An Overview of Methods and Exemplars of the Use of Mendelian Randomisation in Nutritional Research
Abstract
:1. Background
2. Principles of a Mendelian Randomization (MR) Study
3. Threats to the Reliability of MR
3.1. Linkage Disequilibrium
3.2. Population Stratification
3.3. Inadequate Statistical Power
3.4. Weak Instrument Bias
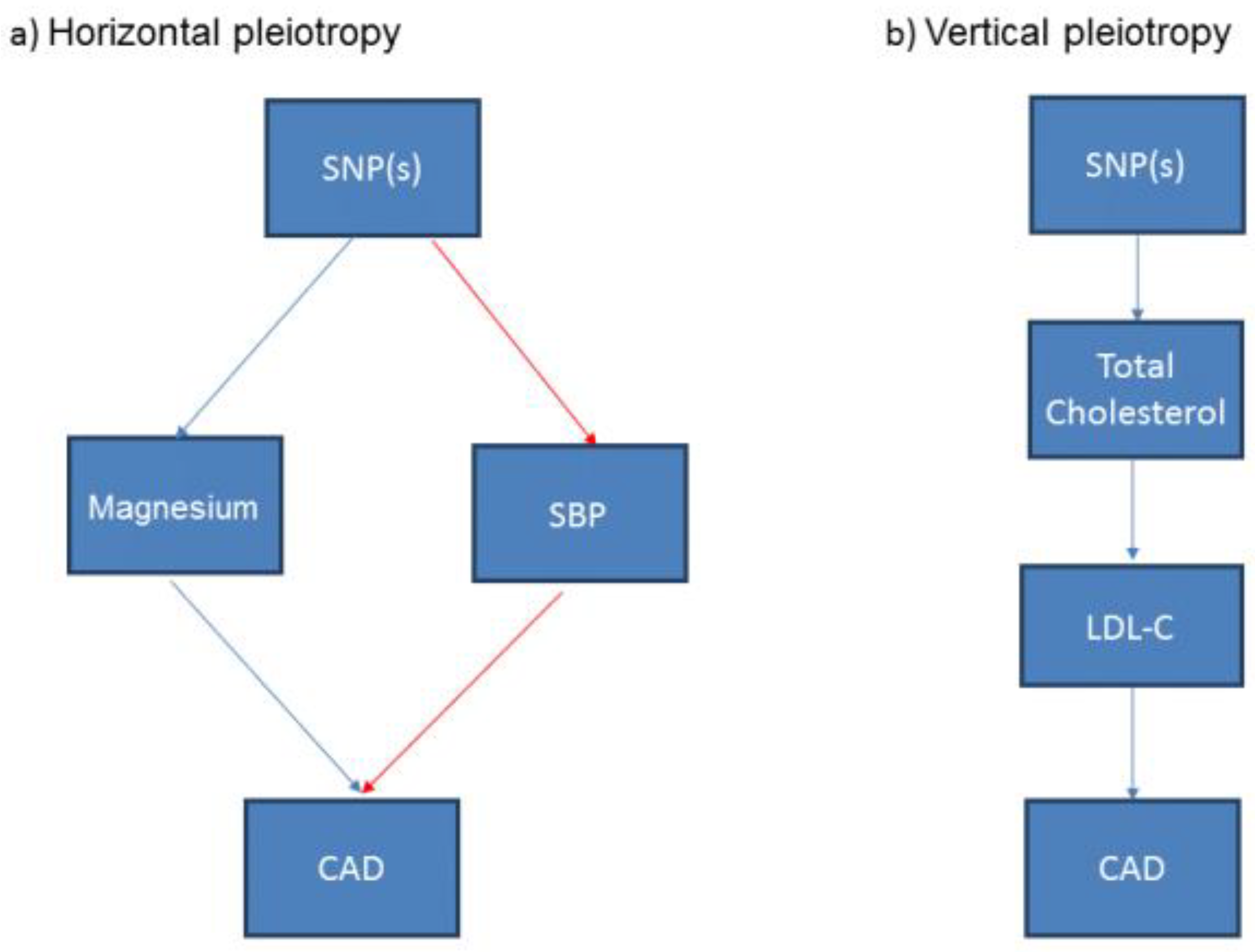
3.5. Associations of the Genetic Variants with Other Traits: Confounding and Pleiotropy
4. Estimating the Causal Effect in MR
5. Extensions to Standard MR Approaches
6. Selected Recent Applications of Mendelian Randomization in Nutritional Epidemiological Studies
6.1. Confirmation or Refutation of an Observational Association
6.2. MR to Overcome Reverse Causality
6.3. MR to Predict Efficacy in the Absence of Trial Evidence
6.4. MR for Hypothesis Generation
6.5. MR to Potentially Repurpose Nutritional Supplementation Strategies
6.6. MR to Inform the Design of an RCT
7. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Willett, W.C.; Stampfer, M.J. Current evidence on healthy eating. Annu. Rev. Public Health 2013, 34, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Afshin, A.; Sur, P.J.; Fay, K.A.; Cornaby, L.; Ferrara, G.; Salama, J.S.; Mullany, E.C.; Abate, K.H.; Abbafati, C.; Abebe, Z.; et al. Health effects of dietary risks in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019, 393, 1958–1972. [Google Scholar] [CrossRef]
- von Ruesten, A.; Feller, S.; Bergmann, M.M.; Boeing, H. Diet and risk of chronic diseases: Results from the first 8 years of follow-up in the EPIC-Potsdam study. Eur. J. Clin. Nutr. 2013, 67, 412–419. [Google Scholar] [CrossRef]
- Zong, G.; Li, Y.; Wanders, A.J.; Alssema, M.; Zock, P.L.; Willett, W.C.; Hu, F.B.; Sun, Q. Intake of individual saturated fatty acids and risk of coronary heart disease in US men and women: Two prospective longitudinal cohort studies. BMJ 2016, 355, i5796. [Google Scholar] [CrossRef] [PubMed]
- Naghshi, S.; Sadeghi, O.; Willett, W.C.; Esmaillzadeh, A. Dietary intake of total, animal, and plant proteins and risk of all cause, cardiovascular, and cancer mortality: Systematic review and dose-response meta-analysis of prospective cohort studies. BMJ 2020, 370, m2412. [Google Scholar] [CrossRef] [PubMed]
- Heath, A.K.; Muller, D.C.; van den Brandt, P.A.; Papadimitriou, N.; Critselis, E.; Gunter, M.; Vineis, P.; Weiderpass, E.; Fagherazzi, G.; Boeing, H.; et al. Nutrient-wide association study of 92 foods and nutrients and breast cancer risk. Breast Cancer Res. 2020, 22, 5. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Davey Smith, G.; Kundu, D.; Bruckdorfer, K.R.; Ebrahim, S. Those confounded vitamins: What can we learn from the differences between observational versus randomised trial evidence? Lancet 2004, 363, 1724–1727. [Google Scholar] [CrossRef]
