Abstract
In addition to sphingomyelin and ceramide, sugar derivatives of ceramides, hexosylceramides (HexCer) are the major circulating sphingolipids. We have shown that silencing of ABCA1 transmembrane protein function for instance in cases of loss of function of ABCA1 gene results in low levels of HDL as well as a concomitant reduction in plasma HexCer levels. However, proteins involved in hepatic synthesis and egress of HexCer from cells is not well known although ABCA1 seems to be indirectly controlling the HexCer plasma levels by supporting HDL synthesis. In this study, we hypothesized that protein(s) other than ABCA1 are involved in the transport of HexCer to HDL. Using an unbiased knockdown approach, we found that ATP-binding cassette transporter protein C10 (ABCC10) participates in the synthesis of HexCer and thereby affects egress to HDL in human hepatoma Huh-7 cells. Furthermore, livers from ABCC10 deficient mice had significantly lower levels of HexCer compared to wild type livers. These studies suggest that ABCC10 partakes in modulating the synthesis and subsequent efflux of HexCer to HDL in liver cells.
1. Introduction
Sphingolipids play a critical role in physiology and are important components of cell membranes. They also act as bioactive lipids in several metabolic and hormonal pathways. The activities of enzymes in sphingolipid synthesis are regulated by fluxes of different substrates and through signaling in response to various stimuli [1,2,3]. Ample information is available about the synthesis, metabolism, and signaling activities of sphingolipids [4,5,6,7,8], but little is known about their transport to circulation.
Ceramide (Cer) is the simplest sphingolipid that is a precursor for the synthesis of complex sphingolipids such as sphingomyelin (SM) and hexosylceramides (HexCer) that include glucosylceramide (GlcCer) and galactosylceramide. HexCer serve as precursors for the synthesis of several complex glycosphingolipids [9] and are present in all cell types [10]. Inhibition of glycosphingolipids has been shown to ameliorate arterial stiffness and atherosclerosis in rabbits and mice [11].
Synthesis of sphingolipids begins at the cytosolic leaflet of the endoplasmic reticulum to generate the precursor Cer that is utilized for the synthesis of complex sphingolipids (Figure 1). Majority of the Cer is transported via vesicular transport or by the protein ceramide transfer protein from its site of synthesis in the endoplasmic reticulum to the Golgi apparatus where it is further modified to SM and glycosphingolipids [12]. During the synthesis of SM in the Golgi, Cer must flip towards lumenal side to serve as the substrate for SM synthases as the active site of these enzymes faces the lumen of the Golgi [13,14]. Similarly, synthesis of GlcCer from Cer and UDP-glucose occurs on the cytosolic side of the Golgi by glucosylceramide synthase [15], a transmembrane protein having its active site facing the cytosol [12,16]. GlcCer synthesis is an important first step for the synthesis of other glycosphingolipids and is regulated by different factors [17]. All the other glycosyltransferases involved in the synthesis of complex glycosphingolipids using GlcCer as a precursor are on the lumenal side of the Golgi and, therefore, necessitates the flipping of GlcCer from the cytosol to the lumen for further glycosylation. It is presumed that this translocation of GlcCer from the cytosol to lumen is mediated by multiple Golgi and trans Golgi network flippases [12,16,18,19]. Recently, Budani et al. [20] showed that multiple ABC transporters potentially act as GlcCer flippases and differentially control glycosphingolipids biosynthesis. It is likely that there are more proteins that regulate HexCer synthesis.
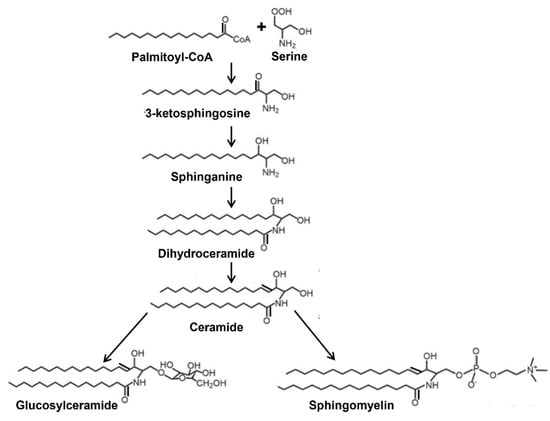
Figure 1.
Simplified schematic representation of the synthesis of glucosylceramide and sphingomyelin from ceramide.
Plasma HexCer are possible biomarkers of disease progression. Li et al. [21] showed that plasma HexCer are high in chronic hepatitis C virus patients with severe fibrosis. They suggested HexCer (d18:1/12:0) as a potential diagnostic marker for severe hepatic steatosis. HexCer (16:1) is also a possible biomarker for disease progression in relapsing multiple sclerosis patients [22]. Furthermore, HexCer levels increase in drug-induced hepatic phospholipidosis [23]. Thus, studies about the origins of plasma HexCer may be useful in understanding progression of different diseases.
To date, a total of 49 different ATP binding cassette (ABC) transporters have been identified in humans [24,25,26]. ABCC1 and ABCA12 were shown to transport GlcCer in vitro [27] and in keratinocytes [10]. We recently demonstrated that ABCA1 is a regulator of plasma glycosphingolipids [28]. However, how HexCer are transported to plasma HDL is unknown. In this study, we used an unbiased siRNA screening against different transporter proteins and identified ABCC10 as an important player in the synthesis and export of HexCer in Huh-7 cells.
