Individual Nutrition Is Associated with Altered Gut Microbiome Composition for Adults with Food Insecurity
Abstract
1. Introduction
2. Methods
2.1. Study Population
2.2. Gut Microbiome Analysis
2.3. My Nutrition Index
2.4. Covariates
3. Statistical Analyses
4. Results
4.1. Study Sample
4.2. Associations between My Nutrition Index and the Gut Microbiome
4.3. Associations between Nutritional Subscales and the Gut Microbiome
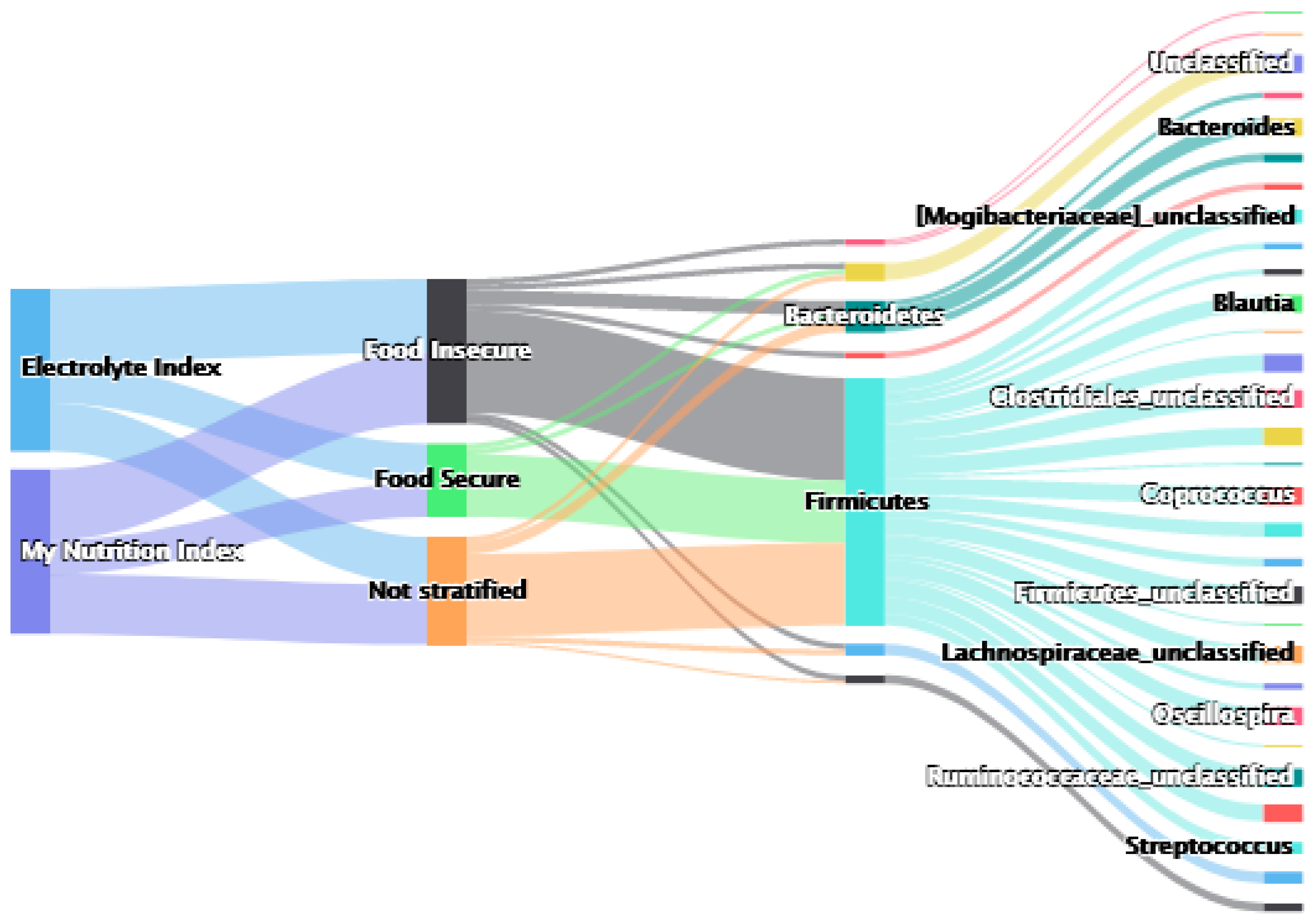
4.4. Associations Stratified by Food Insecurity
4.4.1. My Nutrition Index
4.4.2. Nutritional Subscales
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; DLieber, A.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef] [PubMed]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Upadhyay, V.; Turnbaugh, J.A.; Ly, K.; Turnbaugh, P.J. Meta-Analysis Reveals Reproducible Gut Microbiome Alterations in Response to a High-Fat Diet. Cell Host Microbe 2019, 26, 265–272. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Pereira, R.A.; Hodge, A. Food insecurity: A critical public health nutrition concern. Public Health Nutr. 2015, 18, 2893–2894. [Google Scholar] [CrossRef]
- López-Cepero, A.; Frisard, C.; Bey, G.; Lemon, S.C.; Rosal, M.C. Association between food insecurity and emotional eating in Latinos and the mediating role of perceived stress. Public Health Nutr. 2020, 23, 642–648. [Google Scholar] [CrossRef]
- Gowda, C.; Hadley, C.; Aiello, A.E. The Association between Food Insecurity and Inflammation in the US Adult Population. Am. J. Public Health 2012, 102, 1579–1586. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Filippo, C.D.; Cavalieri, D.; Paola, M.D.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Mitreva, M.; Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria expansion: A microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Simpson, H.L.; Campbell, B.J. Review article: Dietary fibre-microbiota interactions. Aliment. Pharmacol. Ther. 2015, 42, 158–179. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Merra, G.; Noce, A.; Marrone, G.; Cintoni, M.; Tarsitano, M.G.; Capacci, A.; De Lorenzo, A. Influence of Mediterranean Diet on Human Gut Microbiota. Nutrients 2020, 13, 7. [Google Scholar] [CrossRef]
- Pagliai, G.; Russo, E.; Niccolai, E.; Dinu, M.; Di Pilato, V.; Magrini, A.; Bartolucci, G.; Baldi, S.; Menicatti, M.; Giusti, B.; et al. Influence of a 3-month low-calorie Mediterranean diet compared to the vegetarian diet on human gut microbiota and SCFA: The CARDIVEG Study. Eur. J. Nutr. 2020, 59, 2011–2024. [Google Scholar] [CrossRef]