- Trepanowski, J.F.; Ioannidis, J.P.A. Perspective: Limiting Dependence on Nonrandomized Studies and Improving Randomized Trials in Human Nutrition Research: Why and How. Adv. Nutr. 2018, 9, 367–377. [Google Scholar] [CrossRef]
- Grimes, D.A.; Schulz, K.F. Bias and causal associations in observational research. Lancet 2002, 359, 248–252. [Google Scholar] [CrossRef]
- Sattar, N.; Preiss, D. Reverse Causality in Cardiovascular Epidemiological Research. Circulation 2017, 135, 2369–2372. [Google Scholar] [CrossRef]
- Mirmiran, P.; Bahadoran, Z.; Gaeini, Z. Common Limitations and Challenges of Dietary Clinical Trials for Translation into Clinical Practices. Int. J. Endocrinol. Metab. 2021, 19, e108170. [Google Scholar] [CrossRef] [PubMed]
- Rassen, J.A.; Brookhart, M.A.; Glynn, R.J.; Mittleman, M.A.; Schneeweiss, S. Instrumental variables I: Instrumental variables exploit natural variation in nonexperimental data to estimate causal relationships. J. Clin. Epidemiol. 2009, 62, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Rassen, J.A.; Brookhart, M.A.; Glynn, R.J.; Mittleman, M.A.; Schneeweiss, S. Instrumental variables II: Instrumental variable application-in 25 variations, the physician prescribing preference generally was strong and reduced covariate imbalance. J. Clin. Epidemiol. 2009, 62, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [CrossRef]
- McGuire, A.L.; Basford, M.; Dressler, L.G.; Fullerton, S.M.; Koenig, B.A.; Li, R.; McCarty, C.A.; Ramos, E.; Smith, M.E.; Somkin, C.P.; et al. Ethical and practical challenges of sharing data from genome-wide association studies: The eMERGE Consortium experience. Genome Res. 2011, 21, 1001–1007. [Google Scholar] [CrossRef]
- International Cancer Genome, C.; Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabé, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; et al. International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef]
- Consortium, C.A.D.; Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef]
- Bennett, D.A. An Introduction to Instrumental Variables Analysis: Part 1. Neuroepidemiology 2010, 35, 237–240. [Google Scholar] [CrossRef]
- Wehby, G.L.; Ohsfeldt, R.L.; Murray, J.C. ‘Mendelian randomization’ equals instrumental variable analysis with genetic instruments. Stat. Med. 2008, 27, 2745–2749. [Google Scholar] [CrossRef]
- Davey Smith, G.; Ebrahim, S. What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ 2005, 330, 1076–1079. [Google Scholar] [CrossRef]
- Morgan, T.H. (Ed.) Heredity and Sex; Columbia University Press: New York, NY, USA, 1913. [Google Scholar]