ABCC10, also called multidrug resistance protein 7 (MRP7), imparts drug resistance in tumor cells to taxanes [29], such as paclitaxel and docetaxel, which are commonly used in the treatment of common cancers. In addition, ABCC10 might also confer resistance to microtubule alkaloids and certain nucleoside analogues. ABCC10 is predicted to have several substrates with a diverse range including peptides, drugs, phospholipids and sphingolipids. Interestingly, ABCC10 has been shown to selectively recognize glucuronides such as 17-β-estradiol-(17-β-δ-glucuronide) and leukotriene C4 [30,31]. Hopper-Borge et al. [30] generated ABCC10 deficient mice and showed that these mice succumb to early death when exposed to taxanes compared to wild type mice. These studies revealed that ABCC10 affords protection against paclitaxel-induced bone marrow toxicity. Using these mice, we show that ABCC10 deficiency reduces hepatic HexCer levels.
2. Materials and Methods
2.1. Materials
C6-NBD ceramide (6-((N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)caproxyl)sphin- gsine, catalog #6224) was from Setareh Biotech. N-palmitoyl-D-erythro-sphingosine [palmitoyl-1-[14C]-ceramide (C16-[14C]-ceramide, 50–60 mCi/mmol, 1.85–2.22 GBq/mmol, catalog #ARC 0831-50) was purchased from American Radiolabeled Chemicals, Inc. All other chemicals and solvents were from Fisher Scientific (Pittsburgh, PA, USA) or VWR International (Bridgeport, CT, USA).
2.2. Mice
The ABCC10 knockout mice on C57BL6/J background (31) were a kind gift from Dr. Elizabeth Hopper-Borge of Fox Chase Cancer Center, Philadelphia, PA. Mice were housed under a 12:12 h light: dark schedule and bred at Animal Bioresources (SUNY Downstate Medical Center) in accordance with all requirements of SUNY/NIH IACUC. Mice had ad libitum access to chow diet and water.
2.3. Quantification of Lipids
Plasma (100 µL) and liver (2 mg) from Abcc10+/+ and Abcc10−/− mice (n = 4) mice were used to quantify HexCer and SM using HPLC-MS/MS [28,32].
2.4. Identification of Proteins Involved in HexCer Transport and Synthesis
Human hepatoma Huh-7 cells grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), L-glutamine, and antibiotics, were reverse transfected with 50 nM siCTRL or siRNA in triplicate against various membrane transporters (Dharmacon, GE Life Sciences, Lafayette, CO, USA) using RNAiMAX (Thermo Fisher, catalog #13778150) for 72 h.
2.5. Dose Response Effect of siABCC10 on HexCer Export and Synthesis
Human hepatoma Huh-7 cells were reverse transfected with either 50 nM of siCTRL or different concentrations of siABCC10 (12.5, 25, 50, and 100 nM) using RNAiMAX [33].
2.6. HexCer Egress and Synthesis
Huh-7 cells were incubated with 2 µM C6-NBD ceramide in DMEM supplemented with 10% FBS at 37 °C for 3 h. Next, cells were washed 3 times with DMEM plus 0.1% BSA. To study efflux, we added fresh DMEM containing 40 µg/mL of BSA or human serum HDL (catalog #MBS173147, MyBioSource, San Diego, CA, USA). In other experiments, Huh-7 cells were incubated with 0.2 µM [14C]-ceramide to study sphingolipid synthesis and export. Culture media were harvested and centrifuged (600 g, 2500 rpm, 4 °C, 15 min, Heraeus Fresco 21 Centrifuge, 75003424 rotor, ThermoFisher Scientific, Waltham, MA, USA) to pellet cells. Lipids were extracted from media and cells [34], dried and re-suspended (100 µL of isopropanol) for the separation of sphingolipids on thin layer chromatograph (TLC) silica plates (catalog #44931, Analtech, Inc., Newark, NJ, USA) using a CHCl3:CH3OH:C6H5CH3:NH4OH:H2O (40:40:20:0.4:1.6, ratios by volume) solvent system [28,32]. The TLC plates were scanned using a PhosphorImager (GE Healthcare, Chicago, IL, USA). Bands corresponding to Cer, SM and HexCer were quantified using ImageJ.
2.7. Cell-Free GCS Synthesis Assay
A buffer containing 50 mM Tris-HCl, 1 mM EDTA, 5% sucrose, pH 7.4 buffer containing a mixture of protease inhibitors was used to prepare liver homogenates from Abcc10+/+ and Abcc10−/− mice. The homogenate (500 µg of protein) was added to a buffer containing 50 mM Tris-HCl, pH 7.4, 25 mM KCl, phosphatidylcholine (100 µg/mL), C6-NBD ceramide (3.3 µg/mL), and UDP-glucose (500 µM) in a total reaction volume of 200 µL and incubated for 1 h at 37 °C [35,36]. We added 200 µL of CHCl3/CH3OH (2:1, v/v) to stop the reaction. Lipids were extracted and used for TLC.
2.8. Statistics
Biochemical data between Abcc10+/+ and Abcc10−/− mouse models and comparisons amongst different treatments in Huh-7 cells were evaluated using Student t-test (GraphPad Prism).
