Intestinal Taxa Abundance and Diversity in Inflammatory Bowel Disease Patients: An Analysis including Covariates and Confounders
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Sequencing and Bioinformatic Analysis of 16S rRNA Amplicons
2.3. Statistical Analysis
3. Results
3.1. Microbiota Diversity Analysis, Study Cohort Characteristics and Identification of Covariates and Confounders
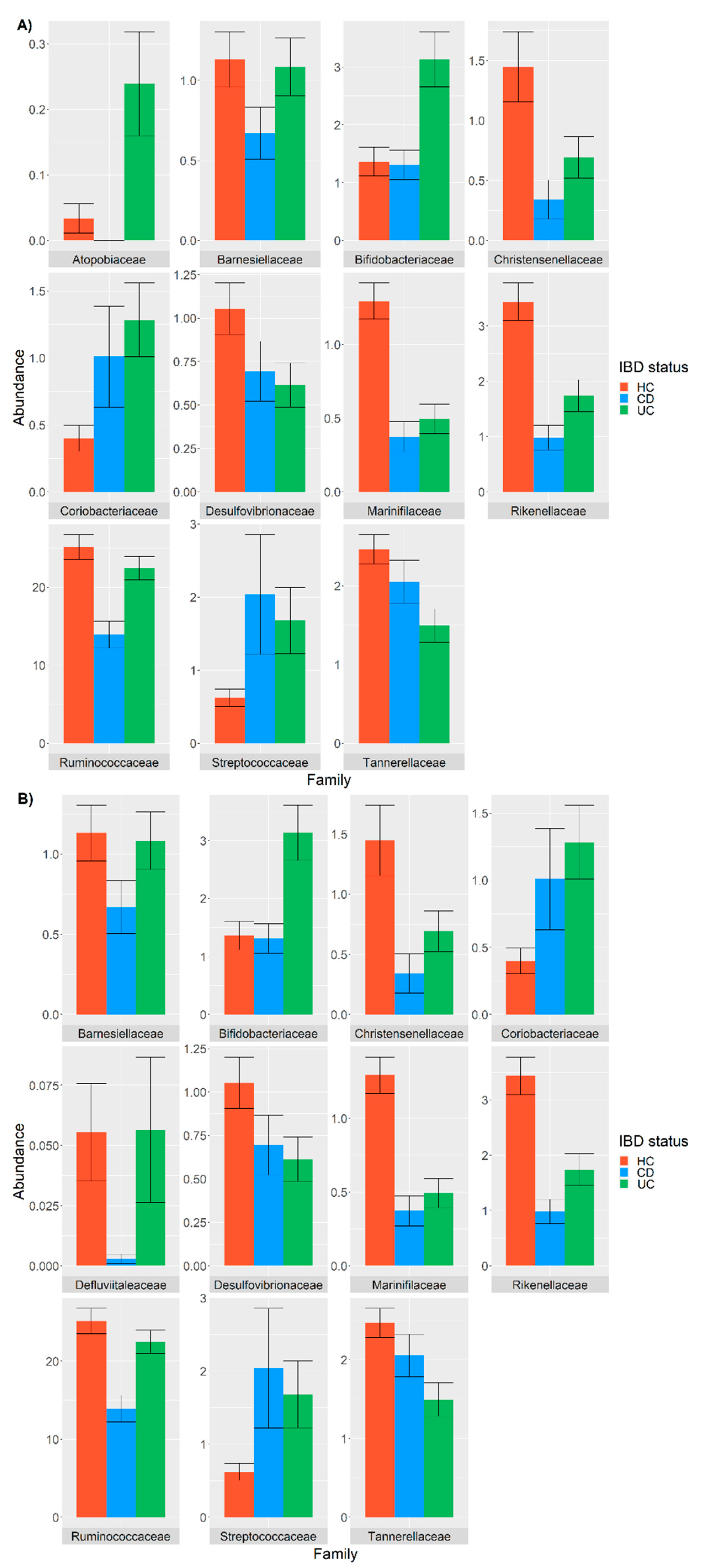
3.2. Differential Abundance Analysis between IBD and HC Taxa
3.3. Analysis of the Covariates and Confounders Influencing Taxa Abundance
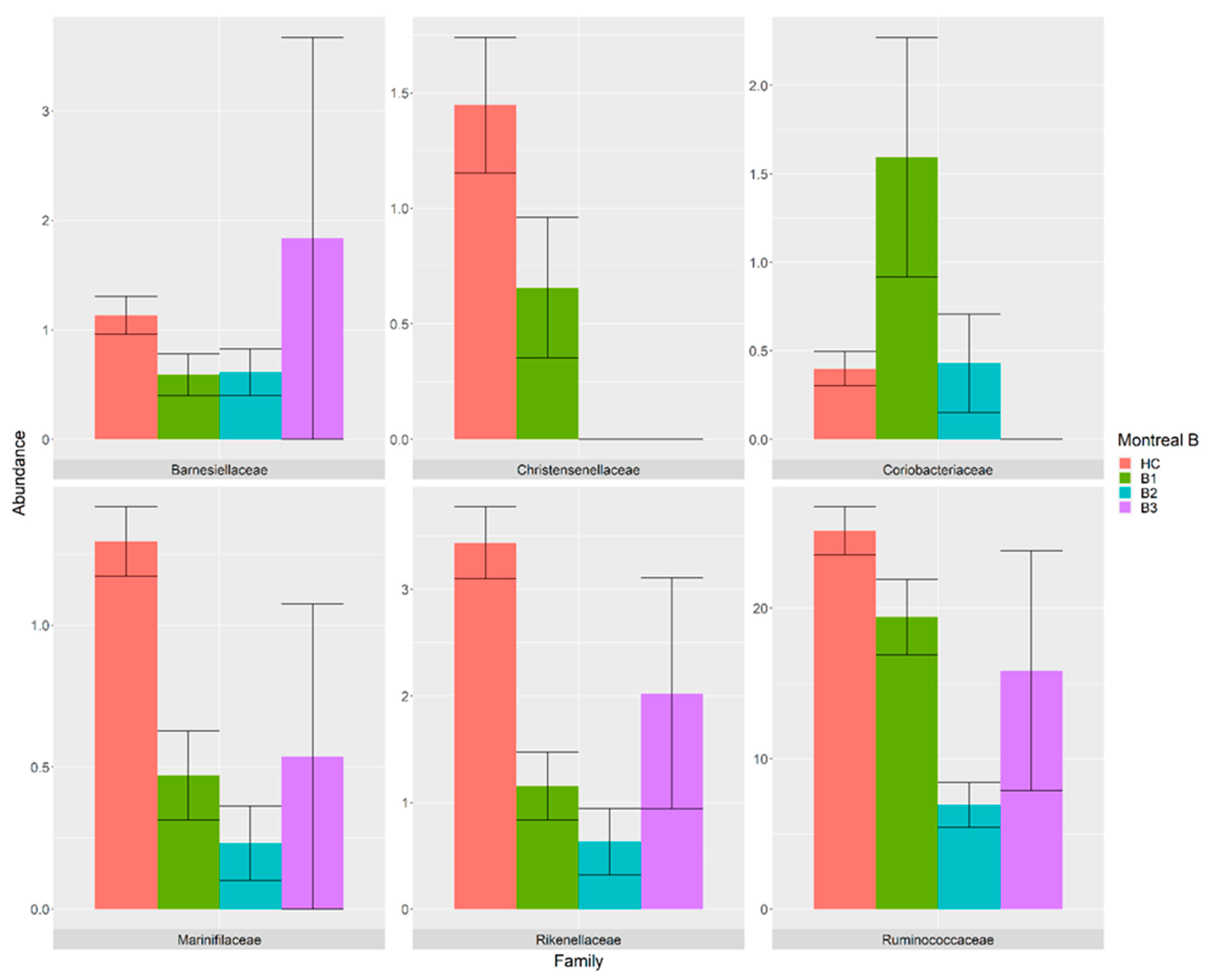
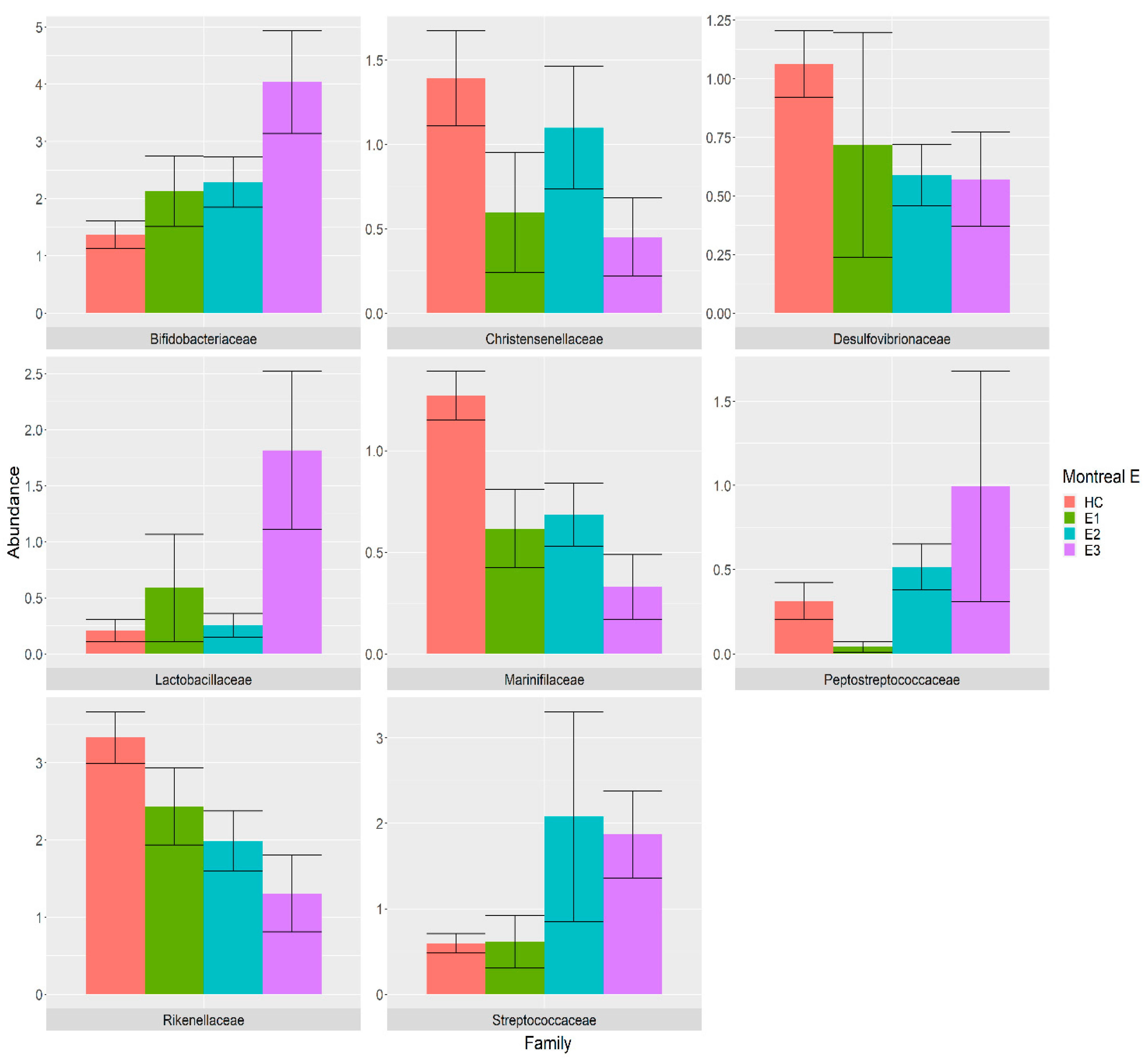
3.4. Association between Gut Microbiota and Montreal Classification
4. Discussion
5. Conclusions and Limitations of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| p-Value | HC | CD | UC | |
|---|---|---|---|---|
| Age | 0.26 | Mean: 38.02 | Mean: 42.42 | Mean: 43.43 |
| Median: 32.5 | Median: 42 | Median: 42 | ||
| Std.Dev: 15.95 | Std.Dev: 15.95 | Std.Dev: 14.04 | ||
| Min: 22 | Min: 19 | Min: 19 | ||
| Max: 72 | Max: 67 | Max: 69 | ||
| Gender | 0.21 | F = 0.64, | F = 0.47, | F = 0.5, |
| M = 0.36 | M = 0.53 | M = 0.5 | ||
| Currently smokes | 0.46 | n = 0.81, | n = 0.91, | n = 0.88, |
| Y = 0.17 | Y = 0.09 | Y = 0.12 | ||
| Height | 0.66 | Mean: 167.33 | Mean: 168.3 | Mean: 170.02 |
| Median: 168 | Median: 173 | Median: 170 | ||
| Std.Dev: 7.78 | Std.Dev: 7.78 | Std.Dev: 10.04 | ||
| Min: 150 | Min: 0 | Min: 150 | ||
| Max: 185 | Max: 186 | Max: 188 | ||
| Caesarean | 0.46 | n = 0.81, Y = 0.17 | n = 0.91, Y = 0.09 | n = 0.88, Y = 0.12 |
| Eat yogurt at least 4 days/week | 0.07 | n = 0.74, Y = 0.26 | n = 0.91, Y = 0.09 | n = 0.78, Y = 0.22 |
| Eat bread at least 4 days/week | 0.15 | n = 0.33, Y = 0.67 | n = 0.19, Y = 0.81 | n = 0.17, Y = 0.83 |
| Eat pasta at least 4 days/week | 0.06 | n = 0.29, Y = 0.71 | n = 0.15, Y = 0.85 | n = 0.34, Y = 0.66 |
| Eat dairy products at least 4 days/week | 0.04 | n = 0.36, Y = 0.64 | n = 0.4, Y = 0.6 | n = 0.59, Y = 0.41 |
| Eat fruit and vegetables at least 4 days/week | 0.01 | n = 0.14, Y = 0.86 | n = 0.34, Y = 0.66 | n = 0.14, Y = 0.86 |
| Eat meat at least 4 days/week | 0.33 | n = 0.24, Y = 0.76 | n = 0.3, Y = 0.7 | n = 0.38, Y = 0.62 |