- Méplan, C. Selenium and chronic diseases: A nutritional genomics perspective. Nutrients 2015, 7, 3621–3651. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, H.; Speckmann, B.; Klotz, L.O. Selenoproteins: Antioxidant selenoenzymes and beyond. Arch. Biochem. Biophys. 2016, 595, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.C.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G. Use of allele scores as instrumental variables for Mendelian randomization. Int. J. Epidemiol. 2013, 42, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, D.I.; Kuchenbaecker, K.B.; Shah, S.; Sofat, R.; Holmes, M.V.; White, J.; Mindell, J.S.; Kivimaki, M.; Brunner, E.J.; Whittaker, J.C.; et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int. J. Epidemiol. 2016, 45, 1600–1616. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.P.; Pestman, W.R.; de Boer, A.; Belitser, S.V.; Klungel, O.H. Instrumental variables: Application and limitations. Epidemiology 2006, 17, 260–267. [Google Scholar] [CrossRef]
- Holmes, M.V.; Asselbergs, F.W.; Palmer, T.M.; Drenos, F.; Lanktree, M.B.; Nelson, C.P.; Dale, C.E.; Padmanabhan, S.; Finan, C.; Swerdlow, D.I.; et al. Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J. 2014, 36, 539–550. [Google Scholar] [CrossRef]
- Burgess, S.; Small, D.S.; Thompson, S.G. A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 2017, 26, 2333–2355. [Google Scholar] [CrossRef]
- Palmer, T.M.; Sterne, J.A.; Harbord, R.M.; Lawlor, D.A.; Sheehan, N.A.; Meng, S.; Granell, R.; Smith, G.D.; Didelez, V. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am. J. Epidemiol. 2011, 173, 1392–1403. [Google Scholar] [CrossRef]
- Hemani, G.; Bowden, J.; Davey Smith, G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 2018, 27, R195–R208. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, F.P.; Davey Smith, G.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Teumer, A. Common Methods for Performing Mendelian Randomization. 2018, 5, 51. [Google Scholar] [CrossRef]
- Cinelli, C.; LaPierre, N.; Hill, B.L.; Sankararaman, S.; Eskin, E. Robust Mendelian randomization in the presence of residual population stratification, batch effects and horizontal pleiotropy. Nat. Commun. 2022, 13, 1093. [Google Scholar] [CrossRef]
- Haneuse, S.; VanderWeele, T.J.; Arterburn, D. Using the E-Value to Assess the Potential Effect of Unmeasured Confounding in Observational Studies. JAMA 2019, 321, 602–603. [Google Scholar] [CrossRef]
- Haycock, P.C.; Burgess, S.; Wade, K.H.; Bowden, J.; Relton, C.; Davey Smith, G. Best (but oft-forgotten) practices: The design, analysis, and interpretation of Mendelian randomization studies. Am. J. Clin. Nutr. 2016, 103, 965–978. [Google Scholar] [CrossRef]
- Zheng, J.; Baird, D.; Borges, M.-C.; Bowden, J.; Hemani, G.; Haycock, P.; Evans, D.M.; Smith, G.D. Recent Developments in Mendelian Randomization Studies. Curr. Epidemiol. Rep. 2017, 4, 330–345. [Google Scholar] [CrossRef]
- Burgess, S.; Davies, N.M.; Thompson, S.G. Instrumental variable analysis with a nonlinear exposure-outcome relationship. Epidemiology 2014, 25, 877–885. [Google Scholar] [CrossRef]