3. Results
3.1. Identification of ABCC10 as a HexCer Transporter
We showed previously that ABCA1 deficiency in humans and mice reduces plasma levels of HexCer [32]. However, induction, knockdown, inhibition or downregulation of ABCA1 in Huh-7 cells had no effect on the efflux of HexCer to HDL [32]. Hence, we concluded that ABCA1 may indirectly affect plasma HexCer levels by participating in the biogenesis of HDL. Next, we hypothesized that other unknown proteins exist that play important roles in the export of HexCer. To identify the HexCer transporter(s), we used the approach that we previously undertook to identify ABCA7 as an important regulator of SM biosynthesis and efflux [33]. Studies to identify the HexCer transporters were performed in parallel with those reported earlier in the identification of SM transporter [33]. Briefly, we knocked down ABC transporter family members using specific siRNAs in Huh-7 cells. These cells were then treated with C6-NBD ceramides and incubated with either bovine serum albumin (BSA) to quantify basal efflux (siCTRL-BSA) or with human HDL to study their transport to HDL (siCTRL-HDL) (Figure 2). HDL significantly increased HexCer efflux when compared to BSA (Figure 2A,B) consistent with our previous studies [32,33] that HDL enhances HexCer efflux. We observed that siABCC10 significantly decreased HexCer export to HDL (Figure 2A,B). A second screening validated these results. siCTRL treated cells showed significantly increased HexCer efflux to HDL when compared to siCTRL-BSA, and this export was significantly reduced after siABCC10 treatment (Figure 2C), suggesting that ABCC10 might participate in the egress of HexCer to HDL.
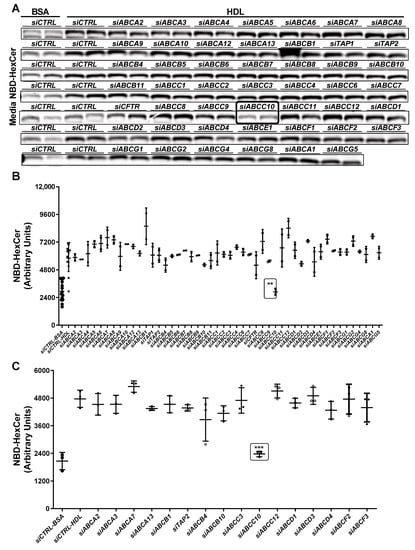
Figure 2.
Identification of transporter(s) involved in HexCer egress to HDL. (A,B) Primary screening. Huh-7 cells were transfected in duplicate with of siCTRL or siRNA against various indicated membrane transporters (50 nM). After 72 h, cells were washed and incubated with 2 µM C6-NBD ceramides in DMEM containing 10% FBS for 3 h. Cells were washed again thrice with DMEM containing 0.1% BSA. Subsequently, cells were incubated with either BSA or HDL (40 µg/mL) in DMEM for 6 h. Media lipids were extracted for separation by TLC. Fluorescence in HexCer bands were photographed with the PhosphorImager (A) and quantified using Image J and plotted (B). (C) A secondary screening for selected transporters was performed in triplicate. Values are replicates (Mean ± SD), ** p < 0.01 and *** p < 0.001 compared with siCTRL-HDL.
3.2. Dose–Response Effect of siABCC10 on HexCer Efflux and Synthesis
Treatment of Huh-7 cells with increasing concentrations of siABCC10 followed by incubation with HDL for 3 h did not affect the export of Cer and SM, respectively (Figure 3A,B). Contrary to Cer or SM, efflux of HexCer to HDL was significantly reduced in a dose-dependent manner in siABCC10-HDL cells compared to siCTRL-HDL treated cells (Figure 3C). At 100 nM of siABCC10, HexCer efflux was reduced by ~67% (Figure 3C). Thus, treatment of cells with siABCC10 is associated with reduced HexCer transport to HDL.
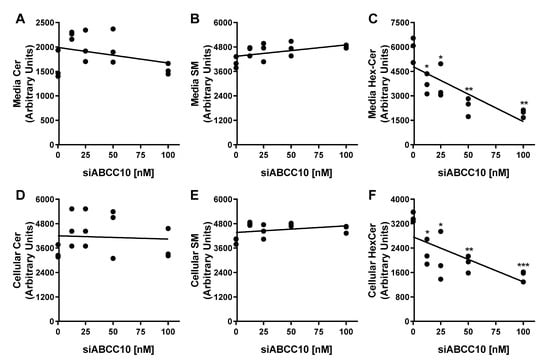
Figure 3.
Dose response effect of siABCC10 on HexCer efflux and synthesis. Huh-7 cells were transfected with 12.5, 25, 50, and 100 nM of either siCTRL or siABCC10 in triplicate for 72 h. Cells were used to study HexCer efflux and synthesis as described in Figure 2. Lipids in the media (A–C) and cells (D–F) were extracted, separated on TLC and fluorescence in Cer, HexCer and SM bands were quantified using Image J. Values are replicates. * p < 0.05, ** p < 0.01 and *** p < 0.001 compared with siCTRL-HDL treated cells.
Next, we quantified the amounts of HexCer remained in the cells (Figure 3D–F). Increasing concentrations of siABCC10 had no significant effect on the amounts of Cer and SM present in cells (Figure 3D,E). However, accumulation of HexCer in the cells was significantly reduced with increasing concentrations of siABCC10 compared to siCTRL (Figure 3F) suggesting that ABCC10 may play a role in HexCer synthesis.
