Transcriptomic Analysis of the Anticancer Effects of Annatto Tocotrienol, Delta-Tocotrienol and Gamma-Tocotrienol on Chondrosarcoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Materials
2.3. Cytotoxicity Assay
2.4. Annexin V-FITC/PI Dual-Labelling Assay
2.5. DNA Content Assay
2.6. RNA Isolation
2.7. RNA Sequencing and Transcriptomic Analysis
2.8. Differential Expression Analysis and Enrichment Analysis
2.9. Quantitative PCR (qPCR) Validation
2.10. Statistical Analysis
3. Results
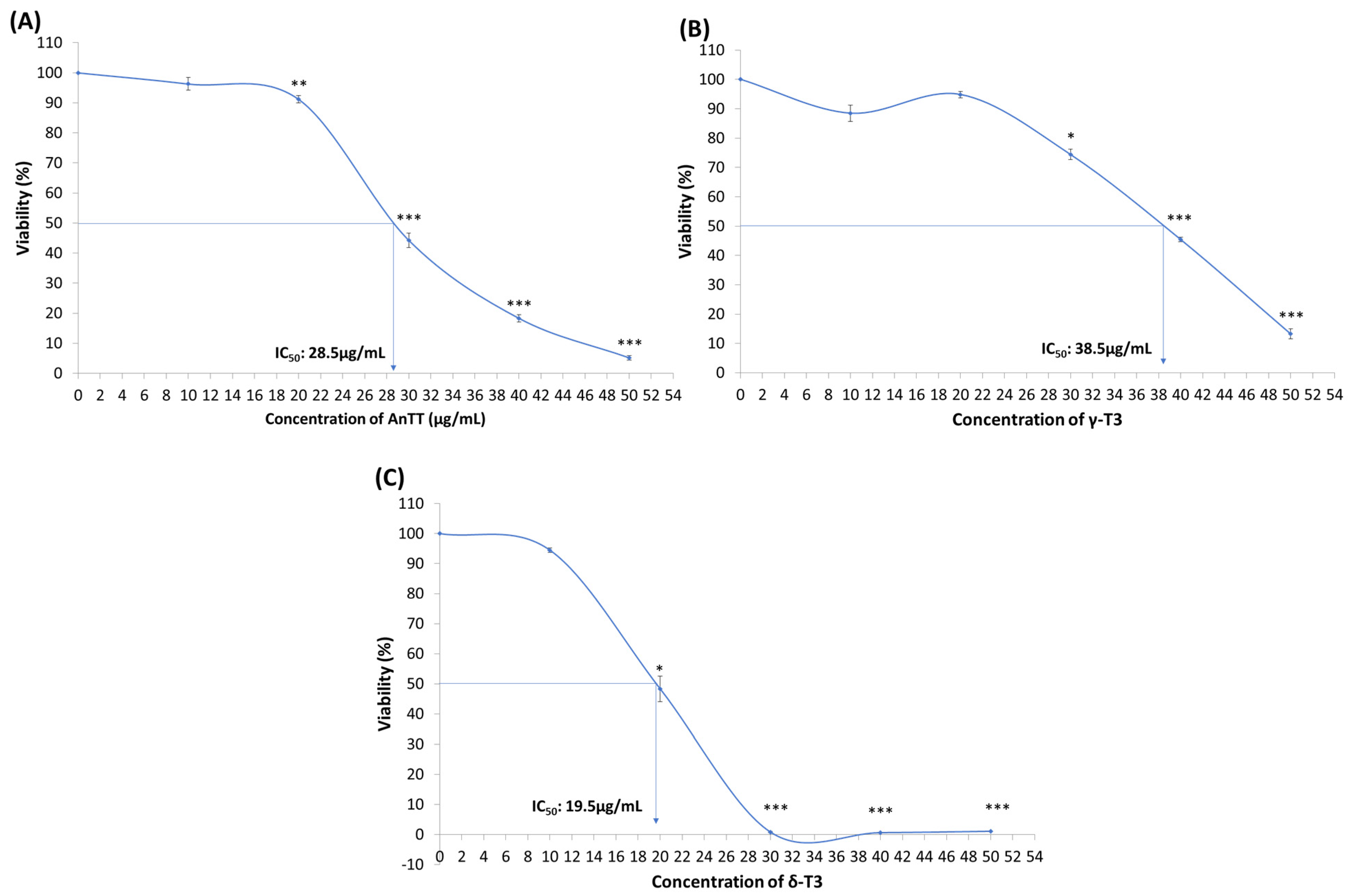
3.1. Cytotoxicity of AnTT, γ-T3 and δ-T3 on Human Chondrosarcoma SW1353 Cells
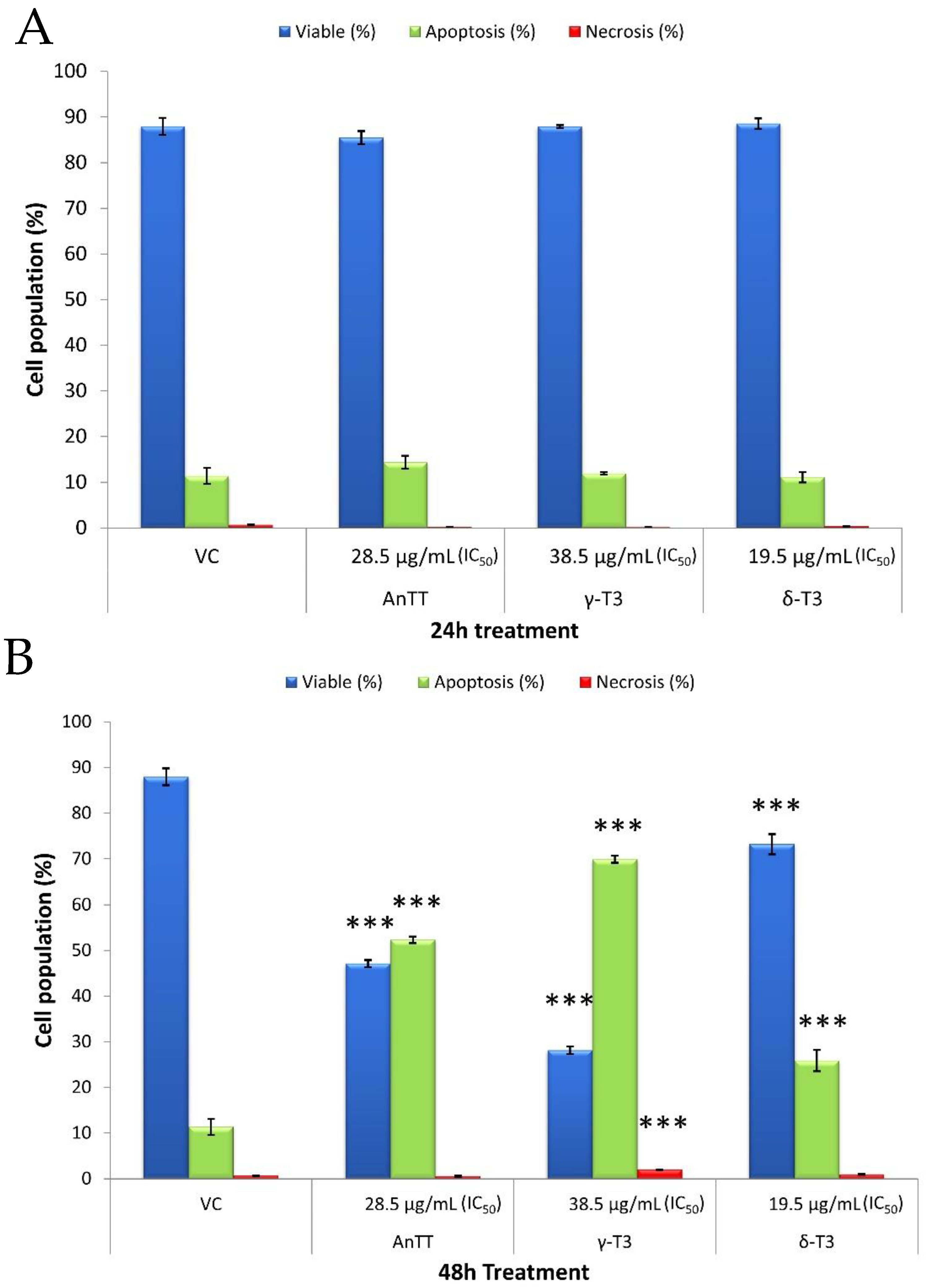
3.2. The Mode of SW1353 Cell Death upon AnTT, γ-T3 and δ-T3 Treatment
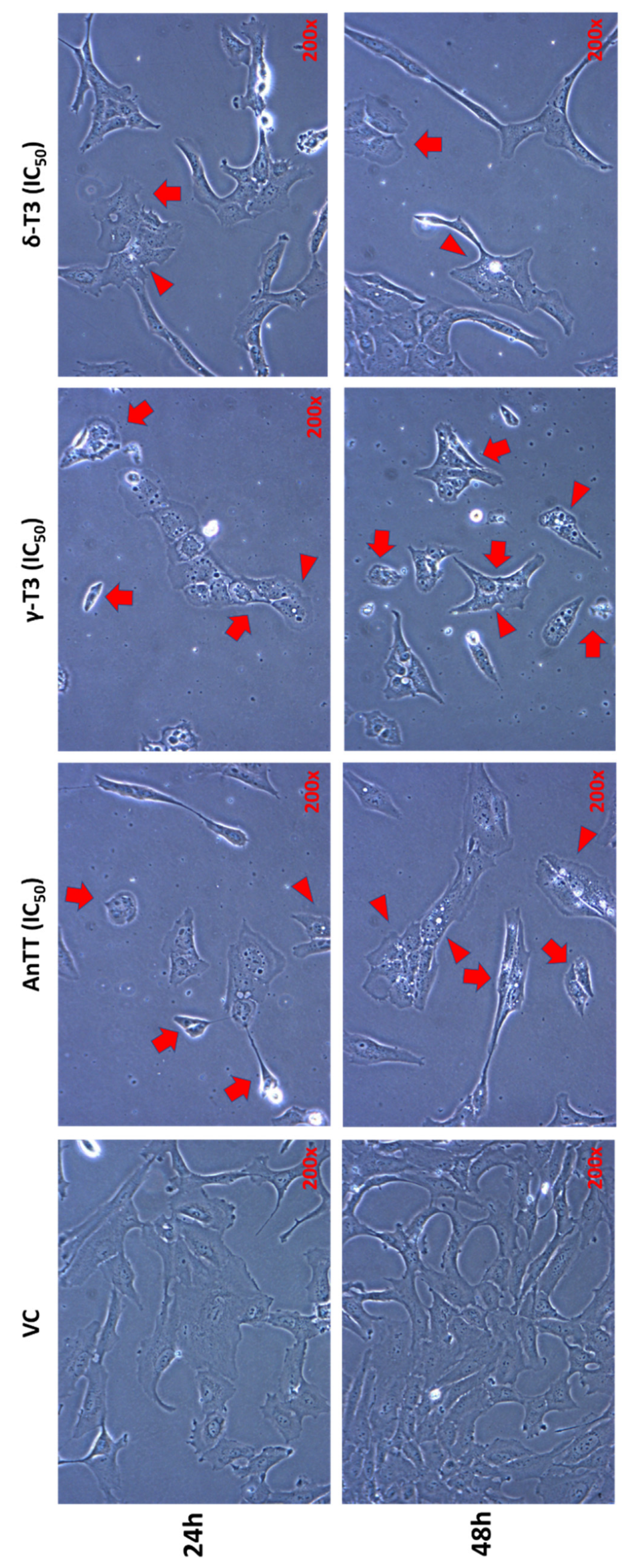
3.3. Morphological Changes of SW1353 Cells upon 24 h and 48 h AnTT, γ-T3 and δ-T3 Treatment
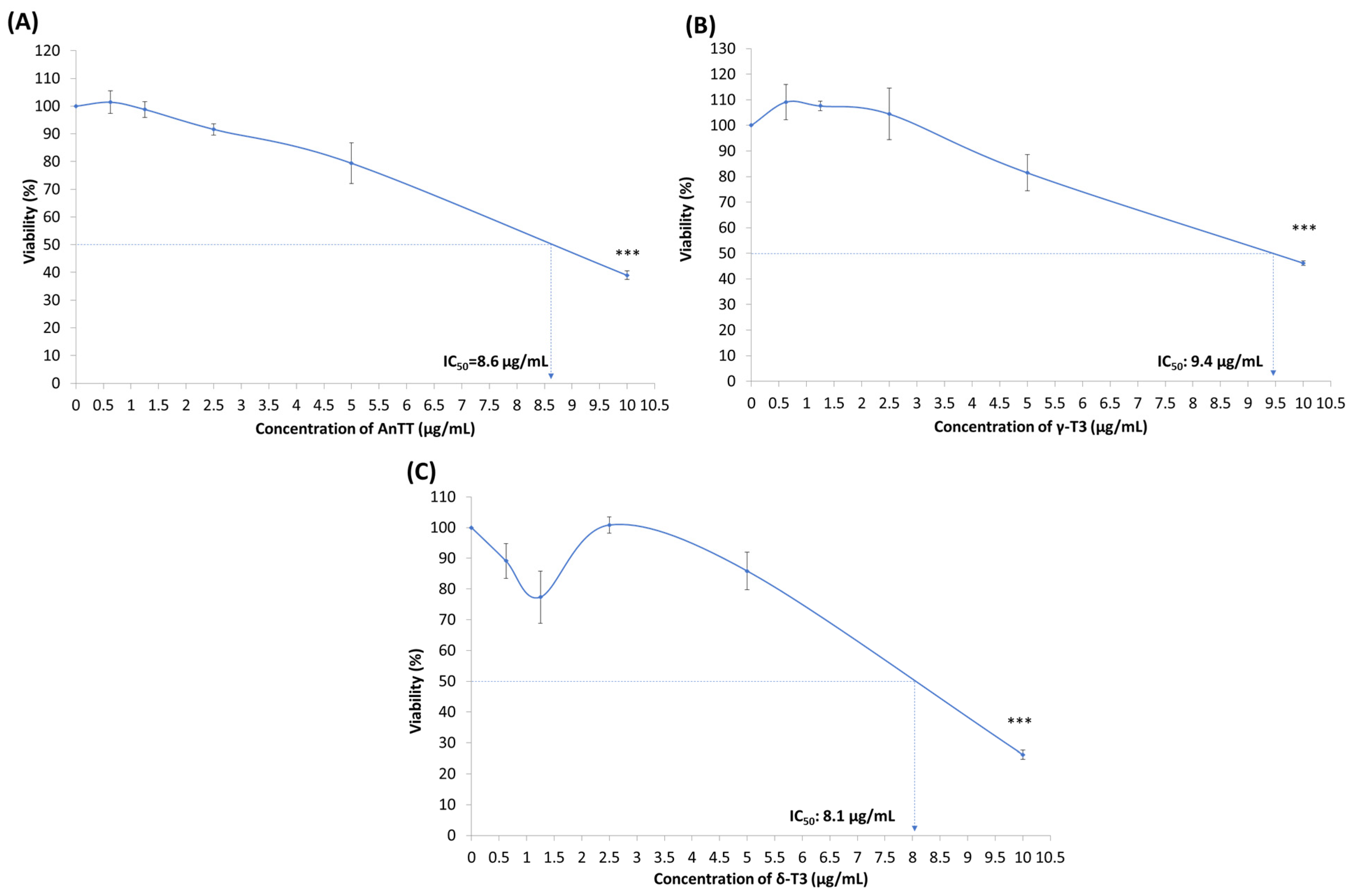
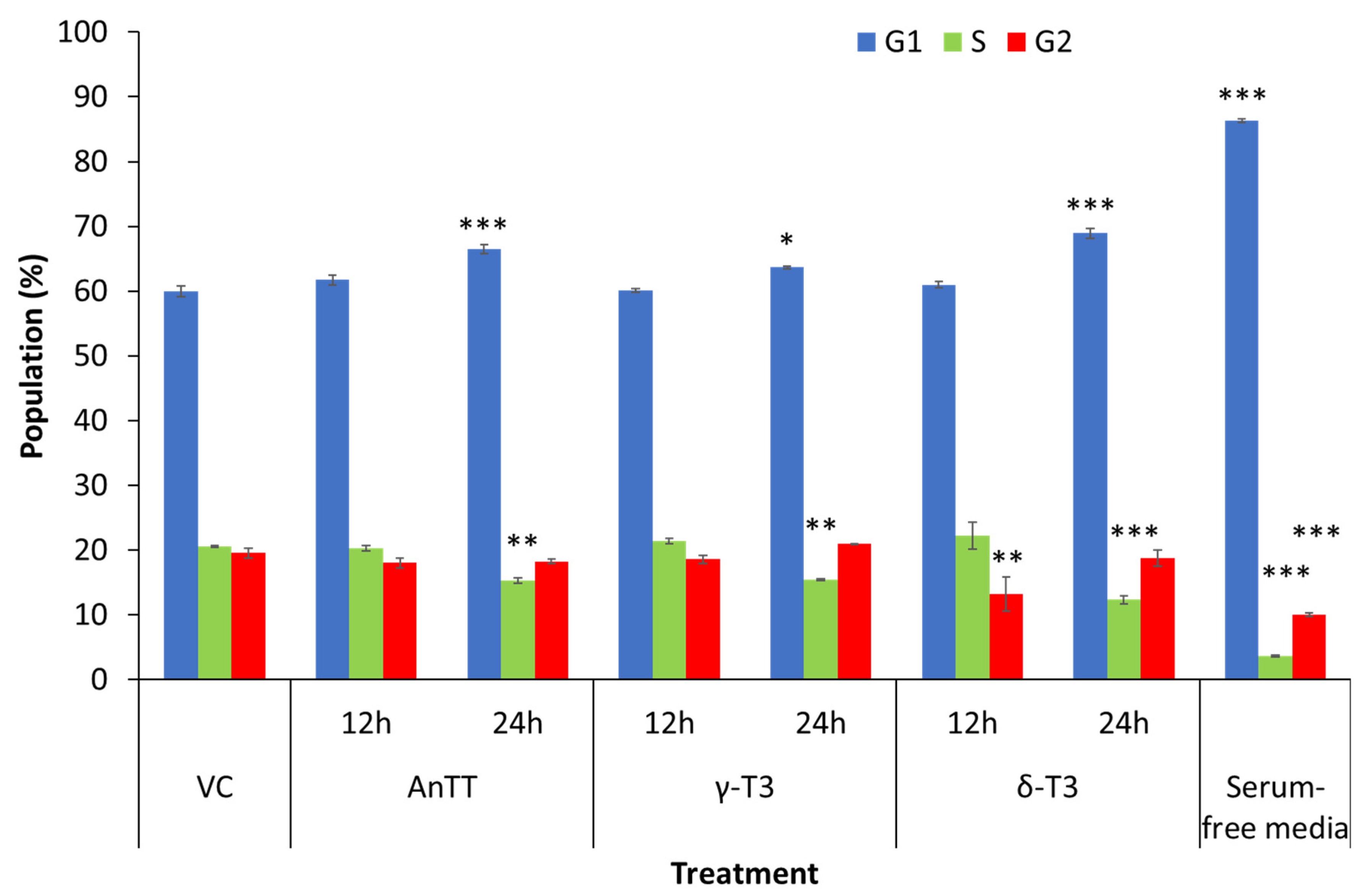
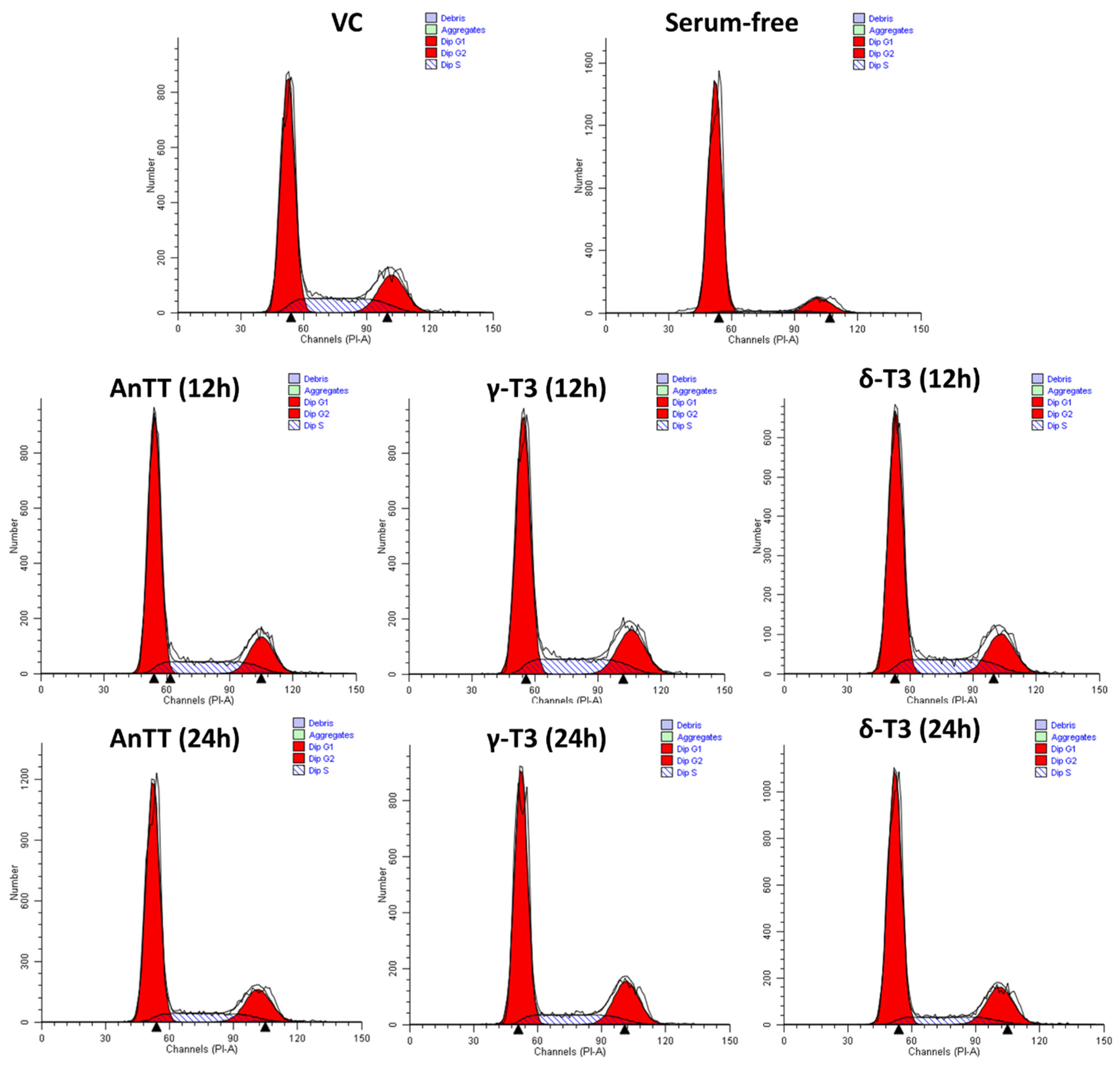
3.4. Antiproliferative and Growth Arrest Action of AnTT, γ-T3 and δ-T3 on SW1353 Cells
3.5. RNA Isolation and Quality
3.6. Data Generation and Quality of RNA Sequencing Reads
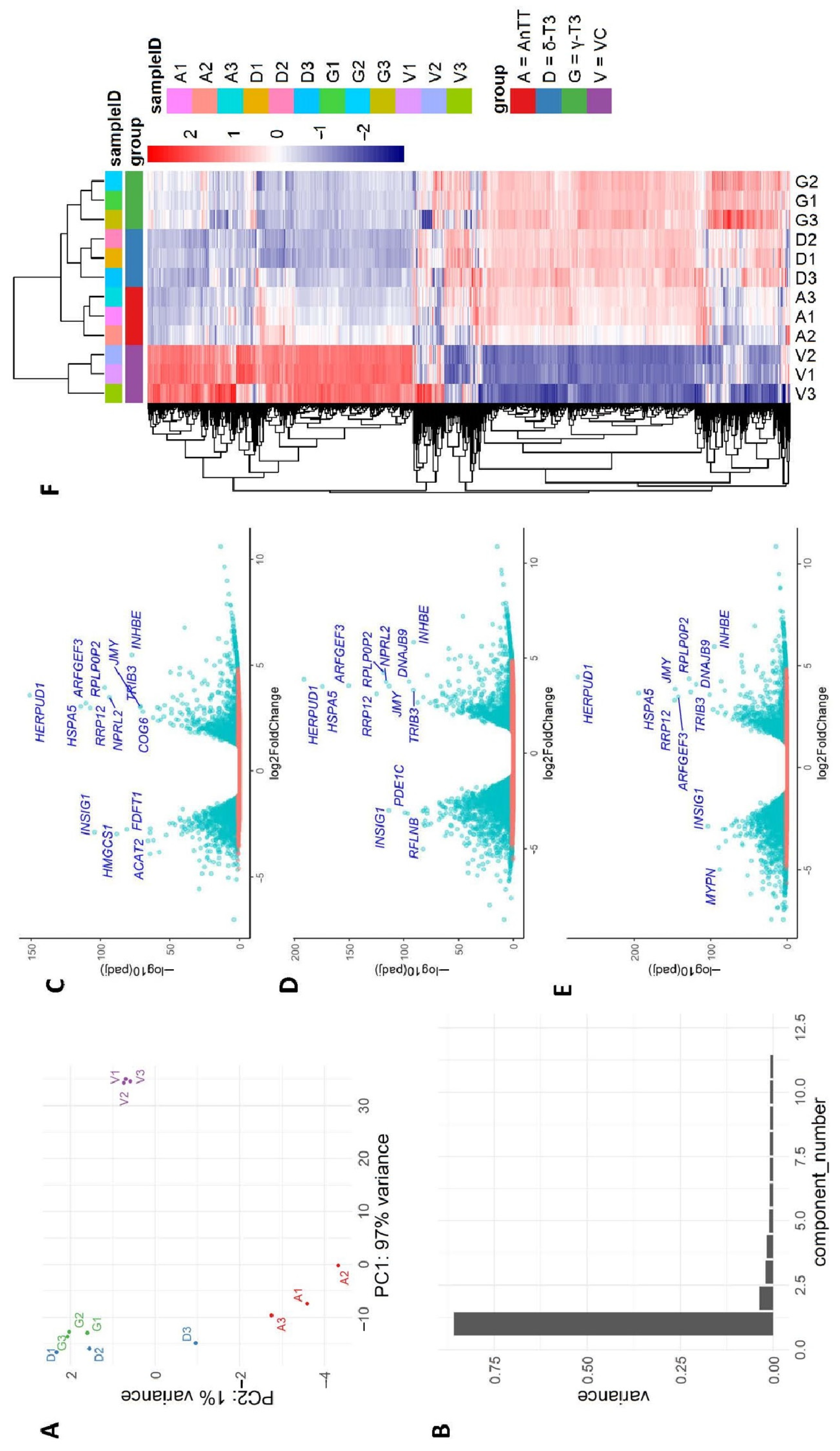
3.7. DEGs Identification upon Tocotrienol Treatment
3.8. GO Enrichment Analysis
3.9. KEGG Pathway Enrichment Analysis
3.10. qPCR Validation
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chin, K.-Y.; Pang, K.-L.; Soelaiman, I.-N. Tocotrienol and Its Role in Chronic Diseases. In Anti-Inflammatory Nutraceuticals and Chronic Diseases; Gupta, S.C., Prasad, S., Aggarwal, B.B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 97–130. [Google Scholar] [CrossRef]
- Chin, K.Y.; Ima-Nirwana, S. The biological effects of tocotrienol on bone: A review on evidence from rodent models. Drug Des. Dev. Ther. 2015, 9, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.Y.; Ima-Nirwana, S. The Role of Tocotrienol in Preventing Male Osteoporosis-A Review of Current Evidence. Int. J. Mol. Sci. 2019, 20, 1355. [Google Scholar] [CrossRef]
- Chin, K.Y.; Ima-Nirwana, S. Vitamin E as an Antiosteoporotic Agent via Receptor Activator of Nuclear Factor Kappa-B Ligand Signaling Disruption: Current Evidence and Other Potential Research Areas. Evid.-Based Complement. Altern. Med. 2012, 2012, 747020. [Google Scholar] [CrossRef] [PubMed]
- Krager, K.J.; Pineda, E.N.; Kharade, S.V.; Kordsmeier, M.; Howard, L.; Breen, P.J.; Compadre, C.M.; Hauer-Jensen, M.; Aykin-Burns, N. Tocotrienol-Rich Fraction from Rice Bran Demonstrates Potent Radiation Protection Activity. Evid.-Based Complement. Altern. Med. 2015, 2015, 148791. [Google Scholar] [CrossRef]