| Eat fish at least 4 days/week | 0.45 | n = 0.74, Y = 0.26 | n = 0.83, Y = 0.17 | n = 0.74, Y = 0.26 |
| Eat cereals at least 4 days/week | 0.02 | n = 0.71, Y = 0.29 | n = 0.89, Y = 0.11 | n = 0.67, Y = 0.33 |
| Eat legumes at least 4 days/week | 0.002 | n = 0.62, Y = 0.38, | n = 0.90, Y = 0.10, | n = 0.84, Y = 0.16, |
| Drink coffee at least 4 days/week | 0.20 | n = 0.14, Y = 0.86 | n = 0.28, Y = 0.72 | n = 0.28, Y = 0.72 |
| Physical activity | 0.10 | n = 0.4, Y = 0.6 | n = 0.56, Y = 0.44 | n = 0.36, Y = 0.64 |
Appendix B
| p-Value | HC | E1 | E2 | E3 | |
|---|---|---|---|---|---|
| Height | 0.39 | Mean: 167.73 | Mean: 173.33 | Mean: 169.28 | Mean: 169.36 |
| Median: 168 | Median: 175 | Median: 168.5 | Median: 171 | ||
| Std.Dev: 7.92 | Std.Dev: 10.69 | Std.Dev: 10.25 | Std.Dev: 9.96 | ||
| Min: 150 | Min: 158 | Min: 155 | Min: 150 | ||
| Max: 185 | Max: 188 | Max: 185 | Max: 188 | ||
| Drink coffee at least 4 days/week | 0.45 | n = 0.16, Y = 0.84 | n = 0.22, Y = 0.78 | n = 0.22, Y = 0.78 | n = 0.32, Y = 0.68 |
| Eat meat at least 4 days/week | 0.33 | n = 0.23, Y = 0.77 | n = 0.33, Y = 0.67 | n = 0.44, Y = 0.56 | n = 0.36, Y = 0.64 |
| Eat cereals at least 4 days/week | 0.47 | n = 0.73, Y = 0.27 | n = 0.56, Y = 0.44 | n = 0.78, Y = 0.22 | n = 0.61, Y = 0.39 |
| Physical activity | 0.72 | n = 0.39, Y = 0.61 | n = 0.56, Y = 0.44 | n = 0.33, Y = 0.67 | n = 0.36, Y = 0.64 |
| Age | 0.13 | Mean: 38.98 | Mean: 39.22 | Mean: 44.28 | Mean: 43.18 |
| Median: 34.5 | Median: 40 | Median: 46.5 | Median: 41 | ||
| Std.Dev: 16.33 | Std.Dev: 10.31 | Std.Dev: 13.37 | Std.Dev: 15.51 | ||
| Min: 22 | Min: 25 | Min: 20 | Min: 19 | ||
| Max: 72 | Max: 58 | Max: 69 | Max: 68 | ||
| Eat fruit and vegetables at least 4 days/week | 0.67 | n = 0.14, Y = 0.86 | Y = 1 | n = 0.17, Y = 0.83 | n = 0.18, Y = 0.82 |
| Currently smokes | 0.49 | n = 0.68, Y = 0.32 | n = 0.89, Y = 0.11 | n = 0.72, Y = 0.28 | n = 0.82, Y = 0.18 |
| Gender | 0.54 | F = 0.64, | F = 0.44, | F = 0.5, | F = 0.5, |
| M = 0.36 | M = 0.56 | M = 0.5 | M = 0.5 | ||
| Eat dairy products at least 4 days/week | 0.01 | n = 0.36, Y = 0.64 | n = 0.56, Y = 0.44 | n = 0.83, Y = 0.17 | n = 0.46, Y = 0.54 |
| Eat legumes at least 4 days/week | 0.07 | n = 0.64, Y = 0.36 | n = 0.78, Y = 0.22 | n = 0.94, Y = 0.06 | n = 0.79, Y = 0.21 |
