Ketone Bodies and SIRT1, Synergic Epigenetic Regulators for Metabolic Health: A Narrative Review
Abstract
:1. Introduction
2. The Epigenetics Regulation at a Glance
3. Ketone Bodies and Epigenetic Regulation
4. SIRT1 and Epigenetic Regulation
5. Ketone Bodies and SIRT1 Share Common Metabolic Outcomes
6. Epigenetic Activity of KBs and SIRT1 in Adipose Tissue Regulation
7. Epigenetic Activity of KBs and SIRT1 against NAFLD
8. Epigenetic Activity of KBs and SIRT1 against Inflammation
9. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Castellana, M.; Conte, E.; Cignarelli, A.; Perrini, S.; Giustina, A.; Giovanella, L.; Giorgino, F.; Trimboli, P. Efficacy and Safety of Very Low Calorie Ketogenic Diet (VLCKD) in Patients with Overweight and Obesity: A Systematic Review and Meta-Analysis. Rev. Endocr. Metab. Disord. 2020, 21, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Norwitz, N.G.; Hu, M.T.; Clarke, K. The Mechanisms by Which the Ketone Body D-β-Hydroxybutyrate May Improve the Multiple Cellular Pathologies of Parkinson’s Disease. Front. Nutr. 2019, 6, 63. [Google Scholar] [CrossRef]
- Sourbron, J.; Thevissen, K.; Lagae, L. The Ketogenic Diet Revisited: Beyond Ketones. Front. Neurol. 2021, 12, 720073. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.R.; Pissios, P.; Otu, H.; Xue, B.; Asakura, K.; Furukawa, N.; Marino, F.E.; Liu, F.-F.; Kahn, B.B.; Libermann, T.A.; et al. A High-Fat, Ketogenic Diet Induces a Unique Metabolic State in Mice. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E1724–E1739. [Google Scholar] [CrossRef] [PubMed]
- Caprio, M.; Infante, M.; Moriconi, E.; Armani, A.; Fabbri, A.; Mantovani, G.; Mariani, S.; Lubrano, C.; Poggiogalle, E.; Migliaccio, S.; et al. Very-Low-Calorie Ketogenic Diet (VLCKD) in the Management of Metabolic Diseases: Systematic Review and Consensus Statement from the Italian Society of Endocrinology (SIE). J. Endocrinol. Investig. 2019, 42, 1365–1386. [Google Scholar] [CrossRef] [PubMed]
- Basciani, S.; Camajani, E.; Contini, S.; Persichetti, A.; Risi, R.; Bertoldi, L.; Strigari, L.; Prossomariti, G.; Watanabe, M.; Mariani, S.; et al. Very-Low-Calorie Ketogenic Diets With Whey, Vegetable, or Animal Protein in Patients With Obesity: A Randomized Pilot Study. J. Clin. Endocrinol. Metab. 2020, 105, 2939–2949. [Google Scholar] [CrossRef]
- Kovács, Z.; Brunner, B.; Ari, C. Beneficial Effects of Exogenous Ketogenic Supplements on Aging Processes and Age-Related Neurodegenerative Diseases. Nutrients 2021, 13, 2197. [Google Scholar] [CrossRef]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sassone-Corsi, P. Physiology. When Metabolism and Epigenetics Converge. Science 2013, 339, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Sahar, S.; Sassone-Corsi, P. The Epigenetic Language of Circadian Clocks. Handb. Exp. Pharmacol. 2013, 217, 29–44. [Google Scholar] [CrossRef]
- Nakagawa, T.; Guarente, L. Sirtuins at a Glance. J. Cell. Sci. 2011, 124, 833–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braud, L.; Pini, M.; Stec, D.F.; Manin, S.; Derumeaux, G.; Stec, D.E.; Foresti, R.; Motterlini, R. Increased Sirt1 Secreted from Visceral White Adipose Tissue Is Associated with Improved Glucose Tolerance in Obese Nrf2-Deficient Mice. Redox Biol. 2021, 38, 101805. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wong, K.; Giles, A.; Jiang, J.; Lee, J.W.; Adams, A.C.; Kharitonenkov, A.; Yang, Q.; Gao, B.; Guarente, L.; et al. Hepatic SIRT1 Attenuates Hepatic Steatosis and Controls Energy Balance in Mice by Inducing Fibroblast Growth Factor 21. Gastroenterology 2014, 146, 539–549.e7. [Google Scholar] [CrossRef]
- Sosnowska, B.; Mazidi, M.; Penson, P.; Gluba-Brzózka, A.; Rysz, J.; Banach, M. The Sirtuin Family Members SIRT1, SIRT3 and SIRT6: Their Role in Vascular Biology and Atherogenesis. Atherosclerosis 2017, 265, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Cohen-Kfir, E.; Artsi, H.; Levin, A.; Abramowitz, E.; Bajayo, A.; Gurt, I.; Zhong, L.; D’Urso, A.; Toiber, D.; Mostoslavsky, R.; et al. Sirt1 Is a Regulator of Bone Mass and a Repressor of Sost Encoding for Sclerostin, a Bone Formation Inhibitor. Endocrinology 2011, 152, 4514–4524. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Gong, Z. The Beneficial Roles of SIRT1 in Neuroinflammation-Related Diseases. Oxid. Med. Cell. Longev. 2020, 2020, 6782872. [Google Scholar] [CrossRef] [PubMed]
- Bordone, L.; Guarente, L. Calorie Restriction, SIRT1 and Metabolism: Understanding Longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef]
- Abduraman, M.A.; Azizan, N.A.; Teoh, S.H.; Tan, M.L. Ketogenesis and SIRT1 as a Tool in Managing Obesity. Obes. Res. Clin. Pract. 2021, 15, 10–18. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Q.; Chang, C. Epigenetics in Health and Disease. Adv. Exp. Med. Biol. 2020, 1253, 3–55. [Google Scholar] [CrossRef] [PubMed]