- Pang, K.-L.; Ghafar, N.A.; Soelaiman, I.N.; Chin, K.-Y. Protective Effects of Annatto Tocotrienol and Palm Tocotrienol-Rich Fraction on Chondrocytes Exposed to Monosodium Iodoacetate. Appl. Sci. 2021, 11, 9643. [Google Scholar] [CrossRef]
- Frega, N.; Mozzon, M.; Bocci, F. Identification and estimation of tocotrienols in the annatto lipid fraction by gas chromatography-mass spectrometry. J. Am. Oil Chem. Soc. 1998, 75, 1723–1727. [Google Scholar] [CrossRef]
- Wong, S.K.; Kamisah, Y.; Mohamed, N.; Muhammad, N.; Masbah, N.; Fahami, N.A.M.; Mohamed, I.N.; Shuid, A.N.; Saad, Q.M.; Abdullah, A.; et al. Potential Role of Tocotrienols on Non-Communicable Diseases: A Review of Current Evidence. Nutrients 2020, 12, 259. [Google Scholar] [CrossRef]
- Ramdas, P.; Rajihuzzaman, M.; Veerasenan, S.D.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienol-treated MCF-7 Human Breast Cancer Cells Show Down-regulation of API5 and Up-regulation of MIG6 Genes. Cancer Genom. Proteom. 2011, 8, 19. [Google Scholar]
- Nesaretnam, K.; Meganathan, P.; Veerasenan, S.D.; Selvaduray, K.R. Tocotrienols and breast cancer: The evidence to date. Genes Nutr. 2012, 7, 3–9. [Google Scholar] [CrossRef]
- Wada, S.; Naito, Y.; Matsushita, Y.; Nouchi, M.; Kawai, M.; Minami, E.; Aoi, W.; Ikeda, S.; Higashi, A.; Yoshikawa, T. δ-Tocotrienol suppresses tumorigenesis by inducing apoptosis and blocking the COX-2/PGE2 pathway that stimulates tumor–stromal interactions in colon cancer. J. Funct. Foods 2017, 35, 428–435. [Google Scholar] [CrossRef]
- Agarwal, M.K.; Agarwal, M.L.; Athar, M.; Gupta, S. Tocotrienol-rich fraction of palm oil activates p53, modulates Bax/Bcl2 ratio and induces apoptosis independent of cell cycle association. Cell Cycle 2004, 3, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K.; Aggarwal, B.B. γ-Tocotrienol suppresses growth and sensitises human colorectal tumours to capecitabine in a nude mouse xenograft model by down-regulating multiple molecules. Br. J. Cancer 2016, 115, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Rajasinghe, L.D.; Pindiprolu, R.H.; Gupta, S.V. Delta-tocotrienol inhibits non-small-cell lung cancer cell invasion via the inhibition of NF-κB, uPA activator, and MMP-9. OncoTargets Ther. 2018, 11, 4301–4314. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.; Atia, A. The role of tocotrienols in liver health and disease. Curr. Top. Pharmacol. 2018, 22, 135–142. [Google Scholar]
- Tang, K.D.; Liu, J.; Russell, P.J.; Clements, J.A.; Ling, M.-T. Gamma-Tocotrienol Induces Apoptosis in Prostate Cancer Cells by Targeting the Ang-1/Tie-2 Signalling Pathway. Int. J. Mol. Sci. 2019, 20, 1164. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.N.; Yap, W.N.; Lee, D.T.; Ling, M.T.; Wong, Y.C.; Yap, Y.L. Evidence of gamma-tocotrienol as an apoptosis-inducing, invasion-suppressing, and chemotherapy drug-sensitizing agent in human melanoma cells. Nutr. Cancer 2009, 61, 357–366. [Google Scholar] [CrossRef]