| Eat bread at least 4 days/week | 0.23 | n = 0.34, Y = 0.66 | n = 0.11, Y = 0.89 | n = 0.22, Y = 0.78 | n = 0.14, Y = 0.86 |
| Caesarean section | 0.96 | n = 0.82, Y = 0.16 | n = 0.89, Y = 0.11 | n = 0.83, Y = 0.17 | n = 0.89, Y = 0.11 |
| Eat pasta at least 4 days/week | 0.71 | n = 0.3, Y = 0.7 | n = 0.44, Y = 0.56 | n = 0.39, Y = 0.61 | n = 0.29, Y = 0.71 |
| Eat fish at least 4 days/week | 0.71 | n = 0.75, Y = 0.25 | n = 0.89, Y = 0.11 | n = 0.72, Y = 0.28 | n = 0.68, Y = 0.32 |
| Eat yogurt at least 4 days/week | 0.95 | n = 0.75, Y = 0.25 | n = 0.78, Y = 0.22 | n = 0.83, Y = 0.17 | n = 0.75, Y = 0.25 |
Appendix C
| p-Value | HC | B1 | B2 | B3 | |
|---|---|---|---|---|---|
| Height | 0.28 | Mean: 167.33 | Mean: 170.07 | Mean: 166.27 | Mean: 167.5 |
| Median: 168 | Median: 173 | Median: 176 | Median: 165 | ||
| Std.Dev: 7.78 | Std.Dev: 9.77 | Std.Dev: 38.34 | Std.Dev: 13 | ||
| Min: 150 | Min: 153 | Min: 0 | Min: 155 | ||
| Max: 185 | Max: 185 | Max: 186 | Max: 185 | ||
| Drink coffee at least 4 days/week | 0.02 | n = 0.14, Y = 0.86 | n = 0.33, Y = 0.67 | n = 0.14, Y = 0.86 | n = 0.75, Y = 0.25 |
| Eat meat at least 4 days/week | 0.33 | n = 0.24, Y = 0.76 | n = 0.26, Y = 0.74 | n = 0.41, Y = 0.59 | Y = 1 |
| Eat cereals at least 4 days/week | 0.09 | n = 0.71, Y = 0.29 | n = 0.85, Y = 0.15 | n = 0.95, Y = 0.05 | n = 0.75, Y = 0.25 |
| Physical activity | 0.03 | n = 0.4, Y = 0.6 | n = 0.67, Y = 0.33 | n = 0.55, Y = 0.45 | Y = 1 |
| Age | 0.03 | Mean: 38.02 | Mean: 42.41 | Mean: 42.82 | Mean: 40.25 |
| Median: 32.5 | Median: 45 | Median: 41.5 | Median: 41 | ||
| Std.Dev: 15.95 | Std.Dev: 13.84 | Std.Dev: 11.39 | Std.Dev: 16.82 | ||
| Min: 22 | Min: 19 | Min: 26 | Min: 21 | ||
| Max: 72 | Max: 67 | Max: 64 | Max: 58 | ||
| Eat fruit and vegetables at least 4 days/week | 0.09 | n = 0.14, Y = 0.86 | n = 0.3, Y = 0.7 | n = 0.41, Y = 0.59 | n = 0.25, Y = 0.75 |
| Currently smokes | 0.08 | n = 0.67, Y = 0.33 | n = 0.48, Y = 0.52 | n = 0.73, Y = 0.23 | n = 1 |
| Gender | 0.07 | F = 0.64, | F = 0.56, | F = 0.32, | F = 0.75, |
| M = 0.36 | M = 0.44 | M = 0.68 | M = 0.25 | ||
| Eat dairy products at least 4 days/week | 0.45 | n = 0.36, Y = 0.64 | n = 0.41, Y = 0.59 | n = 0.32, Y = 0.68 | n = 0.75, Y = 0.25 |