- US Department of Agriculture and US Department of Health and Human Services. Dietary Guidelines for Americans, 2020–2025. 2020. Available online: DietaryGuidelines.gov (accessed on 12 August 2022).
- Coleman-Jensen, A.; Rabbitt, M.P.; Gregory, C.A.; Singh, A. Household Food Security in the United States in 2018. Available online: http://www.ers.usda.gov/publications/pub-details/?pubid=94848 (accessed on 8 February 2022).
- Arenas, D.J.; Thomas, A.; Wang, J.; DeLisser, H.M. A Systematic Review and Meta-analysis of Depression, Anxiety, and Sleep Disorders in US Adults with Food Insecurity. J. Gen. Intern. Med. 2019, 34, 2874–2882. [Google Scholar] [CrossRef]
- Ecklu-Mensah, G.; Gilbert, J.; Devkota, S. Dietary Selection Pressures and Their Impact on the Gut Microbiome. Cell. Mol. Gastroenterol. Hepatol. 2021, 13, 7–18. [Google Scholar] [CrossRef]
- Berry, E.M. Sustainable Food Systems and the Mediterranean Diet. Nutrients 2019, 11, 2229. [Google Scholar] [CrossRef] [PubMed]
- Christian, V.J.; Miller, K.R.; Martindale, R.G. Food Insecurity, Malnutrition, and the Microbiome. Curr. Nutr. Rep. 2020, 9, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Nieto, F.J.; Peppard, P.E.; Engelman, C.D.; McElroy, J.A.; Galvao, L.W.; Friedman, E.M.; Bersch, A.J.; Malecki, K.C. The Survey of the Health of Wisconsin (SHOW), a novel infrastructure for population health research: Rationale and methods. BMC Public Health 2010, 10, 785. [Google Scholar] [CrossRef]
- Eggers, S.; Malecki, K.M.; Peppard, P.; Mares, J.; Shirley, D.; Shukla, S.K.; Poulsen, K.; Gangnon, R.; Duster, M.; Kates, A.; et al. Wisconsin microbiome study, a cross-sectional investigation of dietary fibre, microbiome composition and antibiotic-resistant organisms: Rationale and methods. BMJ Open 2018, 8, e019450. [Google Scholar] [CrossRef]
- Malecki, K.M.C.; Nikodemova, M.; Schultz, A.A.; LeCaire, T.J.; Bersch, A.J.; Cadmus-Bertram, L.; Engelman, C.D.; Hagen, E.; McCulley, L.; Palta, M.; et al. The Survey of the Health of Wisconsin (SHOW) Program: An Infrastructure for Advancing Population Health. Front. Public Health 2022. Available online: https://www.frontiersin.org/articles/10.3389/fpubh.2022.818777 (accessed on 12 July 2022). [CrossRef]
- Eggers, S.; Safdar, N.; Sethi, A.K.; Suen, G.; Peppard, P.E.; Kates, A.E.; Skarlupka, J.H.; Kanarek, M.; Malecki, K.M.C. Urinary lead concentration and composition of the adult gut microbiota in a cross-sectional population-based sample. Environ. Int. 2019, 133, 105122. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- MiSeq SOP—Mothur. Available online: https://www.mothur.org/wiki/MiSeq_SOP (accessed on 12 August 2019).