- Marzagalli, M.; Moretti, R.M.; Messi, E.; Marelli, M.M.; Fontana, F.; Anastasia, A.; Bani, M.R.; Beretta, G.; Limonta, P. Targeting melanoma stem cells with the Vitamin E derivative δ-tocotrienol. Sci. Rep. 2018, 8, 587. [Google Scholar] [CrossRef]
- Montagnani Marelli, M.; Marzagalli, M.; Moretti, R.M.; Beretta, G.; Casati, L.; Comitato, R.; Gravina, G.L.; Festuccia, C.; Limonta, P. Vitamin E δ-tocotrienol triggers endoplasmic reticulum stress-mediated apoptosis in human melanoma cells. Sci. Rep. 2016, 6, 30502. [Google Scholar] [CrossRef]
- Sun, W.; Wang, Q.; Chen, B.; Liu, J.; Liu, H.; Xu, W. gamma-Tocotrienol-induced apoptosis in human gastric cancer SGC-7901 cells is associated with a suppression in mitogen-activated protein kinase signalling. Br. J. Nutr. 2008, 99, 1247–1254. [Google Scholar] [CrossRef]
- Ghanem, P.; Zouein, A.; Mohamad, M.; Hodroj, M.H.; Haykal, T.; Abou Najem, S.; Naim, H.Y.; Rizk, S. The Vitamin E Derivative Gamma Tocotrienol Promotes Anti-Tumor Effects in Acute Myeloid Leukemia Cell Lines. Nutrients 2019, 11, 2808. [Google Scholar] [CrossRef]
- Kumar, K.S.; Raghavan, M.; Hieber, K.; Ege, C.; Mog, S.; Parra, N.; Hildabrand, A.; Singh, V.; Srinivasan, V.; Toles, R.; et al. Preferential radiation sensitization of prostate cancer in nude mice by nutraceutical antioxidant gamma-tocotrienol. Life Sci. 2006, 78, 2099–2104. [Google Scholar] [CrossRef]
- Nesaretnam, K.; Ambra, R.; Selvaduray, K.R.; Radhakrishnan, A.; Reimann, K.; Razak, G.; Virgili, F. Tocotrienol-rich fraction from palm oil affects gene expression in tumors resulting from MCF-7 cell inoculation in athymic mice. Lipids 2004, 39, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-S.; Zhang, S.-J.; Li, Q.; Liu, Y.-H.; He, N.; Zhang, J.; Zhou, P.-H.; Li, M.; Guan, T.; Liu, J.-R. Tocotrienol-rich fraction (TRF) suppresses the growth of human colon cancer xenografts in Balb/C nude mice by the Wnt pathway. PLoS ONE 2015, 10, e0122175. [Google Scholar] [CrossRef] [PubMed]
- Nesaretnam, K.; Selvaduray, K.R.; Abdul Razak, G.; Veerasenan, S.D.; Gomez, P.A. Effectiveness of tocotrienol-rich fraction combined with tamoxifen in the management of women with early breast cancer: A pilot clinical trial. Breast Cancer Res. 2010, 12, R81. [Google Scholar] [CrossRef]
- Wali, V.B.; Bachawal, S.V.; Sylvester, P.W. Endoplasmic reticulum stress mediates gamma-tocotrienol-induced apoptosis in mammary tumor cells. Apoptosis 2009, 14, 1366–1377. [Google Scholar] [CrossRef]