| Eat legumes at least 4 days/week | 0.00 | n = 0.62, Y = 0.38 | n = 0.85, Y = 0.15 | n = 0.95, Y = 0.05 | n = 0.75, Y = 0.25 |
| Eat bread at least 4 days/week | 0.02 | n = 0.33, Y = 0.67 | n = 0.19, Y = 0.81 | n = 0.09, Y = 0.91 | n = 0.75, Y = 0.25 |
| Caesarean section | 0.4 | n = 0.81, Y = 0.17 | n = 0.96, Y = 0.04 | n = 0.82, Y = 0.18 | n = 1 |
| Eat pasta at least 4 days/week | 0.07 | n = 0.29, Y = 0.71 | n = 0.07, Y = 0.93 | n = 0.18, Y = 0.82 | n = 0.5, Y = 0.5 |
| Eat fish at least 4 days/week | 0.65 | n = 0.74, Y = 0.26 | n = 0.85, Y = 0.15 | n = 0.82, Y = 0.18 | n = 0.75, Y = 0.25 |
| Eat yogurt at least 4 days/week | 0.1 | n = 0.74, Y = 0.26 | n = 0.89, Y = 0.11 | n = 0.95, Y = 0.05 | n = 0.75, Y = 0.25 |
References
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef]
- Bouma, G.; Strober, W. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 2003, 3, 521–533. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takaishi, H.; Matsuki, T.; Nakazawa, A.; Takada, T.; Kado, S.; Asahara, T.; Kamada, N.; Sakuraba, A.; Yajima, T.; Higuchi, H.; et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int. J. Med Microbiol. 2008, 298, 463–472. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlan, P.D.; Shanahan, F.; Mahony, C.O.; Marchesi, J.R. Culture-Independent Analyses of Temporal Variation of the Dominant Fecal Microbiota and Targeted Bacterial Subgroups in Crohn’ s Disease. J. Clin. Microbiol. 2006, 44, 3980–3988. [Google Scholar] [CrossRef] [Green Version]
- Lai, S.; Molfino, A.; Testorio, M.; Perrotta, A.M.; Currado, A.; Pintus, G.; Pietrucci, D.; Unida, V.; La Rocca, D.; Biocca, S.; et al. Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease. Nutrients 2019, 11, 3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, A.; Boneh, R.S.; Wine, E. Evolving role of diet in the pathogenesis and treatment of inflammatory bowel diseases. Gut 2018, 67, 1726–1738. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.-H.; Lochhead, P.; Khalili, H.; Song, M.; Tabung, F.K.; Burke, K.E.; Richter, J.M.; Giovannucci, E.L.; Chan, A.T.; Ananthakrishnan, A.N. Dietary Inflammatory Potential and Risk of Crohn’s Disease and Ulcerative Colitis. Gastroenterology 2020, 159, 873–883.e1. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Hoffmann, C.; Sherrill–Mix, S.A.; Keilbaugh, S.A.; Hamady, M.; Chen, Y.