke, A.E.; Balasubramanian, S.; et al. Exome Sequencing and Analysis of 454,787 UK Biobank Participants. Nature 2021, 599, 628–634. [Google Scholar] [CrossRef]
- Barton, A.R.; Sherman, M.A.; Mukamel, R.E.; Loh, P.-R. Whole-Exome Imputation within UK Biobank Powers Rare Coding Variant Association and Fine-Mapping Analyses. Nat. Genet. 2021, 53, 1260–1269. [Google Scholar] [CrossRef]
- Benjamin, E.J.; Dupuis, J.; Larson, M.G.; Lunetta, K.L.; Booth, S.L.; Govindaraju, D.R.; Kathiresan, S.; Keaney, J.F.; Keyes, M.J.; Lin, J.-P.; et al. Genome-Wide Association with Select Biomarker Traits in the Framingham Heart Study. BMC Med. Genet. 2007, 8 (Suppl. 1), S11. [Google Scholar] [CrossRef]
- Locke, A.E.; Steinberg, K.M.; Chiang, C.W.K.; Service, S.K.; Havulinna, A.S.; Stell, L.; Pirinen, M.; Abel, H.J.; Chiang, C.C.; Fulton, R.S.; et al. Exome Sequencing of Finnish Isolates Enhances Rare-Variant Association Power. Nature 2019, 572, 323–328. [Google Scholar] [CrossRef]
- Mbatchou, J.; Barnard, L.; Backman, J.; Marcketta, A.; Kosmicki, J.A.; Ziyatdinov, A.; Benner, C.; O’Dushlaine, C.; Barber, M.; Boutkov, B.; et al. Computationally Efficient Whole-Genome Regression for Quantitative and Binary Traits. Nat. Genet. 2021, 53, 1097–1103. [Google Scholar] [CrossRef]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 Blood and Urine Biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194. [Google Scholar] [CrossRef]
- Sun, Q.; Graff, M.; Rowland, B.; Wen, J.; Huang, L.; Miller-Fleming, T.W.; Haessler, J.; Preuss, M.H.; Chai, J.-F.; Lee, M.P.; et al. Analyses of Biomarker Traits in Diverse UK Biobank Participants Identify Associations Missed by European-Centric Analysis Strategies. J. Hum. Genet. 2022, 67, 87–93. [Google Scholar] [CrossRef]
- Richardson, T.G.; O’Nunain, K.; Relton, C.L.; Smith, G.D. Harnessing Whole Genome Polygenic Risk Scores to Stratify Individuals Based on Cardiometabolic Risk Factors and Biomarkers at Age 10 in the Lifecourse. Arterioscler. Thromb. Vasc. Biol. 2022, 42, ATVBAHA121316650. [Google Scholar] [CrossRef]
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, Molecular Mechanism of Action, and Pleiotropic Effects. Physiol. Rev. 2016, 96, 365–408. [Google Scholar] [CrossRef]
- Aiba, I.; Yamasaki, T.; Shinki, T.; Izumi, S.; Yamamoto, K.; Yamada, S.; Terato, H.; Ide, H.; Ohyama, Y. Characterization of Rat and Human CYP2J Enzymes as Vitamin D 25-Hydroxylases. Steroids 2006, 71, 849–856. [Google Scholar] [CrossRef]
- Zhou, A.; Selvanayagam, J.B.; Hyppönen, E. Non-Linear Mendelian Randomization Analyses Support a Role for Vitamin D Deficiency in Cardiovascular Disease Risk. Eur. Heart J. 2022, 43, 1731–1739. [Google Scholar] [CrossRef]
- Collins, C.S.; Gould, S.J. Identification of a Common PEX1 Mutation in Zellweger Syndrome. Hum. Mutat. 1999, 14, 45–53. [Google Scholar] [CrossRef]
- Rush, E.T.; Goodwin, J.L.; Braverman, N.E.; Rizzo, W.B. Low Bone Mineral Density Is a Common Feature of Zellweger Spectrum Disorders. Mol. Genet. Metab. 2016, 117, 33–37. [Google Scholar] [CrossRef]
- Guerrin, M.; Ishigami, A.; Méchin, M.-C.; Nachat, R.; Valmary, S.; Sebbag, M.; Simon, M.; Senshu, T.; Serre, G. CDNA Cloning, Gene Organization and Expression Analysis of Human Peptidylarginine Deiminase Type I. Biochem. J. 2003, 370, 167–174. [Google Scholar] [CrossRef]
- Méchin, M.C.; Enji, M.; Nachat, R.; Chavanas, S.; Charveron, M.; Ishida-Yamamoto, A.; Serre, G.; Takahara, H.; Simon, M. The Peptidylarginine Deiminases Expressed in Human Epidermis Differ in Their Substrate Specificities and Subcellular Locations. Cell. Mol. Life Sci. CMLS 2005, 62, 1984–1995. [Google Scholar] [CrossRef]
- Jacobs, F.M.J.; van der Heide, L.P.; Wijchers, P.J.E.C.; Burbach, J.P.H.; Hoekman, M.F.M.; Smidt, M.P. FoxO6, a Novel Member of the FoxO Class of Transcription Factors with Distinct Shuttling Dynamics *. J. Biol. Chem. 2003, 278, 35959–35967. [Google Scholar] [CrossRef]
- Zemva, J.; Schilbach, K.; Stöhr, O.; Moll, L.; Franko, A.; Krone, W.; Wiesner, R.J.; Schubert, M. Central FoxO3a and FoxO6 Expression Is Down-Regulated in Obesity Induced Diabetes but Not in Aging. Exp. Clin. Endocrinol. Diabetes Off. J. Ger. Soc. Endocrinol. Ger. Diabetes Assoc. 2012, 120, 340–350. [Google Scholar] [CrossRef]
- Vincent, J.B.; Skaug, J.; Scherer, S.W. The Human Homologue of Flamingo, EGFL2, Encodes a Brain-Expressed Large Cadherin-like Protein with Epidermal Growth Factor-like Domains, and Maps to Chromosome 1p13.3-P21.1. DNA Res. 2000, 7, 233–235. [Google Scholar] [CrossRef][Green Version]
- Ichthyosis Vulgaris Disease: Malacards—Research Articles, Drugs, Genes, Clinical Trials. Available online: https://www.malacards.org/card/ichthyosis_vulgaris (accessed on 27 May 2022).