- Staley, J.R.; Burgess, S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet. Epidemiol. 2017, 41, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Pendergrass, S.A.; Ritchie, M.D. Phenome-Wide Association Studies: Leveraging Comprehensive Phenotypic and Genotypic Data for Discovery. Curr. Genet. Med. Rep. 2015, 3, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D. Commentary: Random Allocation in Observational Data: How Small But Robust Effects Could Facilitate Hypothesis-free Causal Inference. Epidemiology 2011, 22, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Millard, L.A.C.; Davies, N.M.; Timpson, N.J.; Tilling, K.; Flach, P.A.; Smith, G.D. MR-PheWAS: Hypothesis prioritization among potential causal effects of body mass index on many outcomes, using Mendelian randomization. Sci. Rep. 2015, 5, 16645. [Google Scholar] [CrossRef]
- Martens, L.G.; Luo, J.; Dijk, K.W.v.; Jukema, J.W.; Noordam, R.; van Heemst, D. Diet-Derived Antioxidants Do Not Decrease Risk of Ischemic Stroke: A Mendelian Randomization Study in 1 Million People. J. Am. Heart Assoc. 2021, 10, e022567. [Google Scholar] [CrossRef]
- Ay, A.; Alkanli, N.; Ustundag, S. Investigation of the Relationship Between IL-18 (- 607 C/A), IL-18 (- 137 G/C), and MMP-2 (- 1306 C/T) Gene Variations and Serum Copper and Zinc Levels in Patients Diagnosed with Chronic Renal Failure. Biol. Trace Elem. Res. 2022, 200, 2040–2052. [Google Scholar] [CrossRef]
- Niu, Y.Y.; Zhang, Y.Y.; Zhu, Z.; Zhang, X.Q.; Liu, X.; Zhu, S.Y.; Song, Y.; Jin, X.; Lindholm, B.; Yu, C. Elevated intracellular copper contributes a unique role to kidney fibrosis by lysyl oxidase mediated matrix crosslinking. Cell Death Dis. 2020, 11, 211. [Google Scholar] [CrossRef]
- Ahmad, S.; Ärnlöv, J.; Larsson, S.C. Genetically Predicted Circulating Copper and Risk of Chronic Kidney Disease: A Mendelian Randomization Study. Nutrients 2022, 14, 509. [Google Scholar] [CrossRef]
- Nelson, M.R.; Tipney, H.; Painter, J.L.; Shen, J.; Nicoletti, P.; Shen, Y.; Floratos, A.; Sham, P.C.; Li, M.J.; Wang, J.; et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015, 47, 856–860. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683. [Google Scholar] [CrossRef] [PubMed]
- Paterson, J.; Baxter, G.; Lawrence, J.; Duthie, G. Is there a role for dietary salicylates in health? Proc. Nutr. Soc. 2006, 65, 93–96. [Google Scholar] [CrossRef]
- Nounu, A.; Richmond, R.C.; Stewart, I.D.; Surendran, P.; Wareham, N.J.; Butterworth, A.; Weinstein, S.J.; Albanes, D.; Baron, J.A.; Hopper, J.L.; et al. Salicylic Acid and Risk of Colorectal Cancer: A Two-Sample Mendelian Randomization Study. Nutrients 2021, 13, 4164. [Google Scholar] [CrossRef] [PubMed]