3.3. siABCC10 Decreases HexCer Synthesis in Hepatoma Cells
To evaluate the role of ABCC10 in HexCer synthesis, Huh-7 cells were transfected with siCTRL or siABCC10 and incubated with C6-NBD ceramides for shorter time periods (0–60 min) and measured the synthesis of sphingolipids. siABCC10 reduced ABCC10 mRNA (0.18 ± 0.04, n = 3) levels by 82% compared to siCTRL (1.00 ± 0.17, n = 3). Time dependent increases in cellular Cer levels were similar in both siABCC10 and siCTRL treated cells (Figure 4A) indicating similar uptake. At the earliest time point in both siCTRL and siABCC10 treated cells, NBD-Cer was converted to SM and its levels increased with time until 30 min. Subsequently, SM levels reached a similar steady state in siCTRL and siABCC10 treated cells. These studies suggest that Cer uptake and its subsequent conversion to SM were not affected by ABCC10 deficiency. In contrast to SM, synthesis of HexCer showed time dependent increase with time in siCTRL and siABCC10 treated cells and did not reach steady state during this time course. However, these temporal increases were significantly lower in siABCC10 treated cells, thereby, leading to decreased intracellular HexCer levels compared to siCTRL cells (Figure 4A). These studies demonstrate that ABCC10 knockdown reduces HexCer synthesis but has no effect on SM synthesis.
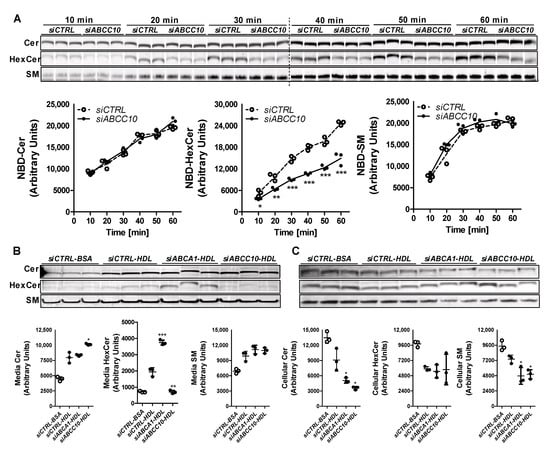
Figure 4.
Role of ABCC10 in HexCer synthesis. (A) Synthesis of HexCer is decreased after treatment with siABCC10. Huh-7 cells were transfected (n = 3) with 50 nM siCTRL or siABCC10. After 72 h, cells were labeled with 2 µM C6-NBD Cer for indicated time points and were washed thrice with 0.1% BSA containing DMEM. Cellular lipids were extracted, separated on TLC plates, fluorescence in Cer, SM, and HexCer bands were scanned using the PhosphorImager (top), and quantified by Image J (bottom). Values are plotted as replicates (Mean ± SD), * p < 0.05, ** p < 0.01 and *** p < 0.001 compared to siCTRL treated cells. (B,C) ABCC10 knockdown diminishes synthesis of [14C]-HexCer and its transport to HDL. Huh-7 cells were transfected (n = 3) with 50 nM siCTRL, siABCA1 or siABCC10. After 72 h, cells were labeled with 0.2 µCi of [14C]-Cer in DMEM and used for efflux studies. Media (B) and cell (C) lipids were separated using TLC and radioactive Cer, SM, and HexCer bands were visualized with the PhosphorImager (top) and quantified using Image J (bottom). Values are replicates (Mean ± SD). * p < 0.05, ** p < 0.01 and *** p < 0.001 compared to siCTRL-HDL.
We considered that reduced HexCer synthesis might be secondary to the use of NBD-labeled C6-Cer. As an additional approach, we labeled siCTRL and siABCC10 treated Huh-7 cells with C16-[14C]-Cer and measured labeled media and cellular sphingolipids. We also knocked down ABCA1 whose deficiency reduces plasma HexCer [32]. We studied efflux of [14C]-sphingolipids to HDL and BSA (Figure 4B). There was a significant increase in Cer, SM and HexCer levels in the media obtained from siCTRL-HDL compared to siCTRL-BSA cells suggesting increased efflux to HDL. Compared to siCTRL, siABCA1 had no effect on Cer and SM efflux to HDL but significantly increased HexCer efflux. Cer egress was significantly increased, whereas HexCer transport to HDL was significant decreased in siABCC10-HDL compared to siCTRL-HDL cells (Figure 4B). However, there was no significant difference in the egress of SM to HDL in siABCC10-HDL cells compared to siCTRL-HDL cells. These studies indicate that siABCC10 significantly reduces [14C]-HexCer efflux to HDL without affecting SM efflux.
We then measured sphingolipids in cells (Figure 4C). siCTRL-HDL cells had reduced levels of Cer, SM and HexCer compared with siCTRL-BSA (Figure 4C) suggesting that increased transport (Figure 4B) is associated with lower cellular levels. siABCA1-HDL did not affect cellular HexCer levels compared to siCTRL-HDL. siABCC10-HDL had lower cellular Cer and SM levels compared to siCTRL-HDL, most likely secondary to their increased efflux to HDL. Since siABCC10-HDL showed decreased HexCer transport to HDL (Figure 4B), we had anticipated an increase in cellular levels of HexCer compared to siCTRL-BSA treated cells. However, our data showed lower levels of radiolabeled HexCer in siABCC10-HDL compared to siCTRL-HDL treated cells (Figure 4C). In short, siABCC10-HDL cells showed increased efflux of Cer and SM and reduced cellular levels. In contrast, siABCC10-HDL cells showed reduced efflux as well as diminished cellular levels. We interpret these data to suggest that ABCC10 deficiency reduces HexCer synthesis and, thereby, results in diminished export.