- Deans, C.; Maggert, K.A. What Do You Mean, “Epigenetic”? Genetics 2015, 199, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Pigeyre, M.; Yazdi, F.T.; Kaur, Y.; Meyre, D. Recent Progress in Genetics, Epigenetics and Metagenomics Unveils the Pathophysiology of Human Obesity. Clin. Sci. 2016, 130, 943–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Chandel, S.; Dey, D.; Ghosh, A.; Roy, S.; Ravichandiran, V.; Ghosh, D. Epigenetic Modification and Therapeutic Targets of Diabetes Mellitus. Biosci. Rep. 2020, 40, BSR20202160. [Google Scholar] [CrossRef] [PubMed]
- Keating, S.T.; El-Osta, A. Epigenetics and Metabolism. Circ. Res. 2015, 116, 715–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haws, S.A.; Leech, C.M.; Denu, J.M. Metabolism and the Epigenome: A Dynamic Relationship. Trends Biochem. Sci. 2020, 45, 731–747. [Google Scholar] [CrossRef]
- Skarke, C. Diet-Epigenome Axis. Circ. Genom. Precis. Med. 2020, 13, e003129. [Google Scholar] [CrossRef]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.-C.; Ried, J.S.; Zhang, W.; Yang, Y.; et al. Epigenome-Wide Association Study of Body Mass Index, and the Adverse Outcomes of Adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Nasser, S.; Vialichka, V.; Biesiekierska, M.; Balcerczyk, A.; Pirola, L. Effects of Ketogenic Diet and Ketone Bodies on the Cardiovascular System: Concentration Matters. World J. Diabetes 2020, 11, 584–595. [Google Scholar] [CrossRef]
- Dabke, P.; Das, A.M. Mechanism of Action of Ketogenic Diet Treatment: Impact of Decanoic Acid and Beta-Hydroxybutyrate on Sirtuins and Energy Metabolism in Hippocampal Murine Neurons. Nutrients 2020, 12, 2379. [Google Scholar] [CrossRef]
- Newman, J.C.; Covarrubias, A.J.; Zhao, M.; Yu, X.; Gut, P.; Ng, C.-P.; Huang, Y.; Haldar, S.; Verdin, E. Ketogenic Diet Reduces Midlife Mortality and Improves Memory in Aging Mice. Cell Metab. 2017, 26, 547–557.e8. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.N.; Wallace, M.A.; Tomilov, A.A.; Zhou, Z.; Marcotte, G.R.; Tran, D.; Perez, G.; Gutierrez-Casado, E.; Koike, S.; Knotts, T.A.; et al. A Ketogenic Diet Extends Longevity and Healthspan in Adult Mice. Cell Metab. 2017, 26, 539–546.e5. [Google Scholar] [CrossRef] [Green Version]
- Zupec-Kania, B.A.; Spellman, E. An Overview of the Ketogenic Diet for Pediatric Epilepsy. Nutr. Clin. Pract. 2008, 23, 589–596. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, J.; Yang, S.; Gao, M.; Cao, L.; Li, X.; Hong, D.; Tian, S.; Sun, C. Effect of the Ketogenic Diet on Glycemic Control, Insulin Resistance, and Lipid Metabolism in Patients with T2DM: A Systematic Review and Meta-Analysis. Nutr. Diabetes 2020, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Tozzi, R.; Risi, R.; Tuccinardi, D.; Mariani, S.; Basciani, S.; Spera, G.; Lubrano, C.; Gnessi, L. Beneficial Effects of the Ketogenic Diet on Nonalcoholic Fatty Liver Disease: A Comprehensive Review of the Literature. Obes. Rev. 2020, 21, e13024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiavo, L.; Pierro, R.; Asteria, C.; Calabrese, P.; Di Biasio, A.; Coluzzi, I.; Severino, L.; Giovanelli, A.; Pilone, V.; Silecchia, G. Low-Calorie Ketogenic Diet with Continuous Positive Airway Pressure to Alleviate Severe Obstructive Sleep Apnea Syndrome in Patients with Obesity Scheduled for Bariatric/Metabolic Surgery: A Pilot, Prospective, Randomized Multicenter Comparative Study. Obes. Surg. 2022, 32, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Ciaffi, J.; Mitselman, D.; Mancarella, L.; Brusi, V.; Lisi, L.; Ruscitti, P.; Cipriani, P.; Meliconi, R.; Giacomelli, R.; Borghi, C.; et al. The Effect of Ketogenic Diet on Inflammatory Arthritis and Cardiovascular Health in Rheumatic Conditions: A Mini Review. Front. Med. (Lausanne) 2021, 8, 792846. [Google Scholar] [CrossRef]
- Simeone, T.A.; Simeone, K.A.; Stafstrom, C.E.; Rho, J.M. Do Ketone Bodies Mediate the Anti-Seizure Effects of the Ketogenic Diet? Neuropharmacology 2018, 133, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.; Brown, N.I.; Williams, S.; Plaisance, E.P.; Fontaine, K.R. Ketogenic Diet for Cancer: Critical Assessment and Research Recommendations. Nutrients 2021, 13, 3562. [Google Scholar] [CrossRef]
- Bandera-Merchan, B.; Boughanem, H.; Crujeiras, A.B.; Macias-Gonzalez, M.; Tinahones, F.J. Ketotherapy as an Epigenetic Modifier in Cancer. Rev. Endocr. Metab. Disord. 2020, 21, 509–519. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-Dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chen, P.; Xiao, W. β-Hydroxybutyrate as an Anti-Aging Metabolite. Nutrients 2021, 13, 3420. [Google Scholar] [CrossRef]