- Millwood, I.Y.; Walters, R.G.; Mei, X.W.; Guo, Y.; Yang, L.; Bian, Z.; Bennett, D.A.; Chen, Y.; Dong, C.; Hu, R.; et al. Conventional and genetic evidence on alcohol and vascular disease aetiology: A prospective study of 500,000 men and women in China. Lancet 2019, 393, 1831–1842. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, J.; Collins, R.; Guo, Y.; Peto, R.; Wu, F.; Li, L. China Kadoorie Biobank of 0.5 million people: Survey methods, baseline characteristics and long-term follow-up. Int. J. Epidemiol. 2011, 40, 1652–1666. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Volpe, S.L. Magnesium in Disease Prevention and Overall Health. Adv. Nutr. 2013, 4, 378S–383S. [Google Scholar] [CrossRef]
- Wark, P.A.; Lau, R.; Norat, T.; Kampman, E. Magnesium intake and colorectal tumor risk: A case-control study and meta-analysis. Am. J. Clin. Nutr. 2012, 96, 622–631. [Google Scholar] [CrossRef]
- Li, L.; Yang, W.; Huang, L.; Feng, X.; Cheng, H.; Ge, X.; Zan, G.; Tan, Y.; Xiao, L.; Liu, C.; et al. MR-PheWAS for the causal effects of serum magnesium on multiple disease outcomes in Caucasian descent. iScience 2021, 24, 103191. [Google Scholar] [CrossRef]
- Forouhi, N.G.; Wareham, N.J. Epidemiology of diabetes. Medicine 2014, 42, 698–702. [Google Scholar] [CrossRef]
- Holick, M.F. Vitamin D deficiency. N. Engl. J. Med. 2007, 357, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am. J. Clin. Nutr. 2004, 80, 1678s–1688s. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Suzuki, G.; Mizuno, S.; Inazu, T.; Kasagi, F.; Kawahara, C.; Okada, Y.; Tanaka, Y. Effect of active vitamin D treatment on development of type 2 diabetes: DPVD randomised controlled trial in Japanese population. BMJ 2022, 377, e066222. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Bennett, D.A.; Millwood, I.Y.; Parish, S.; McCarthy, M.I.; Mahajan, A.; Lin, X.; Bragg, F.; Guo, Y.; Holmes, M.V.; et al. Association of vitamin D with risk of type 2 diabetes: A Mendelian randomisation study in European and Chinese adults. PLoS Med. 2018, 15, e1002566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tan, H.; Tang, J.; Li, J.; Chong, W.; Hai, Y.; Feng, Y.; Lunsford, L.D.; Xu, P.; Jia, D.; et al. Effects of Vitamin D Supplementation on Prevention of Type 2 Diabetes in Patients With Prediabetes: A Systematic Review and Meta-analysis. Diabetes Care 2020, 43, 1650–1658. [Google Scholar] [CrossRef]
- Bouillon, R.; Manousaki, D.; Rosen, C.; Trajanoska, K.; Rivadeneira, F.; Richards, J.B. The health effects of vitamin D supplementation: Evidence from human studies. Nat. Rev. Endocrinol. 2022, 18, 96–110. [Google Scholar] [CrossRef]
- Clarke, R.; Halsey, J.; Bennett, D.; Lewington, S. Homocysteine and vascular disease: Review of published results of the homocysteine-lowering trials. J. Inherit. Metab. Dis. 2011, 34, 83–91. [Google Scholar] [CrossRef]
- Holmes, M.V.; Newcombe, P.; Hubacek, J.A.; Sofat, R.; Ricketts, S.L.; Cooper, J.; Breteler, M.M.; Bautista, L.E.; Sharma, P.; Whittaker, J.C.; et al. Effect modification by population dietary folate on the association between MTHFR genotype, homocysteine, and stroke risk: A meta-analysis of genetic studies and randomised trials. Lancet 2011, 378, 584–594. [Google Scholar] [CrossRef]