-Y.; Knight, R.; Ahima, R.S.; Bushman, F.; Wu, G.D. High-Fat Diet Determines the Composition of the Murine Gut Microbiome Independently of Obesity. Gastroenterology 2009, 137, 1716–1724.e2. [Google Scholar] [CrossRef] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Probert, C.S.; Jayanthi, V.; Hughes, A.O.; Thompson, J.R.; Wicks, A.C.; Mayberry, J.F. 1993. Prevalence and family risk of ulcerative colitis and Crohn’s disease: An epidemiological study among Europeans and south Asians in Leicestershire. Gut 1993, 34, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Acosta, M.B.-D.; Castro, A.A.; Souto, R.; Iglesias, M.; Lorenzo, A.; Dominguez-Muñoz, J. Emigration to western industrialized countries: A risk factor for developing inflammatory bowel disease. J. Crohn’s Coliti 2011, 5, 566–569. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Sundquist, J.; Hemminki, K.; Sundquist, K. Risk of inflammatory bowel disease in first- and second-generation immigrants in Sweden: A nationwide follow-up study. Inflamm Bowel Dis. 2011, 17, 1784–1791. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Waliullah, S.; Godfrey, V.; Khan, A.W.; Ramachandran, R.A.; Cantarel, B.L.; Behrendt, C.; Peng, L.; Hooper, L.V.; Zaki, H. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, P.A.; Morón, B.; Becker, H.M.; Lang, S.; Atrott, K.; Spalinger, M.R.; Scharl, M.; Wojtal, K.A.; Fischbeck-Terhalle, A.; Frey-Wagner, I.; et al. Titanium dioxide nanoparticles exacerbate DSS-induced colitis: Role of the NLRP3 inflammasome. Gut 2017, 66, 1216–1224. [Google Scholar] [CrossRef] [Green Version]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-Schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A.; et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Van De Wiele, T.; De Bodt, J.; Marzorati, M.; Gewirtz, A.T. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017, 66, 1414–1427. [Google Scholar] [CrossRef]
- Laudisi, F.; Stolfi, C.; Monteleone, G. Impact of Food Additives on Gut Homeostasis. Nutrients 2019, 11, 2334. [Google Scholar] [CrossRef] [Green Version]