- Peeling Skin Syndrome 6 Disease: Malacards—Research Articles, Drugs, Genes, Clinical Trials. Available online: https://www.malacards.org/card/peeling_skin_syndrome_6 (accessed on 27 May 2022).
- Kukita, T.; Wada, N.; Kukita, A.; Kakimoto, T.; Sandra, F.; Toh, K.; Nagata, K.; Iijima, T.; Horiuchi, M.; Matsusaki, H.; et al. RANKL-Induced DC-STAMP Is Essential for Osteoclastogenesis. J. Exp. Med. 2004, 200, 941–946. [Google Scholar] [CrossRef]
- Van der Valk, R.J.P.; Kreiner-Møller, E.; Kooijman, M.N.; Guxens, M.; Stergiakouli, E.; Sääf, A.; Bradfield, J.P.; Geller, F.; Hayes, M.G.; Cousminer, D.L.; et al. A Novel Common Variant in DCST2 Is Associated with Length in Early Life and Height in Adulthood. Hum. Mol. Genet. 2015, 24, 1155–1168. [Google Scholar] [CrossRef]
- Borges, C.R.; Jarvis, J.W.; Oran, P.E.; Nelson, R.W. Population Studies of Vitamin D Binding Protein Microheterogeneity by Mass Spectrometry Lead to Characterization of Its Genotype-Dependent O-Glycosylation Patterns. J. Proteome Res. 2008, 7, 4143–4153. [Google Scholar] [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef]
- Ghatnatti, V.; Vastrad, B.; Patil, S.; Vastrad, C.; Kotturshetti, I. Identification of Potential and Novel Target Genes in Pituitary Prolactinoma by Bioinformatics Analysis. AIMS Neurosci. 2021, 8, 254–283. [Google Scholar] [CrossRef] [PubMed]
- PubChem CPS1—Carbamoyl-Phosphate Synthase 1 (Human). Available online: https://pubchem.ncbi.nlm.nih.gov/gene/CPS1/human (accessed on 26 May 2022).
- Meech, R.; Hu, D.G.; McKinnon, R.A.; Mubarokah, S.N.; Haines, A.Z.; Nair, P.C.; Rowland, A.; Mackenzie, P.I. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol. Rev. 2019, 99, 1153–1222. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wong, T.; Hashizume, T.; Dickmann, L.Z.; Scian, M.; Koszewski, N.J.; Goff, J.P.; Horst, R.L.; Chaudhry, A.S.; Schuetz, E.G.; et al. Human UGT1A4 and UGT1A3 Conjugate 25-Hydroxyvitamin D3: Metabolite Structure, Kinetics, Inducibility, and Interindividual Variability. Endocrinology 2014, 155, 2052–2063. [Google Scholar] [CrossRef]
- Biederer, T. Bioinformatic Characterization of the SynCAM Family of Immunoglobulin-like Domain-Containing Adhesion Molecules. Genomics 2006, 87, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wang, Z.; Schmidt, V.; Gauert, A.; Willnow, T.E.; Heinig, M.; Poy, M.N. Cadm2 Regulates Body Weight and Energy Homeostasis in Mice. Mol. Metab. 2018, 8, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Chun, R.F. New Perspectives on the Vitamin D Binding Protein. Cell Biochem. Funct. 2012, 30, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Niederstrasser, H.; Edwards, M.; Jackson, C.E.; Cooper, J.A. Distinct Roles for CARMIL Isoforms in Cell Migration. Mol. Biol. Cell 2009, 20, 5290–5305. [Google Scholar] [CrossRef] [PubMed]
- Canu, N.; Possenti, R.; Ricco, A.S.; Rocchi, M.; Levi, A. Cloning, Structural Organization Analysis, and Chromosomal Assignment of the Human Gene for the Neurosecretory Protein VGF. Genomics 1997, 45, 443–446. [Google Scholar] [CrossRef]
- Benchoula, K.; Parhar, I.S.; Hwa, W.E. The Molecular Mechanism of Vgf in Appetite, Lipids, and Insulin Regulation. Pharmacol. Res. 2021, 172, 105855. [Google Scholar] [CrossRef]