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Gennings, C.; Wolk, A.; Hakansson, N.; Lindh, C.; Bornehag, C.G. Contrasting prenatal nutrition and environmental exposures in association with birth weight and cognitive function in children at 7 years. BMJ Nutr. Prev. Health 2020, 3, 162. [Google Scholar] [CrossRef] [PubMed]
- Gennings, C.; Manning, L.; Keefer, L. Evaluation of My Nutrition Index in an IBD Patient Population. J. Nutr. Med. Diet Care 2021, 7, 49. [Google Scholar]
- Busgang, S.A.; Malin, A.J.; Gennings, C. My Nutrition Index: A method for measuring optimal daily nutrient intake. BMC Nutr. 2022, 8, 1–12. [Google Scholar] [CrossRef]
- Diet History Questionnaire II (DHQ II) for US and Canada. EGRP/DCCPS/NCI/NIH. Available online: https://epi.grants.cancer.gov/dhq2/ (accessed on 12 August 2022).
- DHQ II Diet*Calc Software. EGRP/DCCPS/NCI/NIH. Available online: https://epi.grants.cancer.gov/dhq2/dietcalc/ (accessed on 12 August 2022).
- Curtin, P.; Kellogg, J.; Cech, N.; Gennings, C. A random subset implementation of weighted quantile sum (WQSRS) regression for analysis of high-dimensional mixtures. Commun. Stat.-Simul. Comput. 2021, 50, 1119–1134. [Google Scholar] [CrossRef]
- Tanner, E.M.; Bornehag, C.G.; Gennings, C. Repeated holdout validation for weighted quantile sum regression. MethodsX 2019, 6, 2855–2860. [Google Scholar] [CrossRef]
- Cowell, W.; Colicino, E.; Levin-Schwartz, Y.; Enlow, M.B.; Amarasiriwardena, C.; Andra, S.S.; Gennings, C.; Wright, R.O.; Wright, R.J. Prenatal metal mixtures and sex-specific infant negative affectivity. Environ. Epidemiol. 2021, 5, e147. [Google Scholar] [CrossRef]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef]
- Nagpal, R.; Neth, B.J.; Wang, S.; Craft, S.; Yadav, H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine 2019, 47, 529–542. [Google Scholar] [CrossRef]
- Konikoff, T.; Gophna, U. Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Tims, S.; Derom, C.; Jonkers, D.M.; Vlietinck, R.; Saris, W.H.; Kleerebezem, M.; de Vos, W.M.; Zoetendal, E.G. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013, 7, 707–717. [Google Scholar] [CrossRef]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef]
- Verdam, F.J.; Fuentes, S.; de Jonge, C.; Zoetendal, E.G.; Erbil, R.; Greve, J.W.; Buurman, W.A.; de Vos, W.M.; Rensen, S.S. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity 2013, 21, E607–E615. [Google Scholar] [CrossRef] [PubMed]
- Escobar, J.S.; Klotz, B.; Valdes, B.E.; Agudelo, G.M. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014, 14, 311. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, E.; Kloosterhuis, N.J.; Koster, M.; Dekker, D.C.; Gijbels, M.J.J.; van der Velden, S.; Ríos-Morales, M.; van Faassen, M.J.R.; Loreti, M.G.; de Bruin, A.; et al. A Proinflammatory Gut Microbiota Increases Systemic Inflammation and Accelerates Atherosclerosis. Circ. Res. 2019, 124, 94–100. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef]
- Zeng, H.; Ishaq, S.L.; Zhao, F.Q.; Wright, A.D.G. Colonic inflammation accompanies an increase of β-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J. Nutr. Biochem. 2016, 35, 30–36. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed]