3.4. ABCC10 Deletion Decreases Hepatic HexCer Levels without Affecting Glucosylceramide Synthase Activity in Liver Homogenates
To determine whether ABCC10 plays a role in regulating physiological HexCer levels, we measured HexCer in the plasma and liver of wild type (Abcc10+/+) and ABCC10 knockout (Abcc10−/−) mice fed a chow diet (Figure 5A–D, Table 1). Deletion of ABCC10 in mice had no effect on total plasma SM or HexCer levels (Figure 5A,B, Table 1). Similarly, hepatic SM levels were unaffected by ABCC10 deficiency (Figure 5C, Table 1). However, we observed a significant decrease (~39%) in the levels of hepatic HexCer in Abcc10−/− as compared to Abcc10+/+ mice (Figure 5D). Analysis of individual species of hepatic HexCer (Table 1) suggested a significant decrease in almost all the species of HexCer in Abcc10−/− mice, except for C22_1-HexCer, C22-HexCer and C26_HexCer. We also observed a significant decrease in the plasma levels of C18_1-HexCer, C18-HexCer, and C20-HexCer in Abcc10−/− mice (Table 1). Since these species were minor components of plasma HexCer, their individual reductions had no impact on total plasma HexCer (Figure 5B). These data indicate that ABCC10 regulates the levels of HexCer in the liver.
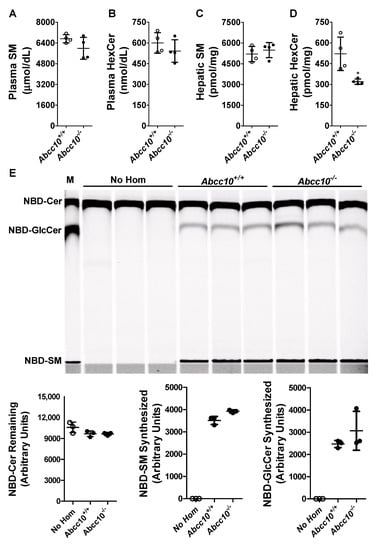
Figure 5.
ABCC10 in hepatic HexCer metabolism. (A–D) Deletion of Abcc10 gene in mice reduces liver HexCer levels. Overnight fasted Abcc10+/+ and Abcc10−/− mice (n = 4, 12-week-old, chow fed) were used to measure plasma (A,B) and liver (C,D) SM and HexCer levels. Values are replicates (Mean ± SD). * p < 0.05 compared with Abcc10+/+ mice. (E) Ablation of ABCC10 does not affect glucosylceramide synthase activity in liver homogenates. Abcc10+/+ and Abcc10−/− liver homogenates (500 µg protein) were incubated (n = 3) with phosphatidylcholine (100 µg/mL), C6-NBD ceramides (3.3 µg/mL) and UDP-glucose (500 µM) at 37 °C for 1 h. As a control, reaction was also carried out in triplicate without the addition of any protein homogenates (No Hom). Lipids were extracted from the reaction mixtures, separated on TLC, fluorescence in Cer, SM, and GlcCer bands were scanned using PhosphorImager (top), and quantified by Image J (bottom). Values are plotted as replicates (Mean ± SD). In lane “M”, NBD-labeled lipids were used to identify different sphingolipids.

Table 1.
Levels of sphingomyelin and hexosylceramide species in the plasma and liver of wild type and ABCC10 knockout mice.
We considered the possibility that decreased levels of hepatic HexCer in ABCC10 knockout mice may be due to a reduction in the glucosylceramide synthase (GCS) activity. To evaluate this possibility, we conducted an in vitro cell-free assay to measure GCS activity, the major contributor of HexCer in liver cells, in the liver homogenates isolated from Abcc10+/+ and Abcc10−/− mice [35,36]. In vitro incubation of NBD-Cer with UDP-glucose for 1 h without the addition of liver homogenates (No Hom) did not result in the synthesis of SM and GlcCer (Figure 5E). However, when the liver homogenates were added to the reaction mixture, there was an increase in the synthesis of both SM and GlcCer. Compared to the liver homogenates isolated from Abcc10+/+ mice, we did not see any significant difference in the synthesis of either SM or GlcCer in the liver homogenates isolated from Abcc10−/− mice (Figure 5E). These data indicate that decrease in HexCer levels in ABCC10 knockout mice liver is likely not due to reductions in GCS activity.
4. Discussion
ABCA1 participates in the transport of cholesterol and phospholipids to HDL and its deficiency in humans and mice has been shown to associate with low levels of plasma HexCer [28]. However, mechanistic studies performed in Huh-7 cells revealed that ABCA1 is not directly involved in the efflux of HexCer. ABCA1 is critical in generating HDL that acts as a potent acceptor for the efflux of HexCer. In this study, using unbiased siRNA screening, we identified ABCC10 as a protein that plays a role in HexCer efflux (Figure 2). Our subsequent experiments showed that ABCC10 also participates in the biosynthesis of HexCer (Figure 3 and Figure 4). Thus, ABCC10 partakes in HexCer synthesis and, thereby, affects its efflux.