- Ruan, H.-B.; Crawford, P.A. Ketone Bodies as Epigenetic Modifiers. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 260–266. [Google Scholar] [CrossRef]
- Newman, J.C.; Verdin, E. Ketone Bodies as Signaling Metabolites. Trends Endocrinol. Metab. 2014, 25, 42–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D.; et al. Suppression of Oxidative Stress by β-Hydroxybutyrate, an Endogenous Histone Deacetylase Inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef] [Green Version]
- Chriett, S.; Dąbek, A.; Wojtala, M.; Vidal, H.; Balcerczyk, A.; Pirola, L. Prominent Action of Butyrate over β-Hydroxybutyrate as Histone Deacetylase Inhibitor, Transcriptional Modulator and Anti-Inflammatory Molecule. Sci. Rep. 2019, 9, 742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Zhang, D.; Chung, D.; Tang, Z.; Huang, H.; Dai, L.; Qi, S.; Li, J.; Colak, G.; Chen, Y.; et al. Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation. Mol. Cell 2016, 62, 194–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolb, H.; Kempf, K.; Röhling, M.; Lenzen-Schulte, M.; Schloot, N.C.; Martin, S. Ketone Bodies: From Enemy to Friend and Guardian Angel. BMC Med. 2021, 19, 313. [Google Scholar] [CrossRef]
- Conboy, K.; Henshall, D.C.; Brennan, G.P. Epigenetic Principles Underlying Epileptogenesis and Epilepsy Syndromes. Neurobiol. Dis. 2021, 148, 105179. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; He, X.; Luan, G.; Li, T. Role of DNA Methylation and Adenosine in Ketogenic Diet for Pharmacoresistant Epilepsy: Focus on Epileptogenesis and Associated Comorbidities. Front. Neurol. 2019, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Landgrave-Gómez, J.; Mercado-Gómez, O.; Guevara-Guzmán, R. Epigenetic Mechanisms in Neurological and Neurodegenerative Diseases. Front. Cell Neurosci. 2015, 9, 58. [Google Scholar] [CrossRef] [Green Version]
- Boison, D. New Insights into the Mechanisms of the Ketogenic Diet. Curr. Opin. Neurol. 2017, 30, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dąbek, A.; Wojtala, M.; Pirola, L.; Balcerczyk, A. Modulation of Cellular Biochemistry, Epigenetics and Metabolomics by Ketone Bodies. Implications of the Ketogenic Diet in the Physiology of the Organism and Pathological States. Nutrients 2020, 12, 788. [Google Scholar] [CrossRef] [Green Version]
- Longo, R.; Peri, C.; Cricrì, D.; Coppi, L.; Caruso, D.; Mitro, N.; De Fabiani, E.; Crestani, M. Ketogenic Diet: A New Light Shining on Old but Gold Biochemistry. Nutrients 2019, 11, 2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 Complex and SIR2 Alone Promote Longevity in Saccharomyces Cerevisiae by Two Different Mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef] [Green Version]
- Rogina, B.; Helfand, S.L. Sir2 Mediates Longevity in the Fly through a Pathway Related to Calorie Restriction. Proc. Natl. Acad. Sci. USA 2004, 101, 15998–16003. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.G.; Rogina, B.; Lavu, S.; Howitz, K.; Helfand, S.L.; Tatar, M.; Sinclair, D. Sirtuin Activators Mimic Caloric Restriction and Delay Ageing in Metazoans. Nature 2004, 430, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative Control of P53 by Sir2alpha Promotes Cell Survival under Stress. Cell 2001, 107, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-Dependent Regulation of FOXO Transcription Factors by the SIRT1 Deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [Green Version]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional Silencing and Longevity Protein Sir2 Is an NAD-Dependent Histone Deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef]
- Vaquero, A.; Scher, M.; Lee, D.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Human SirT1 Interacts with Histone H1 and Promotes Formation of Facultative Heterochromatin. Mol. Cell 2004, 16, 93–105. [Google Scholar] [CrossRef]
- Vaquero, A.; Scher, M.; Erdjument-Bromage, H.; Tempst, P.; Serrano, L.; Reinberg, D. SIRT1 Regulates the Histone Methyl-Transferase SUV39H1 during Heterochromatin Formation. Nature 2007, 450, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-H.; Sengupta, K.; Li, C.; Kim, H.-S.; Cao, L.; Xiao, C.; Kim, S.; Xu, X.; Zheng, Y.; Chilton, B.; et al. Impaired DNA Damage Response, Genome Instability, and Tumorigenesis in SIRT1 Mutant Mice. Cancer Cell 2008, 14, 312–323. [Google Scholar] [CrossRef] [Green Version]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; Machado De Oliveira, R.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 Promotes Fat Mobilization in White Adipocytes by Repressing PPAR-Gamma. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Speakman, J.R.; Talbot, D.A.; Selman, C.; Snart, S.; McLaren, J.S.; Redman, P.; Krol, E.; Jackson, D.M.; Johnson, M.S.; Brand, M.D. Uncoupled and Surviving: Individual Mice with High Metabolism Have Greater Mitochondrial Uncoupling and Live Longer. Aging Cell 2004, 3, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Song, S.; Li, X.; Bian, D.; Wu, X. Association of Metabolic Traits with Occurrence of Nonalcoholic Fatty Liver Disease-Related Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis of Longitudinal Cohort Studies. Saudi. J. Gastroenterol. 2022, 28, 92–100. [Google Scholar] [CrossRef]