- Bier, A.; Braun, T.; Khasbab, R.; Di Segni, A.; Grossman, E.; Haberman, Y.; Leibowitz, A. A High Salt Diet Modulates the Gut Microbiota and Short Chain Fatty Acids Production in a Salt-Sensitive Hypertension Rat Model. Nutrients 2018, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Huang, Z.; Yu, K.; Ding, R.; Ye, K.; Dai, C.; Xu, X.; Zhou, G.; Li, C. High-salt diet has a certain impact on protein digestion and gut microbiota: A sequencing and proteome combined study. Front. Microbiol. 2017, 8, 1838. [Google Scholar] [CrossRef]
- Miranda, P.M.; De Palma, G.; Serkis, V.; Lu, J.; Louis-Auguste, M.P.; McCarville, J.L.; Verdu, E.F.; Collins, S.M.; Bercik, P. High salt diet exacerbates colitis in mice by decreasing Lactobacillus levels and butyrate production. Microbiome 2018, 6, 57. [Google Scholar] [CrossRef]
- Naqvi, S.; Asar, T.O.; Kumar, V.; Al-Abbasi, F.A.; Alhayyani, S.; Kamal, M.A.; Anwar, F. A cross-talk between gut microbiome, salt and hypertension. Biomed. Pharmacother. 2021, 134, 111156. [Google Scholar] [CrossRef] [PubMed]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mähler, A.; Balogh, A.; Markó, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Howard, A.G.; Tsilimigras, M.C.B.; Avery, C.L.; Meyer, K.A.; Sha, W.; Sun, S.; Zhang, J.; Su, C.; et al. Associations of sodium and potassium consumption with the gut microbiota and host metabolites in a population-based study in Chinese adults. Am. J. Clin. Nutr. 2020, 112, 1599–1612. [Google Scholar] [CrossRef]
- Drewnowski, A.; Rehm, C.D.; Maillot, M.; Monsivais, P. The relation of potassium and sodium intakes to diet cost among U.S. adults. J. Hum. Hypertens. 2015, 29, 14–21. [Google Scholar] [CrossRef][Green Version]
- Hanson, K.L.; Connor, L.M. Food insecurity and dietary quality in US adults and children: A systematic review. Am. J. Clin. Nutr. 2014, 100, 684–692. [Google Scholar] [CrossRef]
- Molina-Torres, G.; Rodriguez-Arrastia, M.; Roman, P.; Sanchez-Labraca, N.; Cardona, D. Stress and the gut microbiota-brain axis. Behav. Pharmacol. 2019, 30, 187–200. [Google Scholar] [CrossRef]
- Vlčková, K.; Shutt-Phillips, K.; Heistermann, M.; Pafčo, B.; Petrželková, K.J.; Todd, A.; Modrý, D.; Nelson, K.E.; Wilson, B.A.; Stumpf, R.M.; et al. Impact of stress on the gut microbiome of free-ranging western lowland gorillas. Microbiology 2018, 164, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.T.; Dowd, S.E.; Parry, N.M.A.; Galley, J.D.; Schauer, D.B.; Lyte, M. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 2010, 78, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef]
- Golubeva, A.V.; Crampton, S.; Desbonnet, L.; Edge, D.; O’Sullivan, O.; Lomasney, K.W.; Zhdanov, A.V.; Crispie, F.; Moloney, R.D.; Borre, Y.E.; et al. Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology 2015, 60, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Kamada, K.; Mizushima, K.; Higashimura, Y.; Katada, K.; Uchiyama, K.; Handa, O.; Takagi, T.; Naito, Y.; Itoh, Y. Changes in Intestinal Motility and Gut Microbiota Composition in a Rat Stress Model. Digestion 2017, 95, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Mahbub, R.; Fox, J.G. Xenobiotics: Interaction with the Intestinal Microflora. ILAR J. 2015, 56, 218–227. [Google Scholar] [CrossRef]