- Huo, Y.; Li, J.; Qin, X.; Huang, Y.; Wang, X.; Gottesman, R.F.; Tang, G.; Wang, B.; Chen, D.; He, M.; et al. Efficacy of Folic Acid Therapy in Primary Prevention of Stroke Among Adults With Hypertension in China: The CSPPT Randomized Clinical Trial. JAMA 2015, 313, 1325–1335. [Google Scholar] [CrossRef]
- Larsson, S.C.; Traylor, M.; Markus, H.S. Homocysteine and small vessel stroke: A mendelian randomization analysis. Ann. Neurol. 2019, 85, 495–501. [Google Scholar] [CrossRef]
- Larsson, S.C. Mendelian randomization as a tool for causal inference in human nutrition and metabolism. Curr. Opin. Lipidol. 2021, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wade, K.H.; Yarmolinsky, J.; Giovannucci, E.; Lewis, S.J.; Millwood, I.Y.; Munafò, M.R.; Meddens, F.; Burrows, K.; Bell, J.A.; Davies, N.M.; et al. Applying Mendelian randomization to appraise causality in relationships between nutrition and cancer. Cancer Causes Control. 2022, 33, 631–652. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.A.; Holmes, M.V. Mendelian randomisation in cardiovascular research: An introduction for clinicians. Heart 2017, 103, 1400. [Google Scholar] [CrossRef]
- Benn, M.; Nordestgaard, B.G. From genome-wide association studies to Mendelian randomization: Novel opportunities for understanding cardiovascular disease causality, pathogenesis, prevention, and treatment. Cardiovasc. Res. 2018, 114, 1192–1208. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.A. An introduction to instrumental variable—Part 2: Mendelian randomisation. Neuroepidemiology 2010, 35, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, E.; Glymour, M.M.; Holmes, M.V.; Kang, H.; Morrison, J.; Munafò, M.R.; Palmer, T.; Schooling, C.M.; Wallace, C.; Zhao, Q.; et al. Mendelian randomization. Nat. Rev. Methods Primers 2022, 2, 6. [Google Scholar] [CrossRef]
- de Leeuw, C.; Savage, J.; Bucur, I.G.; Heskes, T.; Posthuma, D. Understanding the assumptions underlying Mendelian randomization. Eur. J. Hum. Genet. 2022, 30, 653–660. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Gagliano Taliun, S.A.; Evans, D.M. Ten simple rules for conducting a mendelian randomization study. PLoS Comput. Biol. 2021, 17, e1009238. [Google Scholar] [CrossRef]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. BMJ 2021, 375, n2233. [Google Scholar] [CrossRef] [PubMed]
- Ohukainen, P.; Virtanen, J.K.; Ala-Korpela, M. Vexed causal inferences in nutritional epidemiology—Call for genetic help. Int. J. Epidemiol. 2021, 51, 6–15. [Google Scholar] [CrossRef] [PubMed]
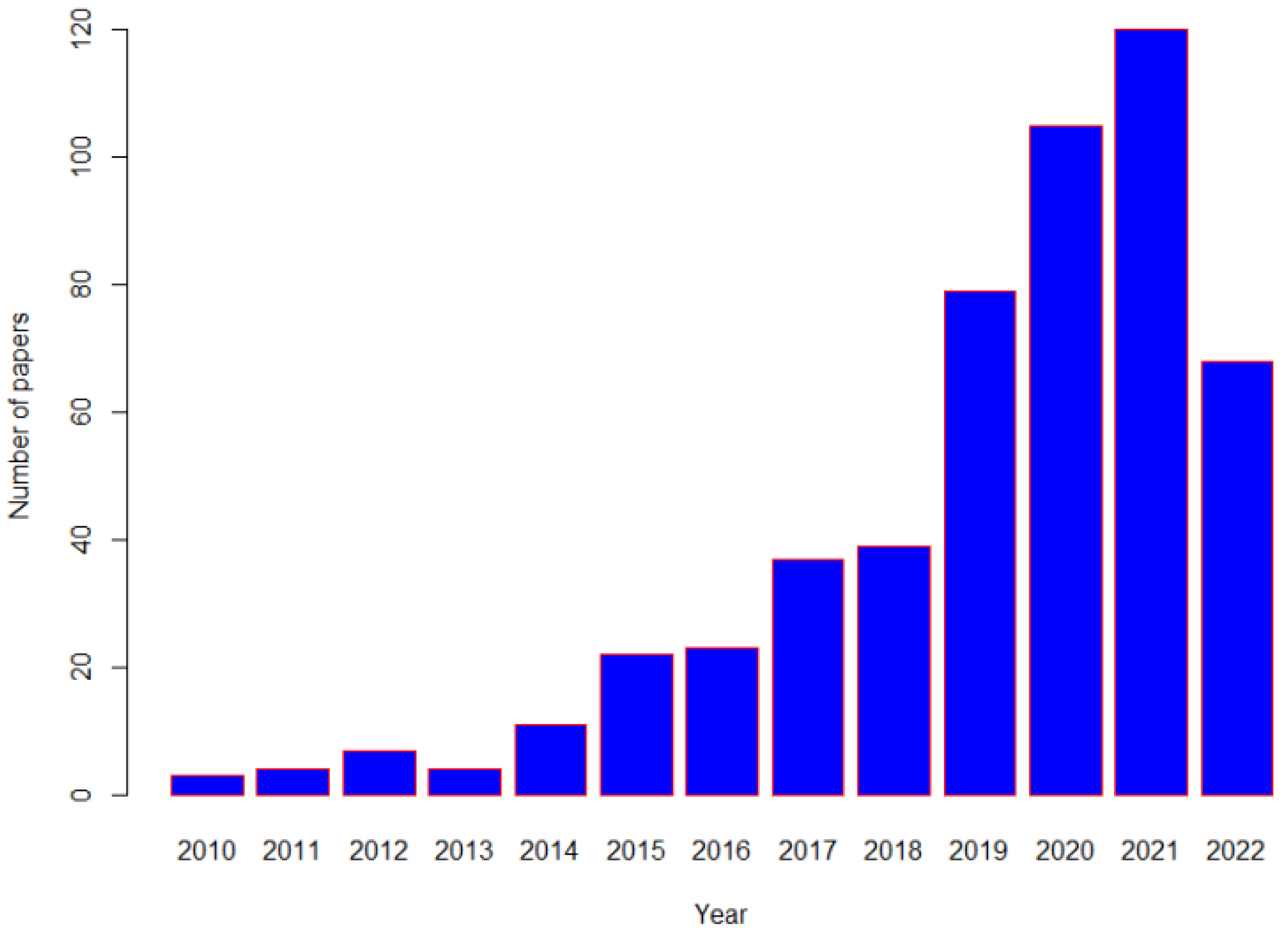


| Proposed Biomarker | Type of Biological Sample | Nutritional Assessment |
|---|---|---|
| Carotenoids | Plasma | Fruit and vegetable intake |
| Creatine | Serum | Meat and fish intake |
| Dyhydrocaeic acid | Urine | Coffee intake |