- Barros, V.J.D.S.; Severo, J.S.; Mendes, P.H.M.; da Silva, A.C.A.; de Oliveira, K.B.V.; Parente, J.M.L.; Lima, M.M.; Neto, E.M.M.; dos Santos, A.A.; Tolentino, M. Effect of dietary interventions on inflammatory biomarkers of inflammatory bowel diseases: A systematic review of clinical trials. Nutrition 2021, 91–92, 111457. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s Disease and Parkinson’s Disease Medications Have Distinct Signatures of the Gut Microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef]
- Pietrucci, D.; Cerroni, R.; Unida, V.; Farcomeni, A.; Pierantozzi, M.; Mercuri, N.B.; Biocca, S.; Stefani, A.; Desideri, A. Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Parkinsonism Relat. Disord. 2019, 65, 124–130. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLOS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- Clooney, A.G.; Eckenberger, J.; Laserna-Mendieta, E.; A Sexton, K.; Bernstein, M.T.; Vagianos, K.; Sargent, M.; Ryan, F.; Moran, C.; Sheehan, D.; et al. Ranking microbiome variance in inflammatory bowel disease: A large longitudinal intercontinental study. Gut 2020, 70, 499–510. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Connolly, M.L.; Tuohy, K.M.; Lovegrove, J.A. Wholegrain oat-based cereals have prebiotic potential and low glycaemic index. Br. J. Nutr. 2012, 108, 2198–2206. [Google Scholar] [CrossRef] [Green Version]
- Satsangi, J.; Silverberg, M.S.; Vermeire, S.; Colombel, J.F. The Montreal classification of inflammatory bowel disease: Controversies, consensus, and implications. Gut 2006, 55, 749–753. [Google Scholar] [CrossRef] [Green Version]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender Bias in Autoimmunity Is Influenced by Microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [Green Version]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [Green Version]
- Mueller, S.; Saunier, K.; Hanisch, C.; Norin, E.; Alm, L.; Midtvedt, T.; Cresci, A.; Silvi, S.; Orpianesi, C.; Verdenelli, M.C.; et al. Differences in Fecal Microbiota in Different European Study Populations in Relation to Age, Gender, and Country: A Cross-Sectional Study. Appl. Environ. Microbiol. 2006, 72, 1027–1033. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Lin, X.; Zhao, F.; Shi, X.; Li, H.; Li, Y.; Zhu, W.; Xu, X.; Li, C.; Zhou, G. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci. Rep. 2015, 5, 15220. [Google Scholar] [CrossRef] [Green Version]