- Taylor, A.; Siddiqui, M.K.; Ambery, P.; Armisen, J.; Challis, B.G.; Haefliger, C.; Pearson, E.R.; Doney, A.S.F.; Dillon, J.F.; Palmer, C.N.A. Metabolic Dysfunction-Related Liver Disease as a Risk Factor for Cancer. BMJ Open Gastroenterol. 2022, 9, e000817. [Google Scholar] [CrossRef] [PubMed]
- Gahete, M.D.; Granata, R.; Luque, R.M. Editorial: Pathophysiological Interrelationship Between Obesity, Metabolic Diseases, and Cancer. Front. Oncol. 2021, 11, 755735. [Google Scholar] [CrossRef]
- Faulds, M.H.; Dahlman-Wright, K. Metabolic Diseases and Cancer Risk. Curr. Opin. Oncol. 2012, 24, 58–61. [Google Scholar] [CrossRef]
- Avgerinos, K.I.; Spyrou, N.; Mantzoros, C.S.; Dalamaga, M. Obesity and Cancer Risk: Emerging Biological Mechanisms and Perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef]
- Cao, Z.; Zheng, X.; Yang, H.; Li, S.; Xu, F.; Yang, X.; Wang, Y. Association of Obesity Status and Metabolic Syndrome with Site-Specific Cancers: A Population-Based Cohort Study. Br. J. Cancer 2020, 123, 1336–1344. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, H.; Yang, M.; Ye, S.; Li, L.; Zhang, H.; Hu, J.; Wang, X.; Xu, J.; Liang, A. SIRT1 Inhibition Impairs Non-Homologous End Joining DNA Damage Repair by Increasing Ku70 Acetylation in Chronic Myeloid Leukemia Cells. Oncotarget 2015, 7, 13538–13550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlando, G.; Khoronenkova, S.V.; Dianova, I.I.; Parsons, J.L.; Dianov, G.L. ARF Induction in Response to DNA Strand Breaks Is Regulated by PARP1. Nucleic Acids Res. 2014, 42, 2320–2329. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Ji, L.; Chen, H.; Li, X.; Zhang, J.; Wang, X.; Wu, W.; Xu, Y.; Huang, F.; Cai, W.; et al. Acetylation of HMOF Modulates H4K16ac to Regulate DNA Repair Genes in Response to Oxidative Stress. Int. J. Biol. Sci. 2017, 13, 923–934. [Google Scholar] [CrossRef] [Green Version]
- Vaziri, H.; Dessain, S.K.; Ng Eaton, E.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. HSIR2(SIRT1) Functions as an NAD-Dependent P53 Deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Firestein, R.; Blander, G.; Michan, S.; Oberdoerffer, P.; Ogino, S.; Campbell, J.; Bhimavarapu, A.; Luikenhuis, S.; de Cabo, R.; Fuchs, C.; et al. The SIRT1 Deacetylase Suppresses Intestinal Tumorigenesis and Colon Cancer Growth. PLoS ONE 2008, 3, e2020. [Google Scholar] [CrossRef] [PubMed]
- Oberdoerffer, P.; Michan, S.; McVay, M.; Mostoslavsky, R.; Vann, J.; Park, S.-K.; Hartlerode, A.; Stegmuller, J.; Hafner, A.; Loerch, P.; et al. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell 2008, 135, 907–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herranz, D.; Muñoz-Martin, M.; Cañamero, M.; Mulero, F.; Martinez-Pastor, B.; Fernandez-Capetillo, O.; Serrano, M. Sirt1 Improves Healthy Ageing and Protects from Metabolic Syndrome-Associated Cancer. Nat. Commun. 2010, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- de Gregorio, E.; Colell, A.; Morales, A.; Marí, M. Relevance of SIRT1-NF-ΚB Axis as Therapeutic Target to Ameliorate Inflammation in Liver Disease. Int. J. Mol. Sci. 2020, 21, 3858. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, J.A.; Gallego-Durán, R.; Gallego, P.; Grande, L. Genetic and Epigenetic Regulation in Nonalcoholic Fatty Liver Disease (NAFLD). Int. J. Mol. Sci. 2018, 19, 911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deleye, Y.; Cotte, A.K.; Hannou, S.A.; Hennuyer, N.; Bernard, L.; Derudas, B.; Caron, S.; Legry, V.; Vallez, E.; Dorchies, E.; et al. CDKN2A/P16INK4a Suppresses Hepatic Fatty Acid Oxidation through the AMPKα2-SIRT1-PPARα Signaling Pathway. J. Biol. Chem. 2020, 295, 17310–17322. [Google Scholar] [CrossRef]
- Ding, R.-B.; Bao, J.; Deng, C.-X. Emerging Roles of SIRT1 in Fatty Liver Diseases. Int. J. Biol. Sci. 2017, 13, 852–867. [Google Scholar] [CrossRef]
- Park, S.; Shin, J.; Bae, J.; Han, D.; Park, S.-R.; Shin, J.; Lee, S.K.; Park, H.-W. SIRT1 Alleviates LPS-Induced IL-1β Production by Suppressing NLRP3 Inflammasome Activation and ROS Production in Trophoblasts. Cells 2020, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, K.; Wakino, S.; Simic, P.; Sakamaki, Y.; Minakuchi, H.; Fujimura, K.; Hosoya, K.; Komatsu, M.; Kaneko, Y.; Kanda, T.; et al. Renal Tubular Sirt1 Attenuates Diabetic Albuminuria by Epigenetically Suppressing Claudin-1 Overexpression in Podocytes. Nat. Med. 2013, 19, 1496–1504. [Google Scholar] [CrossRef] [Green Version]