ABCC10 is expressed in several tissues [29,37,38,39] and has been shown to be present in plasma membrane as well as in intracellular compartments [40,41]. Intracellular localization of ABCC10 was unperturbed by various compounds used in these studies [40,41]. Expression of ABCC10 is elevated in hepatocellular carcinoma compared with adjacent healthy liver tissue [42]. ABCC10 expression is significantly higher in monocyte-derived macrophages and peripheral blood lymphocytes obtained from rheumatoid arthritis patients [43]. A bioinformatics analysis of genes expressed in samples of atherosclerotic lesions and control arteries without atherosclerotic lesions showed an increased expression of ABCC10 in lesion areas [44] suggesting a role of ABCC10 in atherosclerosis.
ABCC10 belongs to the ABCC family that partakes in drug elimination and efflux of other endogenous molecules [29,30,31,45]. Our cell culture studies indicate that ABCC10 plays a role in the biosynthesis and, thereby, in the efflux of HexCer. However, studies using liver homogenates demonstrated that ABCC10 deficiency has no effect on HexCer synthesis by GCS indicating that ABCC10 may not directly modulate the enzyme activity. We speculate that ABCC10 may modulate HexCer synthesis by affecting substrate availability and/or substrate removal from the GCS enzyme.
The overexpression of ABCC10 in HEK293 cells confers resistance to various chemotherapeutic drugs [29,30]. ABCC10 has also been shown to protect cells against paclitaxel toxicity [30]. Our current studies for the first time show that ABCC10 affects HexCer synthesis and export in Huh-7 cells. Thus, it is likely that ABCC10 is a multi-functional protein that plays a role in diverse physiologic functions.
The members of the ABC family are recognized players in the efflux of molecules and drugs. ABCA1 participates in the efflux of phospholipid and cholesterol [46,47]. We have recently shown that ABCA7 affects synthesis and export of SM and its deficiency in mice affects brain SM metabolism causing cognitive defects [33]. The current report indicates that ABCC10 plays a role in the synthesis of HexCer. Consistent with our studies, Budani et al. [20] identified several ABC transporters that may play roles as flippases and affect synthesis. These observations point out to the possibility that, besides being drug/molecule exporters, ABC transporters might be physiologically important modulators of sphingolipid biosynthesis.
HexCer consist of both GlcCer and GalCer. Our TLC approach of separating different sphingolipids does not resolve these two HexCer; therefore, it is unknown whether ABCC10 plays a role in the synthesis of both or not. Another drawback of this study is that we did not look at downstream changes in the biosynthesis of complex sphingolipids in ABCC10 deficient cells and mice.
In summary, we identified a novel role of ABCC10 in the biosynthesis and efflux of HexCer in human hepatoma cells. Furthermore, ABCC10 participates in controlling levels of several HexCer species in mouse liver. Thus, ABCC10 is a regulator of hepatic HexCer homeostasis.
Author Contributions
M.M.H. and J.I. designed and conceptualized the experiments and did all the data analysis. J.I. and M.T.W. performed studies in Huh-7 cells and identified ABCC10 to be involved in HexCer synthesis and export in the laboratory of M.M.H. J.I. made the figures and wrote the first draft of the manuscript which was extensively modified by M.M.H. All authors have read and agreed to the published version of the manuscript.
Funding
U.S. National Institutes of Health (NIH) grants DK121490, HL158054, HL137202, BX004113, and HD094778 to MMH; American Heart Association grant 12GRNT9690010 to JI partly supported this research. The contents of this paper do not represent the views of the U.S. Government. The content is solely the responsibility of the authors. It does not represent the views of NIH.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee of SUNY Downstate Medical Center.
Informed Consent Statement
Not applicable.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
We thank Elizabeth Hopper-Borge of Fox Chase Cancer Center, Philadelphia for providing ABCC10 knockout mice.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shimabukuro, M.; Zhou, Y.T.; Levi, M.; Unger, R.H. Fatty acid-induced beta cell apoptosis: A link between obesity and diabetes. Proc. Natl. Acad. Sci. USA 1998, 95, 2498–2502. [Google Scholar] [CrossRef]
- Hu, W.; Bielawski, J.; Samad, F.; Merrill, A.H., Jr.; Cowart, L.A. Palmitate increases sphingosine-1-phosphate in C2C12 myotubes via upregulation of sphingosine kinase message and activity. J. Lipid Res. 2009, 50, 1852–1862. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ross, J.; Geng, T.; Brice, S.E.; Cowart, L.A. Differential regulation of dihydroceramide desaturase by palmitate versus monounsaturated fatty acids: Implications for insulin resistance. J. Biol. Chem. 2011, 286, 16596–16605. [Google Scholar] [CrossRef] [PubMed]