- Park, S.K.; Sanders, B.G.; Kline, K. Tocotrienols induce apoptosis in breast cancer cell lines via an endoplasmic reticulum stress-dependent increase in extrinsic death receptor signaling. Breast Cancer Res. Treat. 2010, 124, 361–375. [Google Scholar] [CrossRef]
- Xu, W.; Mi, Y.; He, P.; He, S.; Niu, L. γ-Tocotrienol Inhibits Proliferation and Induces Apoptosis Via the Mitochondrial Pathway in Human Cervical Cancer HeLa Cells. Molecules 2017, 22, 1299. [Google Scholar] [CrossRef]
- Ahmed, R.; Alawin, O.; Sylvester, P. γ-Tocotrienol reversal of epithelial-to-mesenchymal transition in human breast cancer cells is associated with inhibition of canonical Wnt signalling. Cell Prolif. 2016, 49, 460–470. [Google Scholar] [CrossRef]
- Zhang, J.-S.; Li, D.-M.; He, N.; Liu, Y.-H.; Wang, C.-H.; Jiang, S.-Q.; Chen, B.-Q.; Liu, J.-R. A paraptosis-like cell death induced by δ-tocotrienol in human colon carcinoma SW620 cells is associated with the suppression of the Wnt signaling pathway. Toxicology 2011, 285, 8–17. [Google Scholar] [CrossRef]
- Shah, S.J.; Sylvester, P.W. Gamma-tocotrienol inhibits neoplastic mammary epithelial cell proliferation by decreasing Akt and nuclear factor kappaB activity. Exp. Biol. Med. 2005, 230, 235–241. [Google Scholar] [CrossRef]
- Srivastava, J.K.; Gupta, S. Tocotrienol-rich fraction of palm oil induces cell cycle arrest and apoptosis selectively in human prostate cancer cells. Biochem. Biophys. Res. Commun. 2006, 346, 447–453. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, B.S.; Briski, K.P.; Tirmenstein, M.A.; Fariss, M.W.; Gapor, A.; Sylvester, P.W. Antiproliferative and apoptotic effects of tocopherols and tocotrienols on normal mouse mammary epithelial cells. Lipids 2000, 35, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Furukawa, F.; Nishikawa, A.; Miyauchi, M.; Son, H.-Y.; Imazawa, T.; Hirose, M. Oral toxicity of a tocotrienol preparation in rats. Food Chem. Toxicol. 2001, 39, 799–805. [Google Scholar] [CrossRef]
- Bendich, A.; Machlin, L.J. Safety of oral intake of vitamin E. Am. J. Clin. Nutr. 1988, 48, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.-L.; Chin, K.-Y. The Role of Tocotrienol in Protecting Against Metabolic Diseases. Molecules 2019, 24, 923. [Google Scholar] [CrossRef] [PubMed]
- Hor, C.P.; Fung, W.Y.; Ang, H.A.; Lim, S.C.; Kam, L.Y.; Sim, S.W.; Lim, L.H.; Choon, W.Y.; Wong, J.W.; Ch’ng, A.S.H.; et al. Efficacy of Oral Mixed Tocotrienols in Diabetic Peripheral Neuropathy: A Randomized Clinical Trial. JAMA Neurol. 2018, 75, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Rasool, A.H.; Yuen, K.H.; Yusoff, K.; Wong, A.R.; Rahman, A.R. Dose dependent elevation of plasma tocotrienol levels and its effect on arterial compliance, plasma total antioxidant status, and lipid profile in healthy humans supplemented with tocotrienol rich vitamin E. J. Nutr. Sci. Vitaminol. 2006, 52, 473–478. [Google Scholar] [CrossRef]