- Karbasforooshan, H.; Karimi, G. The Role of SIRT1 in Diabetic Cardiomyopathy. Biomed. Pharmacother. 2017, 90, 386–392. [Google Scholar] [CrossRef]
- Moreno, C.L.; Mobbs, C.V. Epigenetic Mechanisms Underlying Lifespan and Age-Related Effects of Dietary Restriction and the Ketogenic Diet. Mol. Cell Endocrinol. 2017, 455, 33–40. [Google Scholar] [CrossRef]
- Cunha, G.M.; Guzman, G.; Correa De Mello, L.L.; Trein, B.; Spina, L.; Bussade, I.; Marques Prata, J.; Sajoux, I.; Countinho, W. Efficacy of a 2-Month Very Low-Calorie Ketogenic Diet (VLCKD) Compared to a Standard Low-Calorie Diet in Reducing Visceral and Liver Fat Accumulation in Patients With Obesity. Front. Endocrinol. (Lausanne) 2020, 11, 607. [Google Scholar] [CrossRef]
- Risi, R.; Tozzi, R.; Watanabe, M. Beyond Weight Loss in Nonalcoholic Fatty Liver Disease: The Role of Carbohydrate Restriction. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 349–353. [Google Scholar] [CrossRef]
- Mariani, S.; Fiore, D.; Persichetti, A.; Basciani, S.; Lubrano, C.; Poggiogalle, E.; Genco, A.; Donini, L.M.; Gnessi, L. Circulating SIRT1 Increases After Intragastric Balloon Fat Loss in Obese Patients. Obes. Surg. 2016, 26, 1215–1220. [Google Scholar] [CrossRef]
- Purushotham, A.; Schug, T.T.; Li, X. SIRT1 Performs a Balancing Act on the Tight-Rope toward Longevity. Aging 2009, 1, 669–673. [Google Scholar] [CrossRef] [Green Version]
- Mariani, S.; Di Giorgio, M.R.; Rossi, E.; Tozzi, R.; Contini, S.; Bauleo, L.; Cipriani, F.; Toscano, R.; Basciani, S.; Barbaro, G.; et al. Blood SIRT1 Shows a Coherent Association with Leptin and Adiponectin in Relation to the Degree and Distribution of Adiposity: A Study in Obesity, Normal Weight and Anorexia Nervosa. Nutrients 2020, 12, 3506. [Google Scholar] [CrossRef]
- Liu, Y.; Dentin, R.; Chen, D.; Hedrick, S.; Ravnskjaer, K.; Schenk, S.; Milne, J.; Meyers, D.J.; Cole, P.; Yates, J.; et al. A Fasting Inducible Switch Modulates Gluconeogenesis via Activator/Coactivator Exchange. Nature 2008, 456, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Blander, G.; Tse, J.G.; Krieger, M.; Guarente, L. SIRT1 Deacetylates and Positively Regulates the Nuclear Receptor LXR. Mol. Cell 2007, 28, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Gao, Z.; Zhang, J.; Rivera, C.A.; Yin, J.; Weng, J.; Ye, J. Lack of SIRT1 (Mammalian Sirtuin 1) Activity Leads to Liver Steatosis in the SIRT1+/− Mice: A Role of Lipid Mobilization and Inflammation. Endocrinology 2010, 151, 2504–2514. [Google Scholar] [CrossRef] [Green Version]
- Banks, A.S.; Kon, N.; Knight, C.; Matsumoto, M.; Gutiérrez-Juárez, R.; Rossetti, L.; Gu, W.; Accili, D. SirT1 Gain of Function Increases Energy Efficiency and Prevents Diabetes in Mice. Cell Metab. 2008, 8, 333–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tozzi, R.; Masi, D.; Cipriani, F.; Contini, S.; Gangitano, E.; Spoltore, M.E.; Barchetta, I.; Basciani, S.; Watanabe, M.; Baldini, E.; et al. Circulating SIRT1 and Sclerostin Correlates with Bone Status in Young Women with Different Degrees of Adiposity. Nutrients 2022, 14, 983. [Google Scholar] [CrossRef]
- Matsushima, S.; Sadoshima, J. The Role of Sirtuins in Cardiac Disease. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1375–H1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, Y.; Zhang, C.; Xie, M. Beta-Hydroxybutyrate, Friend or Foe for Stressed Hearts. Front. Aging 2021, 2, 681513. [Google Scholar] [CrossRef]
- Mariani, S.; Costantini, D.; Lubrano, C.; Basciani, S.; Caldaroni, C.; Barbaro, G.; Poggiogalle, E.; Donini, L.M.; Lenzi, A.; Gnessi, L. Circulating SIRT1 Inversely Correlates with Epicardial Fat Thickness in Patients with Obesity. Nutr. Metab. Cardiovasc. Dis. 2016, 26, 1033–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elamin, M.; Ruskin, D.N.; Masino, S.A.; Sacchetti, P. Ketogenic Diet Modulates NAD+-Dependent Enzymes and Reduces DNA Damage in Hippocampus. Front. Cell Neurosci. 2018, 12, 263. [Google Scholar] [CrossRef] [PubMed]
- McCarty, M.F.; DiNicolantonio, J.J.; O’Keefe, J.H. Ketosis May Promote Brain Macroautophagy by Activating Sirt1 and Hypoxia-Inducible Factor-1. Med. Hypotheses 2015, 85, 631–639. [Google Scholar] [CrossRef]