| Homocysteine | Plasma | Folate status |
| Pentadecanoic acid | Plasma/serum | Total dairy fat intake |
| 25-hydroxyvitamin D | Plasma/serum | Vitamin D intake |
| Caffeine | Plasma | Caffeine intake |
| Name of Resource | Notes | Weblink |
|---|---|---|
| One-sampleMR | R package for one-sample MR | https://remlapmot.github.io/OneSampleMR/ (accessed on 12 July 2022) |
| ivmodel | R package that fits instrumental variable analyses for individual data | https://cran.r-project.org/web/packages/ivmodel/ivmodel.pdf (accessed on 12 July 2022) |
| ivonesamplemr | Stata function for implementation of one-sample MR | https://github.com/remlapmot/ivonesamplemr (accessed on 12 July 2022) |
| glsmr | R package that can be used to perform a non-linear (stratified) one-sample MR analysis | https://rdrr.io/github/hughesevoanth/glsmr/man/glsmr.html (accessed on 12 July 2022) |
| Two-sampleMR | R package for two-sample MR analysis, directly links to MR-Base database | https://github.com/MRCIEU/TwoSampleMR/ (accessed on 12 July 2022) |
| MendelianRandomisation | R package for two-sample MR analysis, links to Phenoscanner * database | https://cran.r-project.org/web/packages/MendelianRandomization (accessed on 12 July 2022) |
| MR Robust | Stata package for two-sample MR analysis | https://github.com/remlapmot/mrrobust/ (accessed on 12 July 2022) |
| MR-Base | GWAS summary database of more than 1100 GWAS studies and online platform to automate two-sample MR | http://www.mrbase.org/ (accessed on 12 July 2022) |
| MR-SENSEMAKR | A suite of sensitivity analysis tools that quantify both how much the inferences would have changed under a postulated degree of violation, as well as the minimal strength of violation necessary to overturn a certain conclusion of an MR | https://doi.org/10.5281/zenodo.5635471 (accessed on 12 July 2022) |
| PHEASANT | R package for performing phenome scans in UK Biobank, including MR phenome-wide association studies (MR-pheWAS) | https://github.com/MRCIEU/PHEASANT/ (accessed on 12 July 2022) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bennett, D.A.; Du, H. An Overview of Methods and Exemplars of the Use of Mendelian Randomisation in Nutritional Research. Nutrients 2022, 14, 3408. https://doi.org/10.3390/nu14163408
Bennett DA, Du H. An Overview of Methods and Exemplars of the Use of Mendelian Randomisation in Nutritional Research. Nutrients. 2022; 14(16):3408. https://doi.org/10.3390/nu14163408
Chicago/Turabian StyleBennett, Derrick A., and Huaidong Du. 2022. "An Overview of Methods and Exemplars of the Use of Mendelian Randomisation in Nutritional Research" Nutrients 14, no. 16: 3408. https://doi.org/10.3390/nu14163408
APA StyleBennett, D. A., & Du, H. (2022). An Overview of Methods and Exemplars of the Use of Mendelian Randomisation in Nutritional Research. Nutrients, 14(16), 3408. https://doi.org/10.3390/nu14163408