- Hahm, S.; Fekete, C.; Mizuno, T.M.; Windsor, J.; Yan, H.; Boozer, C.N.; Lee, C.; Elmquist, J.K.; Lechan, R.M.; Mobbs, C.V.; et al. VGF Is Required for Obesity Induced by Diet, Gold Thioglucose Treatment, and Agouti and Is Differentially Regulated in Pro-Opiomelanocortin- and Neuropeptide Y-Containing Arcuate Neurons in Response to Fasting. J. Neurosci. 2002, 22, 6929–6938. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Qin, Z.; Wei, Y.; Deng, Z.; Zhu, C.; Tang, J.; Ma, L. High LINC00536 Expression Promotes Tumor Progression and Poor Prognosis in Bladder Cancer. Exp. Cell Res. 2019, 378, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Fossati, M.; Assendorp, N.; Gemin, O.; Colasse, S.; Dingli, F.; Arras, G.; Loew, D.; Charrier, C. Trans-Synaptic Signaling through the Glutamate Receptor Delta-1 Mediates Inhibitory Synapse Formation in Cortical Pyramidal Neurons. Neuron 2019, 104, 1081–1094.e7. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.B.; Levine, M.A.; Bell, N.H.; Mangelsdorf, D.J.; Russell, D.W. Genetic Evidence That the Human CYP2R1 Enzyme Is a Key Vitamin D 25-Hydroxylase. Proc. Natl. Acad. Sci. USA 2004, 101, 7711–7715. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Y.; Liu, S.; Song, X.; Yang, X.; Fan, Y.; Chen, W.; Akdemir, Z.C.; Yan, Z.; Zuo, Y.; et al. The Coexistence of Copy Number Variations (CNVs) and Single Nucleotide Polymorphisms (SNPs) at a Locus Can Result in Distorted Calculations of the Significance in Associating SNPs to Disease. Hum. Genet. 2018, 137, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.-Y.; Bettany-Saltikov, J.; Cheung, I.Y.K.; Chan, K.K.Y. The Role of Vitamin D in the Pathogenesis of Adolescent Idiopathic Scoliosis. Asian Spine J. 2018, 12, 1127–1145. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed]
- Cases, S.; Stone, S.J.; Zhou, P.; Yen, E.; Tow, B.; Lardizabal, K.D.; Voelker, T.; Farese, R.V. Cloning of DGAT2, a Second Mammalian Diacylglycerol Acyltransferase, and Related Family Members. J. Biol. Chem. 2001, 276, 38870–38876. [Google Scholar] [CrossRef]
- Smith, S.J.; Cases, S.; Jensen, D.R.; Chen, H.C.; Sande, E.; Tow, B.; Sanan, D.A.; Raber, J.; Eckel, R.H.; Farese, R.V. Obesity Resistance and Multiple Mechanisms of Triglyceride Synthesis in Mice Lacking Dgat. Nat. Genet. 2000, 25, 87–90. [Google Scholar] [CrossRef]
- Eckhart, L.; Schmidt, M.; Mildner, M.; Mlitz, V.; Abtin, A.; Ballaun, C.; Fischer, H.; Mrass, P.; Tschachler, E. Histidase Expression in Human Epidermal Keratinocytes: Regulation by Differentiation Status and All-Trans Retinoic Acid. J. Dermatol. Sci. 2008, 50, 209–215. [Google Scholar] [CrossRef]
- Suchi, M.; Sano, H.; Mizuno, H.; Wada, Y. Molecular Cloning and Structural Characterization of the Human Histidase Gene (HAL). Genomics 1995, 29, 98–104. [Google Scholar] [CrossRef]
- Welsh, M.M.; Karagas, M.R.; Applebaum, K.M.; Spencer, S.K.; Perry, A.E.; Nelson, H.H. A Role for Ultraviolet Radiation Immunosuppression in Non-Melanoma Skin Cancer as Evidenced by Gene-Environment Interactions. Carcinogenesis 2008, 29, 1950–1954. [Google Scholar] [CrossRef]
- Landeck, L.; Jakasa, I.; Dapic, I.; Lutter, R.; Thyssen, J.P.; Skov, L.; Braun, A.; Schön, M.P.; John, S.M.; Kezic, S.; et al. The Effect of Epidermal Levels of Urocanic Acid on 25-Hydroxyvitamin D Synthesis and Inflammatory Mediators upon Narrowband UVB Irradiation. Photodermatol. Photoimmunol. Photomed. 2016, 32, 214–223. [Google Scholar] [CrossRef]