- Salles, N. Basic Mechanisms of the Aging Gastrointestinal Tract. Dig. Dis. 2007, 25, 112–117. [Google Scholar] [CrossRef]
- Arias, M.; Cobo, M.; Jaime-Sánchez, P.; Pastor, J.; Marijuan, P.; Pardo, J.; Rezusta, A.; Del Campo, R. Gut microbiota and systemic inflammation changes after bread consumption: The ingredients and the processing influence. J. Funct. Foods 2017, 32, 98–105. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Waters, J.L.; Ley, R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019, 17, 1–11. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Presti, A.L.; Zorzi, F.; Del Chierico, F.; Altomare, A.; Cocca, S.; Avola, A.; De Biasio, F.; Russo, A.; Cella, E.; Reddel, S.; et al. Fecal and Mucosal Microbiota Profiling in Irritable Bowel Syndrome and Inflammatory Bowel Disease. Front. Microbiol. 2019, 10, 1655. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.T.; Amos, G.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1–8. [Google Scholar] [CrossRef]
- Mottawea, W.; Chiang, C.-K.; Mühlbauer, M.; Starr, A.E.; Butcher, J.; Abujamel, T.; Deeke, S.A.; Brandel, A.; Zhou, H.; Shokralla, S.; et al. Altered intestinal microbiota–host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 2016, 7, 13419. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kamada, N. Host-microbial Cross-talk in Inflammatory Bowel Disease. Immune Netw. 2017, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pitcher, M.C.L.; Beatty, E.R.; Cummings, J.H. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 2000, 46, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Papa, E.; Docktor, M.; Smillie, C.; Weber, S.; Preheim, S.P.; Gevers, D.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Ingram, J.; et al. Non-Invasive Mapping of the Gastrointestinal Microbiota Identifies Children with Inflammatory Bowel Disease. PLoS ONE 2012, 7, e39242. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, L.; Zhou, R.; Wang, X.; Song, L.; Huang, S.; Wang, G.; Xia, B.; Forbes, B.A. Increased Proportions of Bifidobacterium and the Lactobacillus Group and Loss of Butyrate-Producing Bacteria in Inflammatory Bowel Disease. J. Clin. Microbiol. 2013, 52, 398–406. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wang, W.; Zhou, R.; Ng, S.C.; Li, J.; Huang, M.; Zhou, F.; Wang, X.; Shen, B.; Kamm, M.A.; et al. Characteristics of Fecal and Mucosa-Associated Microbiota in Chinese Patients With Inflammatory Bowel Disease. Medicine 2014, 93, e51. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Sklar, J.; Jiang, L.; Natarajan, L.; Knight, R.; Belkaid, Y. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454. [Google Scholar] [CrossRef]

| Bray–Curtis | Unweighted Unifrac | Weighted Unifrac | Canberra | |
|---|---|---|---|---|
| IBD | 0.0001 *** | 0.0001 *** | 0.0001 *** | 0.0001 *** |
| Age | 0.0473 * | 0.0265 * | ||
| Gender | 0.0399 * | 0.0052 ** | ||
| Bread | 0.0304 * | |||
| Yogurt | 0.0367 * | |||
| Dairy products | 0.0178 * |
| Model | Atopobiaceae |
|---|---|
| IBD + cereals | N.S. |
| IBD + covariates + confounders (not including cereals) | 0.0236 * |
| IBD + covariates + confounders (including cereals) | N.S. |
| IBD | 0.0147 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teofani, A.; Marafini, I.; Laudisi, F.; Pietrucci, D.; Salvatori, S.; Unida, V.; Biocca, S.; Monteleone, G.; Desideri, A. Intestinal Taxa Abundance and Diversity in Inflammatory Bowel Disease Patients: An Analysis including Covariates and Confounders. Nutrients 2022, 14, 260. https://doi.org/10.3390/nu14020260
Teofani A, Marafini I, Laudisi F, Pietrucci D, Salvatori S, Unida V, Biocca S, Monteleone G, Desideri A. Intestinal Taxa Abundance and Diversity in Inflammatory Bowel Disease Patients: An Analysis including Covariates and Confounders. Nutrients. 2022; 14(2):260. https://doi.org/10.3390/nu14020260
Chicago/Turabian StyleTeofani, Adelaide, Irene Marafini, Federica Laudisi, Daniele Pietrucci, Silvia Salvatori, Valeria Unida, Silvia Biocca, Giovanni Monteleone, and Alessandro Desideri. 2022. "Intestinal Taxa Abundance and Diversity in Inflammatory Bowel Disease Patients: An Analysis including Covariates and Confounders" Nutrients 14, no. 2: 260. https://doi.org/10.3390/nu14020260
APA StyleTeofani, A., Marafini, I., Laudisi, F., Pietrucci, D., Salvatori, S., Unida, V., Biocca, S., Monteleone, G., & Desideri, A. (2022). Intestinal Taxa Abundance and Diversity in Inflammatory Bowel Disease Patients: An Analysis including Covariates and Confounders. Nutrients, 14(2), 260. https://doi.org/10.3390/nu14020260