- Eggers, S.I.; Bixby, M.; Renzetti, S.; Curtin, P.; Gennings, C. Human Microbiome Mixture Analysis using Weighted Quantile Sum Regression. medRxiv 2022. [Google Scholar] [CrossRef]
- TKENNEDYE; Ohls, J.; Carlson, S.; Fleming, K. The healthy eating index: Design and applications. J. Am. Diet. Assoc. 1995, 95, 1103–1108. [Google Scholar]
- Bowman, S.A.; Lino, M.; Gerrior, S.A.; Basiotis, P.P. The Healthy Eating Index 1994–96. In Center for Nutrition Policy and Promotion; United States Department of Agriculture: Washington, DC, USA, 1998. [Google Scholar]
- Mayer-Davis, E.J.; Vitolins, M.Z.; Carmichael, S.L.; Hemphill, S.; Tsaroucha, G.; Rushing, J.; Levin, S. Validity and reproducibility of a food frequency interview in a multi-cultural epidemiologic study. Ann. Epidemiol. 1999, 9, 314–324. [Google Scholar] [CrossRef]
- Kristal, A.R.; Feng, Z.; Coates, R.J.; Oberman, A.; George, V. Associations of race/ethnicity, education, and dietary intervention with the validity and reliability of a food frequency questionnaire: The Women’s Health Trial Feasibility Study in Minority Populations. Am. J. Epidemiol. 1997, 146, 856–869. [Google Scholar] [CrossRef]
- Stallone, D.D.; Brunner, E.J.; Bingham, S.A.; Marmot, M.G. Dietary assessment in Whitehall II: The influence of reporting bias on apparent socioeconomic variation in nutrient intakes. Eur. J. Clin. Nutr. 1997, 51, 815–825. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benezra, A. Race in the Microbiome. Sci. Technol. Hum. Values 2020, 45, 877–902. [Google Scholar] [CrossRef]
- De Wolfe, T.J.; Arefin, M.R.; Benezra, A.; Rebolleda Gómez, M. Chasing Ghosts: Race, Racism, and the Future of Microbiome Research. mSystems 2021, 6, e0060421. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | N | Food Secure 1, N = 489 | Food Insecure 1, N = 144 | p-Value 2 |
|---|---|---|---|---|
| My Nutrition Index | 633 | 62.5 (20.4) | 47.3 (25.6) | <0.001 |
| Age | 633 | 56.4 (16.0) | 49.3 (15.6) | <0.001 |
| Alcohol consumption (g/day) | 633 | 10.4 (28.0) | 15.4 (61.2) | 0.3 |
| Body Mass Index | 624 | 29.8 (7.0) | 33.4 (9.0) | <0.001 |
| Poverty to Income Ratio | 615 | 4.5 (2.8) | 1.6 (1.0) | <0.001 |
| Gender | 633 | 0.2 | ||
| Male | 213 (44%) | 54 (38%) | ||
| Female | 276 (56%) | 90 (62%) | ||
| Race | 632 | <0.001 | ||
| White | 431 (88%) | 90 (62%) | ||
| Other/Non-White | 57 (12%) | 54 (38%) | ||
| Antibiotic use in the Past Year | 633 | 0.2 | ||
| Did not Use | 303 (62%) | 80 (56%) | ||
| Did Use | 161 (33%) | 52 (36%) | ||
| Unknown/Missing | 25 (5%) | 12 (8%) | ||
| Education | 633 | <0.001 | ||
| <High School | 20 (4%) | 19 (13%) | ||
| High School or Associate’s Degree | 251 (51%) | 103 (72%) | ||
| Bachelor’s Degree or Higher | 218 (45%) | 22 (15%) | ||
| Smoking Status | 619 | <0.001 | ||
| Never | 305 (64%) | 60 (42%) | ||
| Current | 37 (8%) | 43 (30%) | ||
| Former | 134 (28%) | 40 (28%) | ||
| Electrolyte Index | 633 | <0.001 | ||
| <Median | 225 (46%) | 93 (65%) | ||
| ≥Median | 264 (54%) | 51 (35%) | ||
| Vitamin Index | 633 | 0.2 | ||
| <90 | 438 (90%) | 134 (93%) | ||