- Simões-costa, M.S.; Azambuja, A.P.; Xavier-Neto, J. The search for non-chordate retinoic acid signaling: Lessons from chordates. J. Exp. Zoolog. B Mol. Dev. Evol. 2008, 310B, 54–72. [Google Scholar] [CrossRef]
- Van Lith, M.; Hartigan, N.; Hatch, J.; Benham, A.M. PDILT, a Divergent Testis-Specific Protein Disulfide Isomerase with a Non-Classical SXXC Motif That Engages in Disulfide-Dependent Interactions in the Endoplasmic Reticulum. J. Biol. Chem. 2005, 280, 1376–1383. [Google Scholar] [CrossRef]
- Kurogi, K.; Sakakibara, Y.; Suiko, M.; Liu, M.-C. Sulfation of Vitamin D3 -Related Compounds-Identification and Characterization of the Responsible Human Cytosolic Sulfotransferases. FEBS Lett. 2017, 591, 2417–2425. [Google Scholar] [CrossRef]
- Wong, T.; Wang, Z.; Chapron, B.D.; Suzuki, M.; Claw, K.G.; Gao, C.; Foti, R.S.; Prasad, B.; Chapron, A.; Calamia, J.; et al. Polymorphic Human Sulfotransferase 2A1 Mediates the Formation of 25-Hydroxyvitamin D3-3-O-Sulfate, a Major Circulating Vitamin D Metabolite in Humans. Drug Metab. Dispos. 2018, 46, 367–379. [Google Scholar] [CrossRef]
- Prassas, I.; Eissa, A.; Poda, G.; Diamandis, E.P. Unleashing the Therapeutic Potential of Human Kallikrein-Related Serine Proteases. Nat. Rev. Drug Discov. 2015, 14, 183–202. [Google Scholar] [CrossRef]
- Jones, G.; Prosser, D.E.; Kaufmann, M. 25-Hydroxyvitamin D-24-Hydroxylase (CYP24A1): Its Important Role in the Degradation of Vitamin D. Arch. Biochem. Biophys. 2012, 523, 9–18. [Google Scholar] [CrossRef]
- Shi, M.; Grabner, A.; Wolf, M. Importance of Extra-Renal CYP24A1 Expression for Maintaining Mineral Homeostasis. J. Endocr. Soc. 2021, 5, A234. [Google Scholar] [CrossRef]
- Murase, R.; Taketomi, Y.; Miki, Y.; Nishito, Y.; Saito, M.; Fukami, K.; Yamamoto, K.; Murakami, M. Group III Phospholipase A2 Promotes Colitis and Colorectal Cancer. Sci. Rep. 2017, 7, 12261. [Google Scholar] [CrossRef]
- Karohl, C.; Su, S.; Kumari, M.; Tangpricha, V.; Veledar, E.; Vaccarino, V.; Raggi, P. Heritability and Seasonal Variability of Vitamin D Concentrations in Male Twins. Am. J. Clin. Nutr. 2010, 92, 1393–1398. [Google Scholar] [CrossRef]
- Engelman, C.D.; Fingerlin, T.E.; Langefeld, C.D.; Hicks, P.J.; Rich, S.S.; Wagenknecht, L.E.; Bowden, D.W.; Norris, J.M. Genetic and Environmental Determinants of 25-Hydroxyvitamin D and 1,25-Dihydroxyvitamin D Levels in Hispanic and African Americans. J. Clin. Endocrinol. Metab. 2008, 93, 3381–3388. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.P.; Zhou, A.; Leach, M.J.; Hyppönen, E. Differences and Determinants of Vitamin D Deficiency among UK Biobank Participants: A Cross-Ethnic and Socioeconomic Study. Clin. Nutr. 2021, 40, 3436–3447. [Google Scholar] [CrossRef] [PubMed]
- Enlund-Cerullo, M.; Koljonen, L.; Holmlund-Suila, E.; Hauta-alus, H.; Rosendahl, J.; Valkama, S.; Helve, O.; Hytinantti, T.; Viljakainen, H.; Andersson, S.; et al. Genetic Variation of the Vitamin D Binding Protein Affects Vitamin D Status and Response to Supplementation in Infants. J. Clin. Endocrinol. Metab. 2019, 104, 5483–5498. [Google Scholar] [CrossRef] [PubMed]