- Parodi, B.; Rossi, S.; Morando, S.; Cordano, C.; Bragoni, A.; Motta, C.; Usai, C.; Wipke, B.T.; Scannevin, R.H.; Mancardi, G.L.; et al. Fumarates Modulate Microglia Activation through a Novel HCAR2 Signaling Pathway and Rescue Synaptic Dysregulation in Inflamed CNS. Acta Neuropathol. 2015, 130, 279–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, A.; Kume, S.; Kondo, M.; Nakazawa, J.; Chin-Kanasaki, M.; Araki, H.; Araki, S.; Koya, D.; Haneda, M.; Chano, T.; et al. Mammalian Autophagy Is Essential for Hepatic and Renal Ketogenesis during Starvation. Sci. Rep. 2016, 6, 18944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liśkiewicz, D.; Liśkiewicz, A.; Nowacka-Chmielewska, M.M.; Grabowski, M.; Pondel, N.; Grabowska, K.; Student, S.; Barski, J.J.; Małecki, A. Differential Response of Hippocampal and Cerebrocortical Autophagy and Ketone Body Metabolism to the Ketogenic Diet. Front. Cell Neurosci. 2021, 15, 733607. [Google Scholar] [CrossRef]
- Purushotham, A.; Schug, T.T.; Xu, Q.; Surapureddi, S.; Guo, X.; Li, X. Hepatocyte-Specific Deletion of SIRT1 Alters Fatty Acid Metabolism and Results in Hepatic Steatosis and Inflammation. Cell Metab. 2009, 9, 327–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellmann, J.; Tang, Y.; Kosuri, M.; Bhatnagar, A.; Spite, M. Resolvin D1 Decreases Adipose Tissue Macrophage Accumulation and Improves Insulin Sensitivity in Obese-Diabetic Mice. FASEB J. 2011, 25, 2399–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nøhr, M.K.; Bobba, N.; Richelsen, B.; Lund, S.; Pedersen, S.B. Inflammation Downregulates UCP1 Expression in Brown Adipocytes Potentially via SIRT1 and DBC1 Interaction. Int. J. Mol. Sci. 2017, 18, 1006. [Google Scholar] [CrossRef]
- Srivastava, S.; Baxa, U.; Niu, G.; Chen, X.; Veech, R.L. A Ketogenic Diet Increases Brown Adipose Tissue Mitochondrial Proteins and UCP1 Levels in Mice. IUBMB Life 2013, 65, 58–66. [Google Scholar] [CrossRef]
- Archer, A.; Venteclef, N.; Mode, A.; Pedrelli, M.; Gabbi, C.; Clément, K.; Parini, P.; Gustafsson, J.-Å.; Korach-André, M. Fasting-Induced FGF21 Is Repressed by LXR Activation via Recruitment of an HDAC3 Corepressor Complex in Mice. Mol. Endocrinol. 2012, 26, 1980–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Gao, Z.; Zhang, J.; Ye, X.; Xu, A.; Ye, J.; Jia, W. Sodium Butyrate Stimulates Expression of Fibroblast Growth Factor 21 in Liver by Inhibition of Histone Deacetylase 3. Diabetes 2012, 61, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Byun, S.; Seok, S.; Kim, Y.-C.; Zhang, Y.; Yau, P.; Iwamori, N.; Xu, H.E.; Ma, J.; Kemper, B.; Kemper, J.K. Fasting-Induced FGF21 Signaling Activates Hepatic Autophagy and Lipid Degradation via JMJD3 Histone Demethylase. Nat. Commun. 2020, 11, 807. [Google Scholar] [CrossRef] [Green Version]
- dos Santos Costa, C.; Hammes, T.O.; Rohden, F.; Margis, R.; Bortolotto, J.W.; Padoin, A.V.; Mottin, C.C.; Guaragna, R.M. SIRT1 Transcription Is Decreased in Visceral Adipose Tissue of Morbidly Obese Patients with Severe Hepatic Steatosis. Obes. Surg. 2010, 20, 633–639. [Google Scholar] [CrossRef]
- Perrini, S.; Porro, S.; Nigro, P.; Cignarelli, A.; Caccioppoli, C.; Genchi, V.A.; Martines, G.; De Fazio, M.; Capuano, P.; Natalicchio, A.; et al. Reduced SIRT1 and SIRT2 Expression Promotes Adipogenesis of Human Visceral Adipose Stem Cells and Associates with Accumulation of Visceral Fat in Human Obesity. Int. J. Obes. 2020, 44, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Mariani, S.; Di Rocco, G.; Toietta, G.; Russo, M.A.; Petrangeli, E.; Salvatori, L. Sirtuins 1–7 Expression in Human Adipose-Derived Stem Cells from Subcutaneous and Visceral Fat Depots: Influence of Obesity and Hypoxia. Endocrine 2017, 57, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Gillum, M.P.; Kotas, M.E.; Erion, D.M.; Kursawe, R.; Chatterjee, P.; Nead, K.T.; Muise, E.S.; Hsiao, J.J.; Frederick, D.W.; Yonemitsu, S.; et al. SirT1 Regulates Adipose Tissue Inflammation. Diabetes 2011, 60, 3235–3245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, A.V.; Beckers, S.; Verrijken, A.; Mertens, I.; Roevens, P.; Peeters, P.J.; Van Hul, W.; Van Gaal, L.F. Association of SIRT1 Gene Variation with Visceral Obesity. Hum. Genet. 2008, 124, 431–436. [Google Scholar] [CrossRef]
- Zillikens, M.C.; van Meurs, J.B.J.; Rivadeneira, F.; Amin, N.; Hofman, A.; Oostra, B.A.; Sijbrands, E.J.G.; Witteman, J.C.M.; Pols, H.A.P.; van Duijn, C.M.; et al. SIRT1 Genetic Variation Is Related to BMI and Risk of Obesity. Diabetes 2009, 58, 2828–2834. [Google Scholar] [CrossRef] [Green Version]