| ≥90 | 51 (10%) | 10 (7%) | ||
| Macro Nutrient Index | 624 | 0.015 | ||
| <90 | 313 (65%) | 105 (76%) | ||
| ≥90 | 172 (35%) | 34 (24%) | ||
| Mineral Index | 633 | 0.11 | ||
| <90 | 224 (46%) | 77 (53%) | ||
| ≥90 | 265 (54%) | 67 (47%) | ||
| Shannon Diversity Index | 624 | 3.3 (0.5) | 3.1 (0.5) | <0.001 |
| Diabetes (Type 1 or 2) | 568 | 0.002 | ||
| Yes | 52 (12%) | 30 (23%) | ||
| No | 384 (88%) | 102 (77%) | ||
| Chronic Conditions | 633 | <0.001 | ||
| Yes | 210 (43%) | 91 (63%) | ||
| No | 279 (57%) | 53 (37%) |
| MNI | Electrolyte Index | |
|---|---|---|
| β (95% CI) | OR (95% CI) | |
| (Intercept) | 47.9 (38.4, 57.4) | 0.15 (0.06, 0.37) |
| WQS | 2.56 (0.52, 4.61) | 1.58 (1.24, 2.02) |
| Antibiotic use in past year: Yes (vs. no) | 0.58 (−2.38, 3.54) | 1.19 (0.89, 1.60) |
| Antibiotic use in past year: Unknown (vs. no) | −2.42 (−10.73, 5.89) | 1.09 (0.58, 2.04) |
| Education: High school/associate’s degree (vs. less than high school degree) | 9.0 (0.77, 17.23) | 3.13 (1.45, 6.75) |
| Education: Bachelor’s degree or higher (vs. less than high school degree) | 13.5 (4.88, 22.24) | 3.94 (1.74, 8.94) |
| Race (non-white vs. white) | −0.1 (−13.2, −4.4) | NA |
| Food insecurity (insecure vs. secure) | −10.02 (−13.85, −6.2) | 0.61 (0.45, 0.83) |
| MNI | Electrolyte Index | |
|---|---|---|
| β (95% CI) | OR (95% CI) | |
| (Intercept) | 49.8 (41, 58.5) | 0.22 (0.10, 0.46) |
| WQS | 7.7 (1.32, 14.1) | 2.86 (1.53, 5.37) |
| Antibiotic use in past year: Yes (vs. no) | 0.32 (−2.44, 3.09) | 1.21 (0.91, 1.61) |
| Antibiotic use in past year: Unknown (vs. no) | −2.5 (−8.8, 3.85) | 1.12 (0.62, 2.05) |
| Education: High school/associate’s degree (vs. less than high school degree) | 8.12 (0.26, 16) | 2.50 (1.23, 5.10) |
| Education: Bachelor’s degree or higher (vs. less than high school degree) | 12.8 (4.45, 21.1) | 3.10 (1.52, 6.30) |
| Race (non-white vs. white) | −8.2 (−12.4, −4.03) | NA |
| Food insecurity (insecure vs. secure) | −15.5 (−21.9, −9.18) | 0.36 (0.21, 0.61) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bixby, M.; Gennings, C.; Malecki, K.M.C.; Sethi, A.K.; Safdar, N.; Peppard, P.E.; Eggers, S. Individual Nutrition Is Associated with Altered Gut Microbiome Composition for Adults with Food Insecurity. Nutrients 2022, 14, 3407. https://doi.org/10.3390/nu14163407
Bixby M, Gennings C, Malecki KMC, Sethi AK, Safdar N, Peppard PE, Eggers S. Individual Nutrition Is Associated with Altered Gut Microbiome Composition for Adults with Food Insecurity. Nutrients. 2022; 14(16):3407. https://doi.org/10.3390/nu14163407
Chicago/Turabian StyleBixby, Moira, Chris Gennings, Kristen M. C. Malecki, Ajay K. Sethi, Nasia Safdar, Paul E. Peppard, and Shoshannah Eggers. 2022. "Individual Nutrition Is Associated with Altered Gut Microbiome Composition for Adults with Food Insecurity" Nutrients 14, no. 16: 3407. https://doi.org/10.3390/nu14163407
APA StyleBixby, M., Gennings, C., Malecki, K. M. C., Sethi, A. K., Safdar, N., Peppard, P. E., & Eggers, S. (2022). Individual Nutrition Is Associated with Altered Gut Microbiome Composition for Adults with Food Insecurity. Nutrients, 14(16), 3407. https://doi.org/10.3390/nu14163407