- Nissen, J.; Vogel, U.; Ravn-Haren, G.; Andersen, E.W.; Madsen, K.H.; Nexø, B.A.; Andersen, R.; Mejborn, H.; Bjerrum, P.J.; Rasmussen, L.B.; et al. Common Variants in CYP2R1 and GC Genes Are Both Determinants of Serum 25-Hydroxyvitamin D Concentrations after UVB Irradiation and after Consumption of Vitamin D₃-Fortified Bread and Milk during Winter in Denmark. Am. J. Clin. Nutr. 2015, 101, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Nissen, J.; Vogel, U.; Ravn-Haren, G.; Andersen, E.W.; Nexø, B.A.; Andersen, R.; Mejborn, H.; Madsen, K.H.; Rasmussen, L.B. Real-Life Use of Vitamin D3-Fortified Bread and Milk during a Winter Season: The Effects of CYP2R1 and GC Genes on 25-Hydroxyvitamin D Concentrations in Danish Families, the VitmaD Study. Genes Nutr. 2014, 9, 413. [Google Scholar] [CrossRef] [PubMed]
- Frost, P. The Problem of Vitamin D Scarcity: Cultural and Genetic Solutions by Indigenous Arctic and Tropical Peoples. Nutrients 2022, 14, 4071. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Smith, G.D. Reading Mendelian Randomisation Studies: A Guide, Glossary, and Checklist for Clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Burgess, S.; Davey Smith, G.; Davies, N.; Dudbridge, F.; Gill, D.; Glymour, M.; Hartwig, F.; Holmes, M.; Minelli, C.; Relton, C.; et al. Guidelines for Performing Mendelian Randomization Investigations [Version 2; Peer Review: 2 Approved]. Wellcome Open Res. 2020, 4, 186. [Google Scholar] [CrossRef]
- Mokry, L.E.; Ross, S.; Ahmad, O.S.; Forgetta, V.; Smith, G.D.; Leong, A.; Greenwood, C.M.T.; Thanassoulis, G.; Richards, J.B. Vitamin D and Risk of Multiple Sclerosis: A Mendelian Randomization Study. PLOS Med. 2015, 12, e1001866. [Google Scholar] [CrossRef]
- Lu, L.; Bennett, D.A.; Millwood, I.Y.; Parish, S.; McCarthy, M.I.; Mahajan, A.; Lin, X.; Bragg, F.; Guo, Y.; Holmes, M.V.; et al. Association of Vitamin D with Risk of Type 2 Diabetes: A Mendelian Randomisation Study in European and Chinese Adults. PLOS Med. 2018, 15, e1002566. [Google Scholar] [CrossRef]
- Vimaleswaran, K.S.; Cavadino, A.; Berry, D.J.; Jorde, R.; Dieffenbach, A.K.; Lu, C.; Alves, A.C.; Heerspink, H.J.L.; Tikkanen, E.; Eriksson, J.; et al. Association of Vitamin D Status with Arterial Blood Pressure and Hypertension Risk: A Mendelian Randomisation Study. Lancet Diabetes Endocrinol. 2014, 2, 719–729. [Google Scholar] [CrossRef]
- Morris, M.C.; Tangney, C.C. A Potential Design Flaw of Randomized Trials of Vitamin Supplements. JAMA 2011, 305, 1348–1349. [Google Scholar] [CrossRef]
- Scragg, R. Limitations of Vitamin D Supplementation Trials: Why Observational Studies Will Continue to Help Determine the Role of Vitamin D in Health. J. Steroid Biochem. Mol. Biol. 2018, 177, 6–9. [Google Scholar] [CrossRef]
- Sutherland, J.P.; Zhou, A.; Hyppönen, E. Vitamin D Deficiency Increases Mortality Risk in the UK Biobank: A Non-Linear Mendelian Randomization Study. Ann. Intern. Med. 2022, in press. [Google Scholar]
- Sofianopoulou, E.; Kaptoge, S.K.; Afzal, S.; Jiang, T.; Gill, D.; Gundersen, T.E.; Bolton, T.R.; Allara, E.; Arnold, M.G.; Mason, A.M.; et al. Estimating Dose-Response Relationships for Vitamin D with Coronary Heart Disease, Stroke, and All-Cause Mortality: Observational and Mendelian Randomisation Analyses. Lancet Diabetes Endocrinol. 2021, 9, 837–846. [Google Scholar] [CrossRef]
- Navale, S.S.; Mulugeta, A.; Zhou, A.; Llewellyn, D.J.; Hyppönen, E. Vitamin D and Brain Health: An Observational and Mendelian Randomization Study. Am. J. Clin. Nutr. 2022, 116, nqac107. [Google Scholar] [CrossRef]