- Jukarainen, S.; Heinonen, S.; Rämö, J.T.; Rinnankoski-Tuikka, R.; Rappou, E.; Tummers, M.; Muniandy, M.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; et al. Obesity Is Associated With Low NAD(+)/SIRT Pathway Expression in Adipose Tissue of BMI-Discordant Monozygotic Twins. J. Clin. Endocrinol. Metab. 2016, 101, 275–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheibye-Knudsen, M.; Mitchell, S.J.; Fang, E.F.; Iyama, T.; Ward, T.; Wang, J.; Dunn, C.A.; Singh, N.; Veith, S.; Hasan-Olive, M.M.; et al. A High-Fat Diet and NAD+ Activate Sirt1 to Rescue Premature Aging in Cockayne Syndrome. Cell Metab. 2014, 20, 840–855. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Jiménez, V.; Cortez-Espinosa, N.; Rodríguez-Varela, E.; Vega-Cárdenas, M.; Briones-Espinoza, M.; Ruíz-Rodríguez, V.M.; López-López, N.; Briseño-Medina, A.; Turiján-Espinoza, E.; Portales-Pérez, D.P. Altered Levels of Sirtuin Genes (SIRT1, SIRT2, SIRT3 and SIRT6) and Their Target Genes in Adipose Tissue from Individual with Obesity. Diabetes Metab. Syndr. 2019, 13, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Mariani, S.; Fiore, D.; Basciani, S.; Persichetti, A.; Contini, S.; Lubrano, C.; Salvatori, L.; Lenzi, A.; Gnessi, L. Plasma Levels of SIRT1 Associate with Non-Alcoholic Fatty Liver Disease in Obese Patients. Endocrine 2015, 49, 711–716. [Google Scholar] [CrossRef]
- Song, Y.M.; Lee, Y.; Kim, J.-W.; Ham, D.-S.; Kang, E.-S.; Cha, B.S.; Lee, H.C.; Lee, B.-W. Metformin Alleviates Hepatosteatosis by Restoring SIRT1-Mediated Autophagy Induction via an AMP-Activated Protein Kinase-Independent Pathway. Autophagy 2015, 11, 46–59. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, T.; Dutchak, P.; Zhao, G.; Ding, X.; Gautron, L.; Parameswara, V.; Li, Y.; Goetz, R.; Mohammadi, M.; Esser, V.; et al. Endocrine Regulation of the Fasting Response by PPARalpha-Mediated Induction of Fibroblast Growth Factor 21. Cell Metab. 2007, 5, 415–425. [Google Scholar] [CrossRef] [Green Version]
- Potthoff, M.J.; Inagaki, T.; Satapati, S.; Ding, X.; He, T.; Goetz, R.; Mohammadi, M.; Finck, B.N.; Mangelsdorf, D.J.; Kliewer, S.A.; et al. FGF21 Induces PGC-1alpha and Regulates Carbohydrate and Fatty Acid Metabolism during the Adaptive Starvation Response. Proc. Natl. Acad. Sci. USA 2009, 106, 10853–10858. [Google Scholar] [CrossRef] [Green Version]
- Liou, C.-J.; Wei, C.-H.; Chen, Y.-L.; Cheng, C.-Y.; Wang, C.-L.; Huang, W.-C. Fisetin Protects Against Hepatic Steatosis Through Regulation of the Sirt1/AMPK and Fatty Acid β-Oxidation Signaling Pathway in High-Fat Diet-Induced Obese Mice. Cell Physiol. Biochem. 2018, 49, 1870–1884. [Google Scholar] [CrossRef]
- Neuhofer, A.; Zeyda, M.; Mascher, D.; Itariu, B.K.; Murano, I.; Leitner, L.; Hochbrugger, E.E.; Fraisl, P.; Cinti, S.; Serhan, C.N.; et al. Impaired Local Production of Proresolving Lipid Mediators in Obesity and 17-HDHA as a Potential Treatment for Obesity-Associated Inflammation. Diabetes 2013, 62, 1945–1956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serhan, C.N.; Levy, B.D. Resolvins in Inflammation: Emergence of the pro-Resolving Superfamily of Mediators. J. Clin. Investig. 2018, 128, 2657–2669. [Google Scholar] [CrossRef]
- Titos, E.; Rius, B.; López-Vicario, C.; Alcaraz-Quiles, J.; García-Alonso, V.; Lopategi, A.; Dalli, J.; Lozano, J.J.; Arroyo, V.; Delgado, S.; et al. Signaling and Immunoresolving Actions of Resolvin D1 in Inflamed Human Visceral Adipose Tissue. J. Immunol. 2016, 197, 3360–3370. [Google Scholar] [CrossRef] [Green Version]
- Graff, E.C.; Fang, H.; Wanders, D.; Judd, R.L. Anti-Inflammatory Effects of the Hydroxycarboxylic Acid Receptor 2. Metabolism 2016, 65, 102–113. [Google Scholar] [CrossRef]
- Stubbs, B.J.; Newman, J.C. Ketogenic Diet and Adipose Tissue Inflammation-a Simple Story? Fat Chance! Nat. Metab. 2020, 2, 3–4. [Google Scholar] [CrossRef]
- Goldberg, E.L.; Shchukina, I.; Asher, J.L.; Sidorov, S.; Artyomov, M.N.; Dixit, V.D. Ketogenesis Activates Metabolically Protective Γδ T Cells in Visceral Adipose Tissue. Nat. Metab. 2020, 2, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Gangitano, E.; Tozzi, R.; Gandini, O.; Watanabe, M.; Basciani, S.; Mariani, S.; Lenzi, A.; Gnessi, L.; Lubrano, C. Ketogenic Diet as a Preventive and Supportive Care for COVID-19 Patients. Nutrients 2021, 13, 1004. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.-P.; Li, S.-N.; Wang, J.-F.; Li, Y.; Xie, S.-S.; Xue, W.-J.; Liu, H.-M.; Huang, B.-X.; Lv, Q.-K.; Lei, L.-C.; et al. BHBA Suppresses LPS-Induced Inflammation in BV-2 Cells by Inhibiting NF-ΚB Activation. Mediat. Inflamm. 2014, 2014, 983401. [Google Scholar] [CrossRef] [Green Version]
- Gambhir, D.; Ananth, S.; Veeranan-Karmegam, R.; Elangovan, S.; Hester, S.; Jennings, E.; Offermanns, S.; Nussbaum, J.J.; Smith, S.B.; Thangaraju, M.; et al. GPR109A as an Anti-Inflammatory Receptor in Retinal Pigment Epithelial Cells and Its Relevance to Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2208–2217. [Google Scholar] [CrossRef]
- Youm, Y.-H.; Nguyen, K.Y.; Grant, R.W.; Goldberg, E.L.; Bodogai, M.; Kim, D.; D’Agostino, D.; Planavsky, N.; Lupfer, C.; Kanneganti, T.D.; et al. The Ketone Metabolite β-Hydroxybutyrate Blocks NLRP3 Inflammasome-Mediated Inflammatory Disease. Nat. Med. 2015, 21, 263–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanikarla-Marie, P.; Jain, S.K. Hyperketonemia (Acetoacetate) Upregulates NADPH Oxidase 4 and Elevates Oxidative Stress, ICAM-1, and Monocyte Adhesivity in Endothelial Cells. Cell Physiol. Biochem. 2015, 35, 364–373. [Google Scholar] [CrossRef]