- Shen, C.-L.; Wang, S.; Yang, S.; Tomison, M.D.; Abbasi, M.; Hao, L.; Scott, S.; Khan, M.S.; Romero, A.W.; Felton, C.K.; et al. A 12-week evaluation of annatto tocotrienol supplementation for postmenopausal women: Safety, quality of life, body composition, physical activity, and nutrient intake. BMC Complement. Altern. Med. 2018, 18, 198. [Google Scholar] [CrossRef] [PubMed]
- Leddy, L.R.; Holmes, R.E. Chondrosarcoma of bone. In Orthopaedic Oncology; Springer: Cham, Switzerland, 2014; pp. 117–130. [Google Scholar]
- van Praag Veroniek, V.M.; Rueten-Budde, A.J.; Ho, V.; Dijkstra, P.D.S.; Fiocco, M.; van de Sande, M.A.J. Incidence, outcomes and prognostic factors during 25 years of treatment of chondrosarcomas. Surg. Oncol. 2018, 27, 402–408. [Google Scholar] [CrossRef]
- Zając, A.E.; Kopeć, S.; Szostakowski, B.; Spałek, M.J.; Fiedorowicz, M.; Bylina, E.; Filipowicz, P.; Szumera-Ciećkiewicz, A.; Tysarowski, A.; Czarnecka, A.M.; et al. Chondrosarcoma-from Molecular Pathology to Novel Therapies. Cancers 2021, 13, 2390. [Google Scholar] [CrossRef] [PubMed]
- Verdegaal, S.H.M.; Bovée, J.V.M.G.; Pansuriya, T.C.; Grimer, R.J.; Ozger, H.; Jutte, P.C.; San Julian, M.; Biau, D.J.; van der Geest, I.C.M.; Leithner, A.; et al. Incidence, predictive factors, and prognosis of chondrosarcoma in patients with Ollier disease and Maffucci syndrome: An international multicenter study of 161 patients. Oncologist 2011, 16, 1771–1779. [Google Scholar] [CrossRef]
- Gelderblom, H.; Hogendoorn, P.C.; Dijkstra, S.D.; Van Rijswijk, C.S.; Krol, A.D.; Taminiau, A.H.; Bovee, J.V. The clinical approach towards chondrosarcoma. Oncologist 2008, 13, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Kim, H.-J. Biomarkers of chondrosarcoma. J. Clin. Pathol. 2018, 71, 579. [Google Scholar] [CrossRef] [PubMed]
- Yokota, K.; Sakamoto, A.; Matsumoto, Y.; Matsuda, S.; Harimaya, K.; Oda, Y.; Iwamoto, Y. Clinical outcome for patients with dedifferentiated chondrosarcoma: A report of 9 cases at a single institute. J. Orthop. Surg. Res. 2012, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, G.; Chen, X.; Huang, X.; Liu, M.; Pan, W.; Yan, X.; Lin, N.; Ye, Z. Predictors of the survival of patients with chondrosarcoma of bone and metastatic disease at diagnosis. J. Cancer 2019, 10, 2457–2463. [Google Scholar] [CrossRef] [PubMed]