- Gault, C.R.; Obeid, L.M.; Hannun, Y.A. An overview of sphingolipid metabolism: From synthesis to breakdown. Adv. Exp. Med. Biol 2010, 688, 1–23. [Google Scholar] [PubMed]
- Lucki, N.C.; Sewer, M.B. Nuclear sphingolipid metabolism. Annu. Rev. Physiol. 2012, 74, 131–151. [Google Scholar] [CrossRef]
- Bikman, B.T.; Summers, S.A. Ceramides as modulators of cellular and whole-body metabolism. J. Clin. Investig. 2011, 121, 4222–4230. [Google Scholar] [CrossRef]
- Morad, S.A.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Hla, T.; Dannenberg, A.J. Sphingolipid signaling in metabolic disorders. Cell Metab. 2012, 16, 420–434. [Google Scholar] [CrossRef]
- Sandhoff, R.; Sandhoff, K. Emerging concepts of ganglioside metabolism. FEBS Lett. 2018, 592, 3835–3864. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Kohyama-Koganeya, A.; Hirabayashi, Y. New insights on glucosylated lipids: Metabolism and functions. Biochim. Biophys. Acta 2013, 1831, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Bedja, D.; Mishra, S.; Amuzie, C.; Avolio, A.; Kass, D.A.; Berkowitz, D.; Renehan, M. Inhibition of glycosphingolipid synthesis ameliorates atherosclerosis and arterial stiffness in apolipoprotein E-/- mice and rabbits fed a high-fat and -cholesterol diet. Circulation 2014, 129, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Futerman, A.H.; Stieger, B.; Hubbard, A.L.; Pagano, R.E. Sphingomyelin synthesis in rat liver occurs predominantly at the cis and medial cisternae of the Golgi apparatus. J. Biol. Chem. 1990, 265, 8650–8657. [Google Scholar] [CrossRef]
- Ichikawa, S.; Sakiyama, H.; Suzuki, G.; Hidari, K.I.; Hirabayashi, Y. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Natl. Acad. Sci. USA 1996, 93, 4638–4643. [Google Scholar] [CrossRef]
- Ichikawa, S.; Hirabayashi, Y. Glucosylceramide synthase and glycosphingolipid synthesis. Trends Cell Biol. 1998, 8, 198–202. [Google Scholar] [CrossRef]
- Abe, A.; Radin, N.S.; Shayman, J.A. Induction of glucosylceramide synthase by synthase inhibitors and ceramide. Biochim Biophys. Acta 1996, 1299, 333–341. [Google Scholar] [CrossRef]
- Lannert, H.; Bünning, G.; Jeckel, D.; Wieland, F.T. Lactosylceramide is synthesized in the lumen of the Golgi apparatus. FEBS Lett. 1994, 342, 91–96. [Google Scholar] [CrossRef]
- Burger, K.N.; van der Bijl, P.; van Meer, G. Topology of sphingolipid galactosyltransferases in ER and Golgi: Transbilayer movement of monohexosyl sphingolipids is required for higher glycosphingolipid biosynthesis. J. Cell Biol. 1996, 133, 15–28. [Google Scholar] [CrossRef]
- Budani, M.; Auray-Blais, C.; Lingwood, C. ATP-binding cassette transporters mediate differential biosynthesis of glycosphingolipid species. J. Lipid Res. 2021, 62, 100128. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Qu, F.; Zheng, S.J.; Ren, F.; Wu, H.L.; Liu, M.; Ren, J.Y.; Chen, Y.; Duan, Z.P.; Zhang, J.L. Plasma sphingolipids: Potential biomarkers for severe hepatic fibrosis in chronic hepatitis C. Mol. Med. Rep. 2015, 12, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Checa, A.; Khademi, M.; Sar, D.G.; Haeggstrom, J.Z.; Lundberg, J.O.; Piehl, F.; Olsson, T.; Wheelock, C.E. Hexosylceramides as intrathecal markers of worsening disability in multiple sclerosis. Mult. Scler. 2015, 21, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Maekawa, K.; Ishikawa, M.; Senoo, Y.; Urata, M.; Murayama, M.; Nakatsu, N.; Yamada, H.; Saito, Y. Glucosylceramide and lysophosphatidylcholines as potential blood biomarkers for drug-induced hepatic phospholipidosis. Toxicol. Sci. 2014, 141, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Dassa, E.; Bouige, P. The ABC of ABCS: A phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001, 152, 211–229. [Google Scholar] [CrossRef]
- Kimura, Y.; Morita, S.Y.; Matsuo, M.; Ueda, K. Mechanism of multidrug recognition by MDR1/ABCB1. Cancer Sci. 2007, 98, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Sharom, F.J. ABC multidrug transporters: Structure, function and role in chemoresistance. Pharmacogenomics 2008, 9, 105–127. [Google Scholar] [CrossRef] [PubMed]
- Raggers, R.J.; van Helvoort, A.; Evers, R.; van Meer, G. The human multidrug resistance protein MRP1 translocates sphingolipid analogs across the plasma membrane. J. Cell Sci. 1999, 112, 415–422. [Google Scholar] [CrossRef]
- Iqbal, J.; Walsh, M.T.; Hammad, S.M.; Cuchel, M.; Tarugi, P.; Hegele, R.A.; Davidson, N.O.; Rader, D.J.; Klein, R.L.; Hussain, M.M. Microsomal Triglyceride Transfer Protein Transfers and Determines Plasma Concentrations of Ceramide and Sphingomyelin but Not Glycosylceramide. J. Biol. Chem. 2015, 290, 25863–25875. [Google Scholar] [CrossRef] [PubMed]