- Kurepa, D.; Pramanik, A.K.; Kakkilaya, V.; Caldito, G.; Groome, L.J.; Bocchini, J.A.; Jain, S.K. Elevated Acetoacetate and Monocyte Chemotactic Protein-1 Levels in Cord Blood of Infants of Diabetic Mothers. Neonatology 2012, 102, 163–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Han, P.; Tang, Z.; Liu, Q.; Shi, J. Sirtuin 3 Mediates Neuroprotection of Ketones against Ischemic Stroke. J. Cereb. Blood Flow Metab. 2015, 35, 1783–1789. [Google Scholar] [CrossRef] [Green Version]
- Gangitano, E.; Tozzi, R.; Mariani, S.; Lenzi, A.; Gnessi, L.; Lubrano, C. Ketogenic Diet for Obese COVID-19 Patients: Is Respiratory Disease a Contraindication? A Narrative Review of the Literature on Ketogenic Diet and Respiratory Function. Front. Nutr. 2021, 8, 771047. [Google Scholar] [CrossRef]
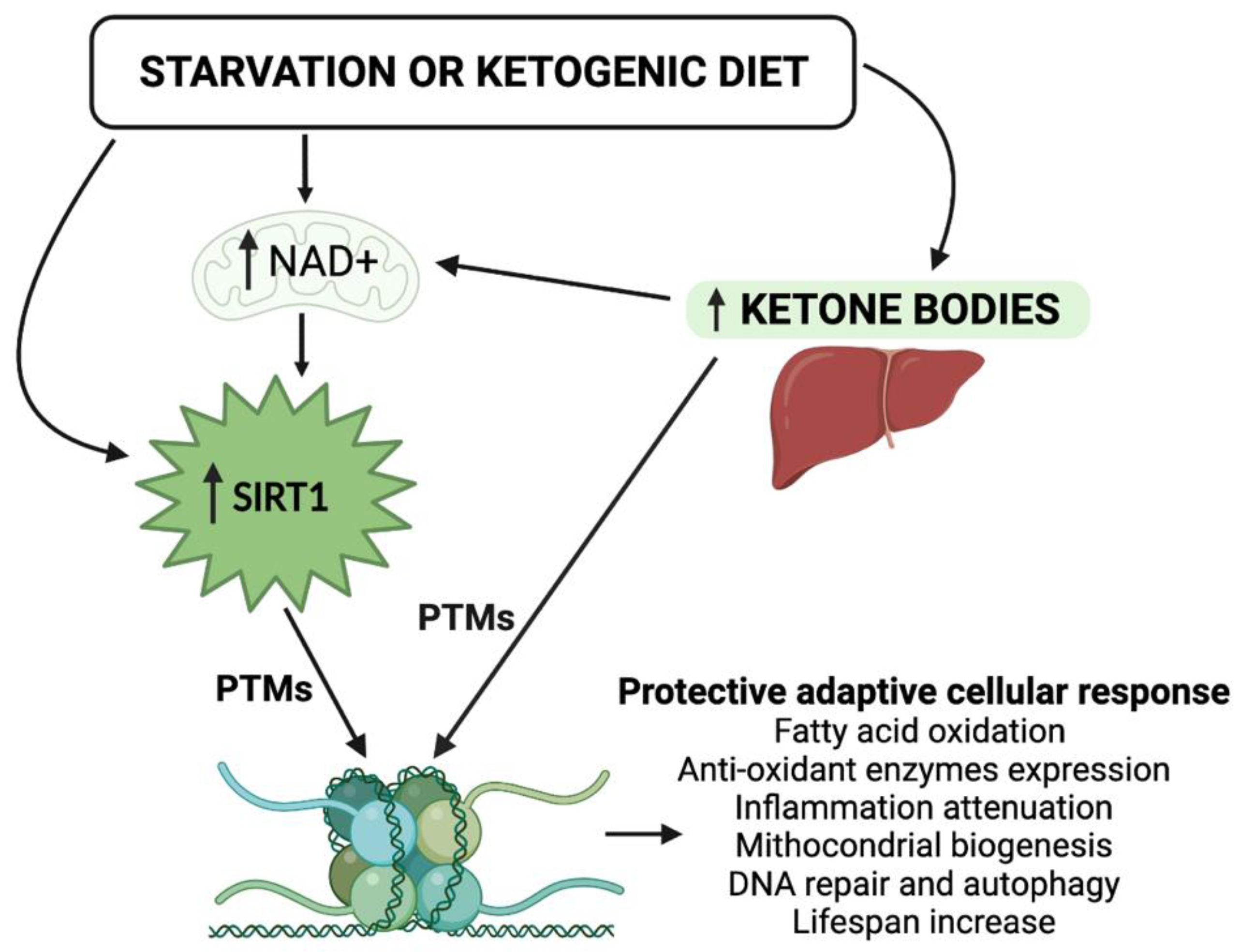
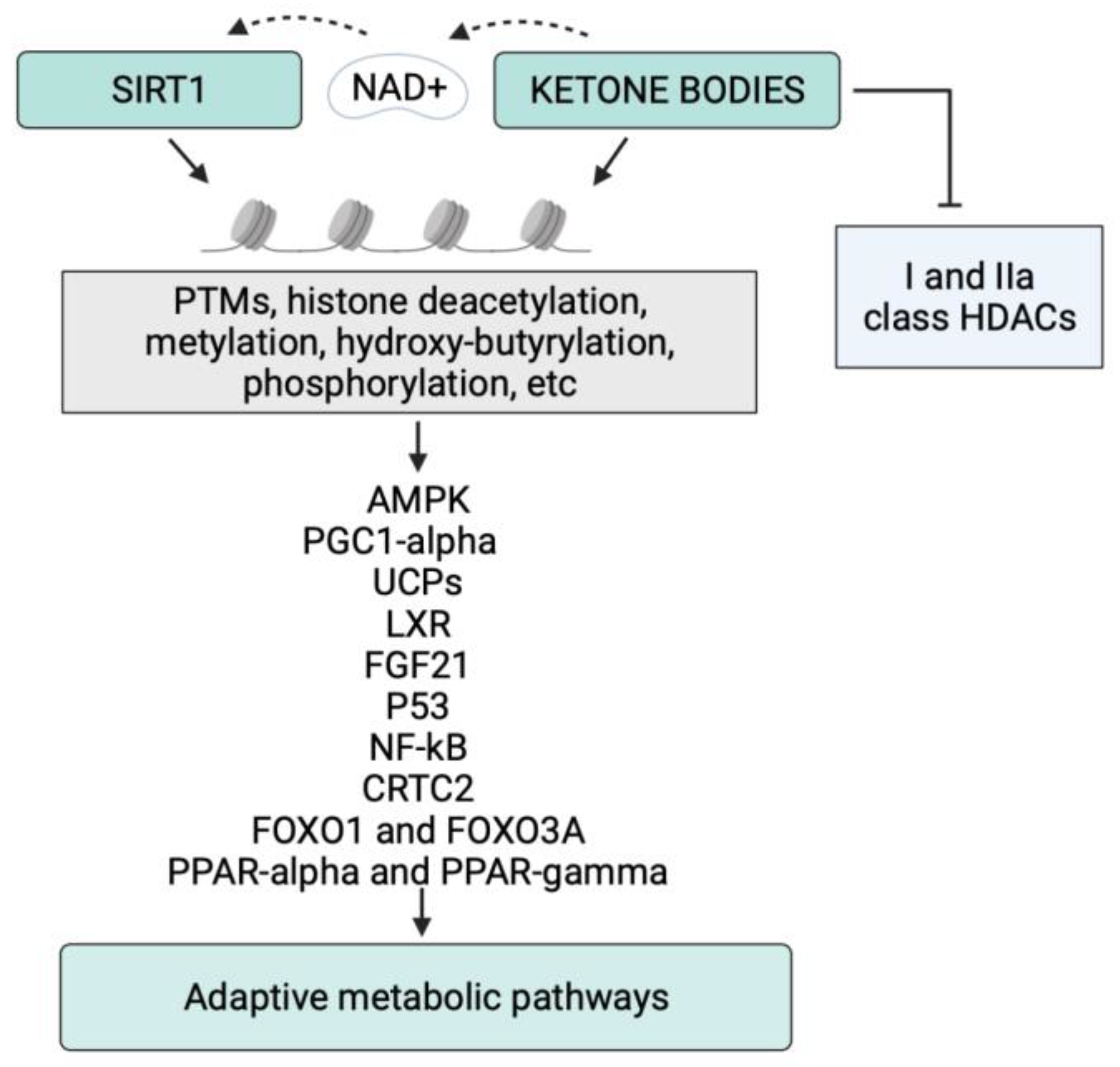
| Target | Main SIRT1 Effects | Main KBs Effects | Common Metabolic Outcomes |
|---|---|---|---|
| AMPK | Increase. Induction of FOXO3 and NF-kB evoking the expression of antioxidant genes and autophagy [2,100]. | Activation through G-protein-coupled-HCAR2 leading to NAD+ increase [101]. Disposal of inflammation while improving intrahepatic fat discharge. | Anti-inflammatory effects; neuroprotection; protection against ischemic stroke. |
| PGC1-α | Deacetylation and activation. Support to the late phase of gluconeogenesis and fatty acid oxidation [91]. Autophagy [102]. | Activation. Promotion of the oxidation of fatty acids with a metabolic shift from glucose homeostasis [103]. | Adaptive starvation response. Increase in autophagic flux. Fat loss. |
| PPAR-α | Activation also through AMPK pathway. Improved β-oxidation of fatty acid, better response to HFD with decreased hepatic inflammation, endoplasmic reticulum stress, and NAFLD [104]. | Increase. Reduction in inflammatory interleukins IL-1β and IL-6 [3,105]. | Reduction in hepatic inflammation and NAFLD. |
| PPAR-γ | Suppression by docking to the negative cofactors of the nuclear receptor. Fat mobilization into the blood stream [63]. | Repression. Reduction in inflammatory interleukins IL-1β and IL-6 mitigating neuroinflammation [3]. | Fat loss. |
| UCP1 | Increase. Induction in BAT after caloric restriction and nutrient deprivation [17,106]. | Induction in BAT [4,107]. Expenditure of heat instead of ATP production. | Improved thermogenesis and energy expenditure. |
| LXR | Deacetylation and activation in the nucleus. Increase in the liver cholesterol efflux [92]. | Activation after FGF21 induction. Glucose and lipid metabolism improvement [108]. | Reduction in NAFLD. |
| FGF-21 | Activation of transcriptional activity of the FGF21 promoter. Fatty acid oxidation and energy expenditure, decreased fasting-induced steatosis, promoted browning of WAT [13]. | Increase in FGF-21 through the inhibition of class I HDAC3 [109]. Upregulation of global autophagy-network genes, inhibition of de novo lipogenesis. | Weight loss and reduction in liver fat content [110]. |
| FOXO | Deacetylation and induction of FOXO1 in the liver with modulation of gluconeogenesis genes when fasting is prolonged [91]. Overexpression of FOXO3 with antioxidant effects [111]. | Inhibition class I and II HDACs with the upregulation of FOXO3A transcription factor network genes [44] and modulation of DNA transcription [30,43]. | Improved insulin signaling pathway and regulation of longevity. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tozzi, R.; Cipriani, F.; Masi, D.; Basciani, S.; Watanabe, M.; Lubrano, C.; Gnessi, L.; Mariani, S. Ketone Bodies and SIRT1, Synergic Epigenetic Regulators for Metabolic Health: A Narrative Review. Nutrients 2022, 14, 3145. https://doi.org/10.3390/nu14153145
Tozzi R, Cipriani F, Masi D, Basciani S, Watanabe M, Lubrano C, Gnessi L, Mariani S. Ketone Bodies and SIRT1, Synergic Epigenetic Regulators for Metabolic Health: A Narrative Review. Nutrients. 2022; 14(15):3145. https://doi.org/10.3390/nu14153145
Chicago/Turabian StyleTozzi, Rossella, Fiammetta Cipriani, Davide Masi, Sabrina Basciani, Mikiko Watanabe, Carla Lubrano, Lucio Gnessi, and Stefania Mariani. 2022. "Ketone Bodies and SIRT1, Synergic Epigenetic Regulators for Metabolic Health: A Narrative Review" Nutrients 14, no. 15: 3145. https://doi.org/10.3390/nu14153145
APA StyleTozzi, R., Cipriani, F., Masi, D., Basciani, S., Watanabe, M., Lubrano, C., Gnessi, L., & Mariani, S. (2022). Ketone Bodies and SIRT1, Synergic Epigenetic Regulators for Metabolic Health: A Narrative Review. Nutrients, 14(15), 3145. https://doi.org/10.3390/nu14153145