- Mokry, L.E.; Ross, S.; Forgetta, V.; Morris, J.A.; Manousaki, D.; Richards, J.B. Genetically Decreased Vitamin D and Risk of Alzheimer Disease. Neurology 2016, 87, 2567–2574. [Google Scholar] [CrossRef]
- Wang, L.; Qiao, Y.; Zhang, H.; Zhang, Y.; Hua, J.; Jin, S.; Liu, G. Circulating Vitamin D Levels and Alzheimer’s Disease: A Mendelian Randomization Study in the IGAP and UK Biobank. J. Alzheimers Dis. JAD 2020, 73, 609–618. [Google Scholar] [CrossRef]
- Larsson, S.C.; Traylor, M.; Malik, R.; Dichgans, M.; Burgess, S.; Markus, H.S. Modifiable Pathways in Alzheimer’s Disease: Mendelian Randomisation Analysis. BMJ 2017, 359, j5375. [Google Scholar] [CrossRef]
- Bjelakovic, G.; Gluud, L.L.; Nikolova, D.; Whitfield, K.; Wetterslev, J.; Simonetti, R.G.; Bjelakovic, M.; Gluud, C. Vitamin D Supplementation for Prevention of Mortality in Adults. Cochrane Database Syst. Rev. 2014, CD007470. [Google Scholar] [CrossRef]
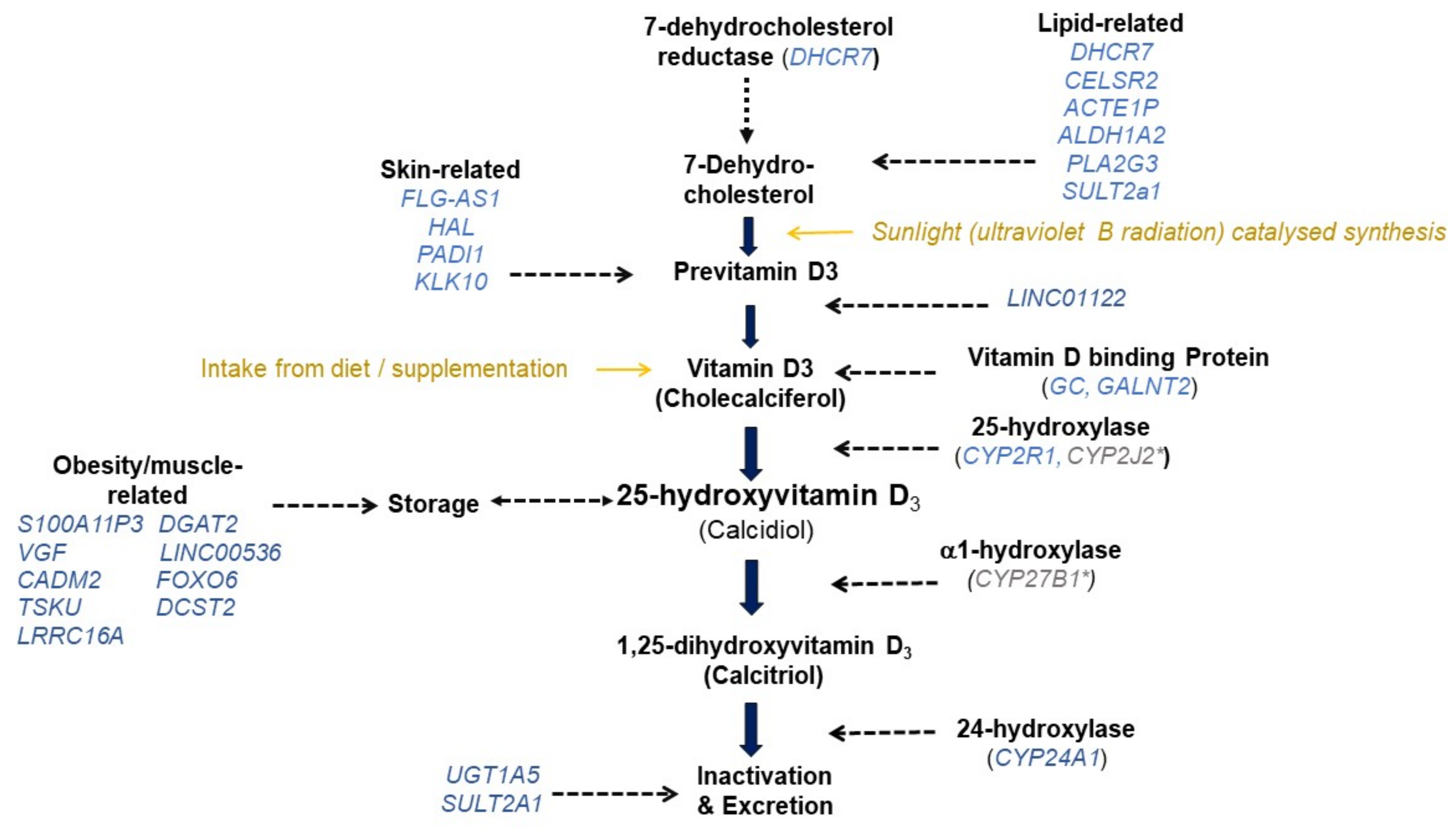
| Gene (SNP) | CHR | |
|---|---|---|
| PEX10 (rs6671730) | 1 | PEX10 encodes a protein involved in import of peroxisomal matrix proteins. Mutations in PEX10 gene have led to Zellweger syndrome [40] and osteopenia [41], for which vitamin D supplementation has been the treatment. |
| PADI1 (rs35408430) | 1 | PADI1 encodes an enzyme, which catalyses the post-translational deimination of proteins by converting arginine residues into citrullines in the presence of calcium ions [42]. Deimination by PADIs occurs during epidermal differentiation [43], with possible influence on skin properties [20]. |
| FOXO6 (rs7522116) | 1 | FOXO6 encodes a protein that has been predicted to enable DNA-binding transcription factor activity, and RNA polymerase related DNA binding activity [44]. FoxO6 expression is downregulated in the brain of dietary obese mice [45]. |
| CELSR2 (rs7528419) | 1 | CELSR2 encodes the cadherin EGF LAG seven-pass G-type receptor 2 that is involved in contact-mediated communication, with cadherin domains acting as homophilic binding regions and the EGF-like domains involved in cell adhesion and receptor-ligand interactions [46]. |
| FLG-AS1 (rs1933064) | 1 | FLG antisense RNA 1 (FLG-AS1) is an RNA Gene that is affiliated with the long non-coding RNA class. Skin pigmentation-related diseases such as Ichthyosis Vulgaris [47] and Peeling Skin Syndrome 6 [48] have been shown to be associated with FLG-AS1. |
| DCST2 (rs76798800) | 1 | DCST2 gene encodes the DC-STAMP domain containing 2 protein that has been shown to be an important regulator of osteoclast cell-fusion in bone homeostasis [49]. DCST2 gene is associated with early length and adult height [50]. |
| GALNT2 (rs6672758) | 1 | GALNT2 gene encodes the polypeptide N-acetylgalactosaminyltransferase 2 which is a member of the glycosyltransferase 2 protein family and which has been linked to post-translational modification of vitamin D-binding protein [51]. |
| LINC01122 (rs727857) | 2 | LINC01122 gene is an RNA gene that is affiliated with the lncRNA class [52]. LINC01122 was one of the 989 differentially expressed genes which was significantly enriched in vitamin D3 biosynthesis [53]. |
| CPS1 (rs1047891) | 2 | CPS1 gene encodes the carbamoyl-phosphate synthase 1 which is a mitochondrial enzyme that catalyses synthesis of carbamoyl phosphate from ammonia and bicarbonate [54]. |