- Thorkildsen, J.; Taksdal, I.; Bjerkehagen, B.; Haugland, H.K.; Børge Johannesen, T.; Viset, T.; Norum, O.J.; Bruland, Ø.; Zaikova, O. Chondrosarcoma in Norway 1990-2013; an epidemiological and prognostic observational study of a complete national cohort. Acta Oncol. 2019, 58, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Damron, T.A.; Ward, W.G.; Stewart, A. Osteosarcoma, chondrosarcoma, and Ewing’s sarcoma: National Cancer Data Base Report. Clin. Orthop. Relat. Res. 2007, 459, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Andreou, D.; Ruppin, S.; Fehlberg, S.; Pink, D.; Werner, M.; Tunn, P.-U. Survival and prognostic factors in chondrosarcoma: Results in 115 patients with long-term follow-up. Acta Orthop. 2011, 82, 749–755. [Google Scholar] [CrossRef]
- Donati, D.; Colangeli, S.; Colangeli, M.; Di Bella, C.; Bertoni, F. Surgical treatment of grade I central chondrosarcoma. Clin. Orthop. Relat. Res. 2010, 468, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Wan Hasan, W.N.; Abd Ghafar, N.; Chin, K.Y.; Ima-Nirwana, S. Annatto-derived tocotrienol stimulates osteogenic activity in preosteoblastic MC3T3-E1 cells: A temporal sequential study. Drug Des. Dev. Ther. 2018, 12, 1715–1726. [Google Scholar] [CrossRef] [PubMed]
- Wan Hasan, W.N.; Chin, K.Y.; Abd Ghafar, N.; Soelaiman, I.N. Annatto-Derived Tocotrienol Promotes Mineralization of MC3T3-E1 Cells by Enhancing BMP-2 Protein Expression via Inhibiting RhoA Activation and HMG-CoA Reductase Gene Expression. Drug Des. Dev. Ther. 2020, 14, 969–976. [Google Scholar] [CrossRef]
- Pang, K.L.; Chow, Y.Y.; Leong, L.M.; Law, J.X.; Ghafar, N.A.; Soelaiman, I.N.; Chin, K.Y. Establishing SW1353 Chondrocytes as a Cellular Model of Chondrolysis. Life 2021, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Leong, L.M.; Chan, K.M.; Hamid, A.; Latip, J.; Rajab, N.F. Herbal Formulation C168 Attenuates Proliferation and Induces Apoptosis in HCT 116 Human Colorectal Carcinoma Cells: Role of Oxidative Stress and DNA Damage. Evid.-Based Complement. Altern. Med. 2016, 2016, 2091085. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.-L.; Chin, K.-Y. Comment on: Food for Bone: Evidence for a Role for Delta-Tocotrienol in the Physiological Control of Osteoblast Migration. Int. J. Mol. Sci. 2020, 21, 4661. Int. J. Mol. Sci. 2020, 21, 6674. [Google Scholar] [CrossRef]
- Darzynkiewicz, Z. Critical aspects in analysis of cellular DNA content. Curr. Protoc. Cytom. 2010, 52, 7.2.1–7.2.8. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. Available online: https://www.bioconductor.org/packages/devel/bioc/manuals/enrichplot/man/enrichplot.pdf (accessed on 4 September 2022).
- Kolde, R. Heatmap: Pretty Heatmaps. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 4 September 2022).
- Wickham, H.; Navarro, D.; Pedersen, T.L. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- ADAMTSL1 ADAMTS Like 1 [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene?Cmd=DetailsSearch&Term=92949 (accessed on 5 July 2022).