- Hopper-Borge, E.; Chen, Z.S.; Shchaveleva, I.; Belinsky, M.G.; Kruh, G.D. Analysis of the drug resistance profile of multidrug resistance protein 7 (ABCC10): Resistance to docetaxel. Cancer Res. 2004, 64, 4927–4930. [Google Scholar] [CrossRef]
- Hopper-Borge, E.A.; Churchill, T.; Paulose, C.; Nicolas, E.; Jacobs, J.D.; Ngo, O.; Kuang, Y.; Grinberg, A.; Westphal, H.; Chen, Z.S.; et al. Contribution of Abcc10 (Mrp7) to in vivo paclitaxel resistance as assessed in Abcc10(-/-) mice. Cancer Res. 2011, 71, 3649–3657. [Google Scholar] [CrossRef]
- Chen, Z.S.; Hopper-Borge, E.; Belinsky, M.G.; Shchaveleva, I.; Kotova, E.; Kruh, G.D. Characterization of the transport properties of human multidrug resistance protein 7 (MRP7, ABCC10). Mol. Pharmacol. 2003, 63, 351–358. [Google Scholar] [CrossRef]
- Iqbal, J.; Walsh, M.T.; Hammad, S.M.; Cuchel, M.; Rader, D.J.; Hussain, M.M. ATP binding cassette family A protein 1 determines hexosylceramide and sphingomyelin levels in human and mouse plasma. J. Lipid Res. 2018, 59, 2084–2097. [Google Scholar] [CrossRef]
- Iqbal, J.; Suarez, M.D.; Yadav, P.K.; Walsh, M.T.; Li, Y.; Wu, Y.; Huang, Z.; James, A.W.; Escobar, V.; Mokbe, A.; et al. ATP-binding cassette protein ABCA7 deficiency impairs sphingomyelin synthesis, cognitive discrimination, and synaptic plasticity in the entorhinal cortex. J. Biol. Chem. 2022, 298, 102411. [Google Scholar] [CrossRef]
- BLIGH, E.G.; DYER, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Jeckel, D.; Karrenbauer, A.; Burger, K.N.; van Meer, G.; Wieland, F. Glucosylceramide is synthesized at the cytosolic surface of various Golgi subfractions. J. Cell Biol. 1992, 117, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Futerman, A.H.; Pagano, R.E. Determination of the intracellular sites and topology of glucosylceramide synthesis in rat liver. Biochem. J. 1991, 280, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Kruh, G.D.; Guo, Y.; Hopper-Borge, E.; Belinsky, M.G.; Chen, Z.S. ABCC10, ABCC11, and ABCC12. Pflügers Arch. Eur. J. Physiol. 2007, 453, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, S.; Kataoka, T.; Ohara, O.; Oishi, M.; Kuo, M.T.; Ishikawa, T. Human ATP-binding cassette transporter ABCC10: Expression profile and p53-dependent upregulation. J. Exp. Ther. Oncol. 2004, 4, 239–246. [Google Scholar]
- Bleasby, K.; Castle, J.C.; Roberts, C.J.; Cheng, C.; Bailey, W.J.; Sina, J.F.; Kulkarni, A.V.; Hafey, M.J.; Evers, R.; Johnson, J.M.; et al. Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: A resource for investigations into drug disposition. Xenobiotica 2006, 36, 963–988. [Google Scholar] [CrossRef]
- Zhang, H.; Kathawala, R.J.; Wang, Y.J.; Zhang, Y.K.; Patel, A.; Shukla, S.; Robey, R.W.; Talele, T.T.; Ashby, C.R., Jr.; Ambudkar, S.V.; et al. Linsitinib (OSI-906) antagonizes ATP-binding cassette subfamily G member 2 and subfamily C member 10-mediated drug resistance. Int. J. Biochem. Cell Biol. 2014, 51, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Wang, B.; Teng, Q.X.; Lei, Z.N.; Li, Y.D.; Shi, Z.; Ma, L.Y.; Liu, H.M.; Liu, Z.; Chen, Z.S. CMP25, a synthetic new agent, targets multidrug resistance-associated protein 7 (MRP7/ABCC10). Biochem. Pharmacol. 2021, 190, 114652. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Han, R.; Visser, A.; Petry, H.; van Deventer, S.J.; Jansen, P.L.; Konstantinova, P.; Reseau Centre de Ressources Biologiques; Foie, F. Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology 2012, 55, 821–832. [Google Scholar] [CrossRef]
- Oerlemans, R.; Blits, M.; Dijkmans, B.A.; van der Heijden, J.W.; Lems, W.F.; Scheffer, G.L.; van de Ven, R.; Peters, G.J.; Assaraf, Y.G.; Scheper, R.J.; et al. Expression profiling of ABC transporters in peripheral blood lymphocytes and monocyte-derived macrophages of rheumatoid arthritis patients. J. Mol. Clin. Med. 2020, 3, 47–60. [Google Scholar] [CrossRef]
- Kotlyarov, S.; Kotlyarova, A. Analysis of ABC Transporter Gene Expression in Atherosclerosis. Cardiogenetics 2021, 11, 204–218. [Google Scholar] [CrossRef]
- Malofeeva, E.V.; Domanitskaya, N.; Gudima, M.; Hopper-Borge, E.A. Modulation of the ATPase and transport activities of broad-acting multidrug resistance factor ABCC10 (MRP7). Cancer Res. 2012, 72, 6457–6467. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, G.; Langmann, T. Structure, function and regulation of the ABC1 gene product. Curr. Opin. Lipidol. 2001, 12, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.A.; Knopp, R.H.; Oram, J.F. Defective removal of cellular cholesterol and phospholipids by apolipoprotein A-I in Tangier Disease. J. Clin. Investig. 1995, 96, 78–87. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).