| UGT1A5 * (rs2012736) | 2 | UGT1A5 gene encodes the UDP glucuronosyltransferase family 1 member A5 which has been shown to transform small lipophilic molecules, into water-soluble, excretable metabolites [55]. Related isoenzymes have been identified as catalysts for 25(OH)D3 glucuronidation in the human liver [56]. |
| CADM2 (rs6782190) | 3 | CADM2 gene encodes the cell adhesion molecule 2 which is a member of the synaptic cell adhesion molecule 1 family [57]. In animal studies, CADM2 is associated with metabolic traits [58], suggesting possible influence on vitamin D concentrations through its effect on obesity and storage capacity of 25(OH)D. |
| GC (rs705117, rs1352846) | 4 | GC gene encodes the vitamin D binding protein which binds to vitamin D and its plasma metabolites and transports them to target tissues [59]. |
| CARMIL1/LRRC16A (rs78151190) | 6 | CARMIL1 gene encodes the capping protein regulator and myosin 1 linker 1 with a role in actin filament network formation [60]. Approximately 10% of muscle tissue consists of actin, providing a possible link with 25(OH)D through storage capacity. |
| VGF (rs75741381) | 7 | VGF gene encodes a protein that is expressed in neuroendocrine cells and is upregulated by nerve growth factor [61]. VGF has been linked with appetite control [62], and diet-induced obesity [63], with a possible link through storage capacity. |
| LINC00536 (rs12056768) | 8 | LINC00536 gene interacts with Wnt3a/β-Catenin signalling [64]. Wnt/β -Catenin signalling is an important signalling pathway in regulating adipose tissue lipogenesis with a possible link with 25(O)D through storage capacity. |
| GRID1 (rs77532868) | 10 | GRID1 gene encodes the glutamate ionotropic receptor delta type subunit 1 which is a subunit of glutamate receptor channels that mediate the fast excitatory synaptic transmission in the central nervous system [65]. |
| CYP2R1 (rs12794714) | 11 | CYP2R1 gene encodes the cytochrome P450 family 2 subfamily R member 1 which acts as 25-hydroxylase of vitamin D [66]. |
| TMEM151A (rs61891388) | 11 | TMEM151A has been predicted to be an integral component of membrane and CD248 enables extracellular matrix binding activity and regulates endothelial cell apoptotic process. |
| AP002387.1/ACTE1P (rs1660839, rs12803256) | 11 | ACTE1P gene is an RNA gene. ACTE1P [67] and vitamin D [68] are both involved in adolescent idiopathic scoliosis (abnormal curvature of the spine), suggesting a possible role of ACTE1P in bone health. |
| S100A11P3 (rs12798050) | 11 | S100A11P3 gene encodes the S100 calcium binding protein A11 pseudogene 3. It has multiple roles in buffering calcium ion concentration, participating in energy metabolism, regulating cell proliferation and differentiation [69]. |
| DGAT2 (rs72997623) | 11 | DGAT2 encodes the diacylglycerol O-acyltransferase 2, catalysing the synthesis of triglycerides [70]. Affects adipose tissue formation [71] with possible link to 25(OH)D storage. |
| GUCY2EP/TSKU (rs1149605) | 11 | GUCY2EP gene encodes guanylate cyclase 2E that is involved in chemosensation and TSKU gene encodes tsukushi, small leucine rich proteoglycan that has been predicted to act upstream/within several processes, including negative regulation of Wnt signaling pathway. |
| HAL (rs10859995) | 12 | HAL gene is upregulated during the differentiation of keratinocytes [72]. HAL deaminates L-histidine to trans-uronic acid [73], which in the stratum corneum absorbs UVB [74] and reduce the production 25(OH)D [75]. |