- Pierpaoli, E.; Viola, V.; Barucca, A.; Orlando, F.; Galli, F.; Provinciali, M. Effect of annatto-tocotrienols supplementation on the development of mammary tumors in HER-2/neu transgenic mice. Carcinogenesis 2013, 34, 1352–1360. [Google Scholar] [CrossRef]
- Sato, C.; Kaneko, S.; Sato, A.; Virgona, N.; Namiki, K.; Yano, T. Combination Effect of δ-Tocotrienol and γ-Tocopherol on Prostate Cancer Cell Growth. J. Nutr. Sci. Vitaminol. 2017, 63, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, R.; Sato, A.; Uchida, A.; Shiozawa, S.; Sato, C.; Virgona, N.; Yano, T. Annatto Tocotrienol Induces a Cytotoxic Effect on Human Prostate Cancer PC3 Cells via the Simultaneous Inhibition of Src and Stat3. J. Nutr. Sci. Vitaminol. 2015, 61, 497–501. [Google Scholar] [CrossRef]
- Loganathan, R.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienols promote apoptosis in human breast cancer cells by inducing poly(ADP-ribose) polymerase cleavage and inhibiting nuclear factor kappa-B activity. Cell Prolif. 2013, 46, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Yap, W.N.; Chang, P.N.; Han, H.Y.; Lee, D.T.W.; Ling, M.T.; Wong, Y.C.; Yap, Y.L. γ-Tocotrienol suppresses prostate cancer cell proliferation and invasion through multiple-signalling pathways. Br. J. Cancer 2008, 99, 1832–1841. [Google Scholar] [CrossRef]
- Lim, S.-W.; Loh, H.-S.; Ting, K.-N.; Bradshaw, T.D.; Zeenathul, N.A. Cytotoxicity and apoptotic activities of alpha-, gamma- and delta-tocotrienol isomers on human cancer cells. BMC Complement. Altern. Med. 2014, 14, 469. [Google Scholar] [CrossRef]
- Wada, S.; Satomi, Y.; Murakoshi, M.; Noguchi, N.; Yoshikawa, T.; Nishino, H. Tumor suppressive effects of tocotrienol in vivo and in vitro. Cancer Lett. 2005, 229, 181–191. [Google Scholar] [CrossRef]
- Yang, Z.; Xiao, H.; Jin, H.; Koo, P.T.; Tsang, D.J.; Yang, C.S. Synergistic actions of atorvastatin with gamma-tocotrienol and celecoxib against human colon cancer HT29 and HCT116 cells. Int. J. Cancer 2010, 126, 852–863. [Google Scholar] [CrossRef]
- Fernandes, N.V.; Guntipalli, P.K.; Mo, H. d-δ-Tocotrienol-mediated Cell Cycle Arrest and Apoptosis in Human Melanoma Cells. Anticancer Res. 2010, 30, 4937. [Google Scholar] [PubMed]
- Wang, C.; Husain, K.; Zhang, A.; Centeno, B.A.; Chen, D.-T.; Tong, Z.; Sebti, S.M.; Malafa, M.P. EGR-1/Bax pathway plays a role in vitamin E δ-tocotrienol-induced apoptosis in pancreatic cancer cells. J. Nutr. Biochem. 2015, 26, 797–807. [Google Scholar] [CrossRef]
- Idriss, M.; Hodroj, M.H.; Fakhoury, R.; Rizk, S. Beta-Tocotrienol Exhibits More Cytotoxic Effects than Gamma-Tocotrienol on Breast Cancer Cells by Promoting Apoptosis via a P53-Independent PI3-Kinase Dependent Pathway. Biomolecules 2020, 10, 577. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, P.W.; Nachnani, A.; Shah, S.; Briski, K.P. Role of GTP-binding proteins in reversing the antiproliferative effects of tocotrienols in preneoplastic mammary epithelial cells. Asia Pac. J. Clin. Nutr. 2002, 11 (Suppl. 7), S452–S459. [Google Scholar] [CrossRef]
- Husain, K.; Francois, R.A.; Hutchinson, S.Z.; Neuger, A.M.; Lush, R.; Coppola, D.; Sebti, S.; Malafa, M.P. Vitamin E delta-tocotrienol levels in tumor and pancreatic tissue of mice after oral administration. Pharmacology 2009, 83, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Hiura, Y.; Tachibana, H.; Arakawa, R.; Aoyama, N.; Okabe, M.; Sakai, M.; Yamada, K. Specific accumulation of gamma- and delta-tocotrienols in tumor and their antitumor effect in vivo. J. Nutr. Biochem. 2009, 20, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Bradlow, B.A.; Brace, L.; Manganello, J.; Peterson, D.M.; Pearce, B.C.; Wright, J.J.; Gapor, A.; Elson, C.E. Response of hypercholesterolemic subjects to administration of tocotrienols. Lipids 1995, 30, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Pearce, B.C.; Nor, R.M.; Gapor, A.; Peterson, D.M.; Elson, C.E. Dietary alpha-tocopherol attenuates the impact of gamma-tocotrienol on hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in chickens. J. Nutr. 1996, 126, 389–394. [Google Scholar] [CrossRef]
- Khalili, M.; Radosevich, J.A. Paraptosis. In Apoptosis and Beyond; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 343–366. [Google Scholar] [CrossRef]
- Zhang, J.-S.; Li, D.-M.; Ma, Y.; He, N.; Gu, Q.; Wang, F.-S.; Jiang, S.-Q.; Chen, B.-Q.; Liu, J.-R. γ-Tocotrienol induces paraptosis-like cell death in human colon carcinoma SW620 cells. PLoS ONE 2013, 8, e57779. [Google Scholar] [CrossRef]
- Raimondi, M.; Fontana, F.; Marzagalli, M.; Audano, M.; Beretta, G.; Procacci, P.; Sartori, P.; Mitro, N.; Limonta, P. Ca2+ overload- and ROS-associated mitochondrial dysfunction contributes to δ-tocotrienol-mediated paraptosis in melanoma cells. Apoptosis 2021, 26, 277–292. [Google Scholar] [CrossRef]
- Fontana, F.; Moretti, R.M.; Raimondi, M.; Marzagalli, M.; Beretta, G.; Procacci, P.; Sartori, P.; Montagnani Marelli, M.; Limonta, P. δ-Tocotrienol induces apoptosis, involving endoplasmic reticulum stress and autophagy, and paraptosis in prostate cancer cells. Cell Prolif. 2019, 52, e12576. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, F.; Yang, Y.; Wang, P.; Zhang, Z.; Tang, Y.; Shen, Y.; Wang, K. Paraptosis-Inducing Nanomedicine Overcomes Cancer Drug Resistance for a Potent Cancer Therapy. Small 2018, 14, 1702446. [Google Scholar] [CrossRef]
- Ram, B.M.; Ramakrishna, G. Endoplasmic reticulum vacuolation and unfolded protein response leading to paraptosis like cell death in cyclosporine A treated cancer cervix cells is mediated by cyclophilin B inhibition. Biochim. Biophys. Acta 2014, 1843, 2497–2512. [Google Scholar] [CrossRef]
- Comitato, R.; Guantario, B.; Leoni, G.; Nesaretnam, K.; Ronci, M.B.; Canali, R.; Virgili, F. Tocotrienols induce endoplasmic reticulum stress and apoptosis in cervical cancer cells. Genes Nutr. 2016, 11, 32. [Google Scholar] [CrossRef]
- Adams, C.J.; Kopp, M.C.; Larburu, N.; Nowak, P.R.; Ali, M.M.U. Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front. Mol. Biosci. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Patacsil, D.; Tran, A.T.; Cho, Y.S.; Suy, S.; Saenz, F.; Malyukova, I.; Ressom, H.; Collins, S.P.; Clarke, R.; Kumar, D. Gamma-tocotrienol induced apoptosis is associated with unfolded protein response in human breast cancer cells. J. Nutr. Biochem. 2012, 23, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.V.; Parajuli, P.; Sylvester, P.W. γ-Tocotrienol-induced endoplasmic reticulum stress and autophagy act concurrently to promote breast cancer cell death. Biochem. Cell Biol. 2015, 93, 306–320. [Google Scholar] [CrossRef] [PubMed]




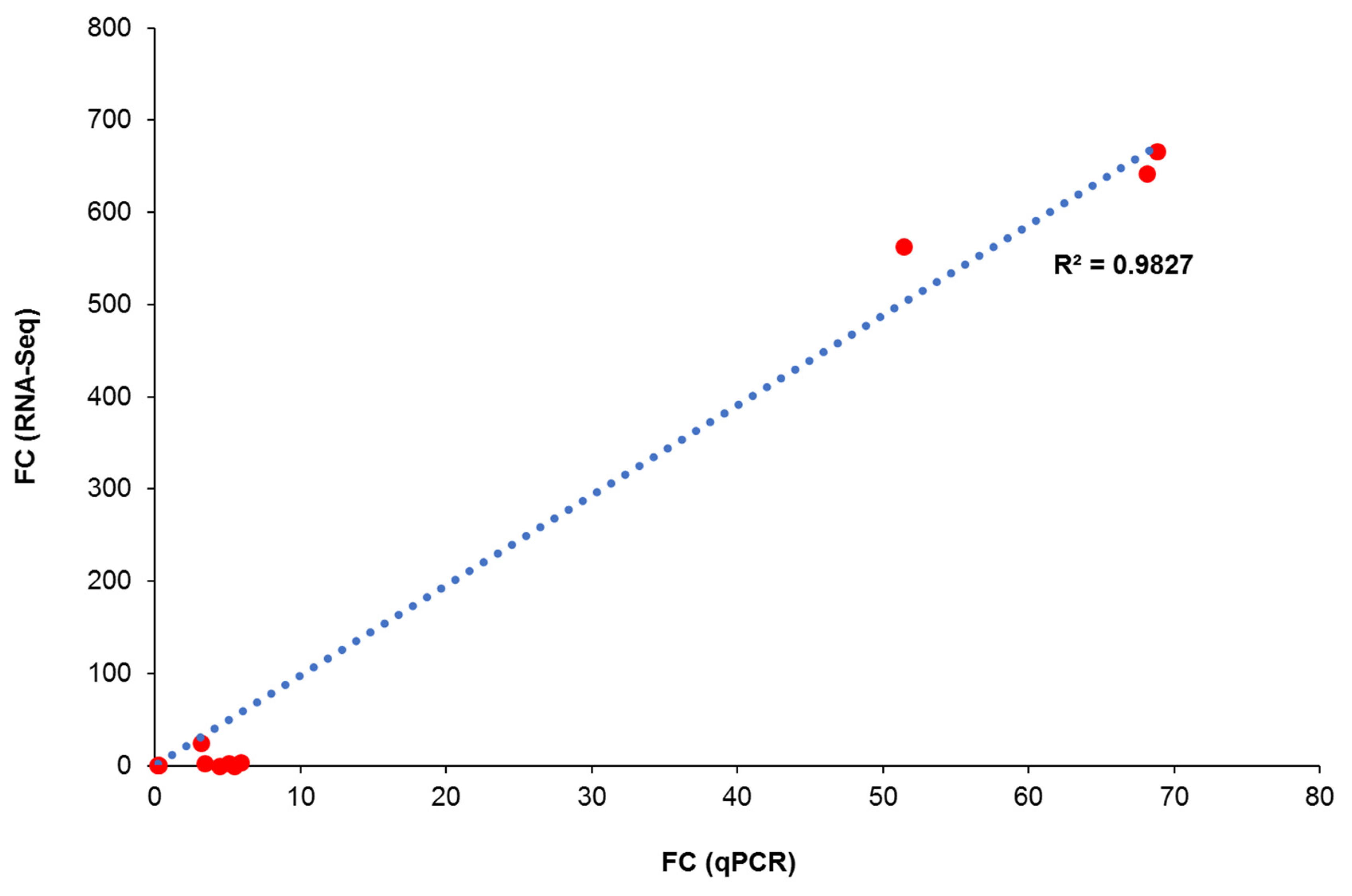
| Gene ID | AnTT vs. VC | δ-T3 vs. VC | γ-T3 vs. VC | AnTT vs. δ-T3 | AnTT vs. γ-T3 | δ-T3 vs. γ-T3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FC (RNA-seq) | FC (qPCR) | FC (RNA-seq) | FC (qPCR) | FC (RNA-seq) | FC (qPCR) | FC (RNA-seq) | FC (qPCR) | FC (RNA-seq) | FC (qPCR) | FC (RNA-seq) | FC (qPCR) | |
| ARMCX3 | 2.42 | 3.42 | 2.98 | 5.02 | 3.55 | 5.92 | N.A. | 0.68 | N.A. | 0.58 | N.A. | 0.85 |
| HMGCS1 | 0.13 | 0.02 | 0.14 | 0.21 | 0.15 | 0.23 | N.A. | 0.94 | N.A. | 0.85 | N.A. | 0.91 |
| ADAMTSL1 | 0.04 | 3.06 | 0.03 | 4.33 | 0.01 | 5.20 | N.A. | 0.70 | N.A. | 0.59 | N.A. | 0.83 |
| KLHDC7B | 562.64 | 45.92 | 666.12 | 66.26 | 642.02 | 62.92 | N.A. | 0.69 | N.A. | 0.73 | N.A. | 1.05 |
| Tocotrienols | Cells | IC50 Values (μg/mL) | References |
|---|---|---|---|
| AnTT | Human breast cancer SKBR3 cells | 24 h: 14.33 * | [65] |
| Human prostate cancer LNCaP cells | 24 h: <1.99 * | [66] | |
| Human prostate cancer PC-3 cells | 48 h: <10 | [67] | |
| δ-T3 | Human breast cancer SKBR3 cells | 24 h: 8.73 | [65] |
| Human breast cancer MDA-MB-231 cells | 72 h: 6.9 | [68] | |
| Human breast cancer MCF-7 cells | 72 h: 6.8 | ||
| Human prostate cancer LNCap cells | 24 h: 22.61 | [69] | |
| 48 h: 21.02 | |||
| 72 h: 13.48 | |||
| Human prostate cancer PC-3 cells | 24 h: 23.40 | ||
| 48 h: 7.14 | |||
| 72 h: 3.57 | |||
| Human lung cancer A549 cells | 24 h: 0.68 | [70] | |
| 48 h: 0.63 | |||
| 72 h: 0.50 | |||
| Human glioma U87MG cells | 24 h: 0.58 | ||
| 48 h: 0.51 | |||
| 72 h: 0.45 | |||
| Human liver cancer HepG2 cells | 72 h: 3.81 | [71] | |
| Human liver cancer HCT116 cells | 48 h: 7.93 | [72] | |
| Human liver cancer HT29 cells | 48 h: 11.90 | ||
| Human melanoma A2058 cells | 72 h: 14.87 | [73] | |
| Human melanoma A375 cells | 72 h: 8.84 | ||
| Human pancreatic cancer MiaPaCa-2 cells | 72 h: 19.83 | [74] | |
| γ-T3 | Human breast cancer MDA-MB-231 | 24 h: 16.01 48 h: 12.73 | [75] |
| 72 h: 4.7 | [68] | ||
| Human breast cancer MCF-7 cells | 24 h: 16.88 48 h: 13.51 | [75] | |
| 72 h: 6.35 | [68] | ||
| Human prostate cancer LNCap cells | 24 h: 24.23 | [69] | |
| 48 h: 20.12 | |||
| 72 h: 18.48 | |||
| Human prostate cancer PC-3 cells | 24 h: 21.76 | ||
| 48 h: 4.11 | |||
| 72 h: 3.28 | |||
| Human lung cancer A549 cells | 24 h: 1.17 | [70] | |
| 48 h: 1.05 | |||
| 72 h: 0.71 | |||
| Human glioma U87MG cells | 24 h: 2.06 | ||
| 48 h: 1.54 | |||
| 72 h: 1.36 | |||
| Human liver cancer HepG2 cells | 72 h: 10.14 | [71] | |
| Human liver cancer HCT116 cells | 48 h: 7.19 | [72] | |
| Human liver cancer HT29 cells | 48 h: 12.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, K.-L.; Foong, L.-C.; Abd Ghafar, N.; Soelaiman, I.N.; Law, J.X.; Leong, L.M.; Chin, K.-Y. Transcriptomic Analysis of the Anticancer Effects of Annatto Tocotrienol, Delta-Tocotrienol and Gamma-Tocotrienol on Chondrosarcoma Cells. Nutrients 2022, 14, 4277. https://doi.org/10.3390/nu14204277
Pang K-L, Foong L-C, Abd Ghafar N, Soelaiman IN, Law JX, Leong LM, Chin K-Y. Transcriptomic Analysis of the Anticancer Effects of Annatto Tocotrienol, Delta-Tocotrienol and Gamma-Tocotrienol on Chondrosarcoma Cells. Nutrients. 2022; 14(20):4277. https://doi.org/10.3390/nu14204277
Chicago/Turabian StylePang, Kok-Lun, Lian-Chee Foong, Norzana Abd Ghafar, Ima Nirwana Soelaiman, Jia Xian Law, Lek Mun Leong, and Kok-Yong Chin. 2022. "Transcriptomic Analysis of the Anticancer Effects of Annatto Tocotrienol, Delta-Tocotrienol and Gamma-Tocotrienol on Chondrosarcoma Cells" Nutrients 14, no. 20: 4277. https://doi.org/10.3390/nu14204277
APA StylePang, K.-L., Foong, L.-C., Abd Ghafar, N., Soelaiman, I. N., Law, J. X., Leong, L. M., & Chin, K.-Y. (2022). Transcriptomic Analysis of the Anticancer Effects of Annatto Tocotrienol, Delta-Tocotrienol and Gamma-Tocotrienol on Chondrosarcoma Cells. Nutrients, 14(20), 4277. https://doi.org/10.3390/nu14204277