| SEC23A (rs8018720) | 14 | SEC23A gene encodes the Sec23 homolog A, coat complex II component which plays a role in the ER-Golgi protein trafficking. |
| ALDH1A2 (rs261291) | 15 | ALDH1A2 gene encodes aldehyde dehydrogenase 1 family member A2 which catalyses the synthesis of retinoic acid (RA) from retinaldehyde [76]. |
| PDILT (rs77924615) | 16 | PDILT/PDIA7 gene encodes the protein disulphide isomerase like, testis expressed which catalyses protein folding and thiol-disulphide interchange reactions [77]. |
| SULT2A1 (rs212100) | 19 | SULT2A1 gene encodes a liver- and intestine-expressed sulpho-conjugating enzyme that is responsible for the inactivation by sulphonation of 25(OH)D [78,79]. |
| KLK10 (rs10426) | 19 | KLK10 gene encodes the kallikrein related peptidase 10 that has been shown to play a role in dermal integrity [80]. |
| CYP24A1† | 20 | CYP24A1 gene encodes cytochrome P450 family 24 subfamily A member 1 which is an important candidate for vitamin D metabolic pathway given that it initiates the degradation of 1,25-dihydroxyvitamin D3 by hydroxylation of the side chain [81]. In addition, this enzyme also plays a role in calcium homeostasis and vitamin D endocrine system [82]. |
| PLA2G3 (rs2074735) | 22 | PLA2G3 gene encodes the phospholipase A2 group III which functions in lipid metabolism and catalyses the calcium-dependent hydrolysis of the sn-2 acyl bond of phospholipids to release arachidonic acid and lysophospholipids [83]. |
| Vitamin D Winter (n = 176,577) | Vitamin D Summer (n = 130,855) | |||||
|---|---|---|---|---|---|---|
| 25(OH)D Mean (SD) | <25 nmol/L OR (95% CI) | <50 nmol/L OR (95% CI) | 25(OH)D Mean (SD) | <25 nmol/L OR (95% CI) | <50 nmol/L OR (95% CI) | |
| Quintile 1 (Lowest 20%) | 39.16 (17.41) | Reference | Reference | 52.13 (17.37) | Reference | Reference |
| Quintile 2 | 41.84 (18.47) | 0.79 (0.76–0.82) | 0.75 (0.73–0.78) | 56.16 (18.51) | 0.72 (0.66–0.79) | 0.68 (0.66–0.71) |
| Quintile 3 | 43.73 (19.36) | 0.68 (0.66–0.71) | 0.64 (0.61–0.66) | 58.50 (19.20) | 0.63 (0.57–0.69) | 0.56 (0.54–0.58) |
| Quintile 4 | 45.40 (20.14) | 0.60 (0.58–0.62) | 0.54 (0.53–0.56) | 60.65 (20.05) | 0.53 (0.48–0.58) | 0.49 (0.47–0.51) |
| Quintile 5 | 47.51 (21.25) | 0.52 (0.50–0.54) | 0.47 (0.45–0.48) | 64.05 (21.17) | 0.42 (0.38–0.47) | 0.39 (0.37–0.40) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hyppönen, E.; Vimaleswaran, K.S.; Zhou, A. Genetic Determinants of 25-Hydroxyvitamin D Concentrations and Their Relevance to Public Health. Nutrients 2022, 14, 4408. https://doi.org/10.3390/nu14204408
Hyppönen E, Vimaleswaran KS, Zhou A. Genetic Determinants of 25-Hydroxyvitamin D Concentrations and Their Relevance to Public Health. Nutrients. 2022; 14(20):4408. https://doi.org/10.3390/nu14204408
Chicago/Turabian StyleHyppönen, Elina, Karani S. Vimaleswaran, and Ang Zhou. 2022. "Genetic Determinants of 25-Hydroxyvitamin D Concentrations and Their Relevance to Public Health" Nutrients 14, no. 20: 4408. https://doi.org/10.3390/nu14204408
APA StyleHyppönen, E., Vimaleswaran, K. S., & Zhou, A. (2022). Genetic Determinants of 25-Hydroxyvitamin D Concentrations and Their Relevance to Public Health. Nutrients, 14(20), 4408. https://doi.org/10.3390/nu14204408







