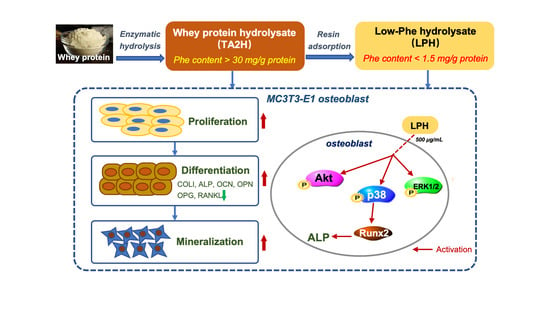
A Low-Phenylalanine-Containing Whey Protein Hydrolysate Stimulates Osteogenic Activity through the Activation of p38/Runx2 Signaling in Osteoblast Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of Whey Protein Hydrolysate (TA2H) and Its Derived Low-Phe Hydrolysate (LPH)
2.3. Cell Culture
2.4. Cell Proliferation Assay
2.5. ALP Activity Assay
2.6. ELISA Assay
2.7. Real-Time Polymerase Chain Reaction (RT-PCR) Assay
2.8. Western Blot Analysis
2.9. Extracellular Matrix Mineralization Assay
2.10. Detection of Signaling Pathway
2.11. Pathway Inhibitors Assay
2.12. Small Interfering RNA (siRNA) Knockdown of Runx2 Expression
2.13. Statistical Analysis
3. Results
3.1. Amino-Acid Compositions of Whey Protein Hydrolysates
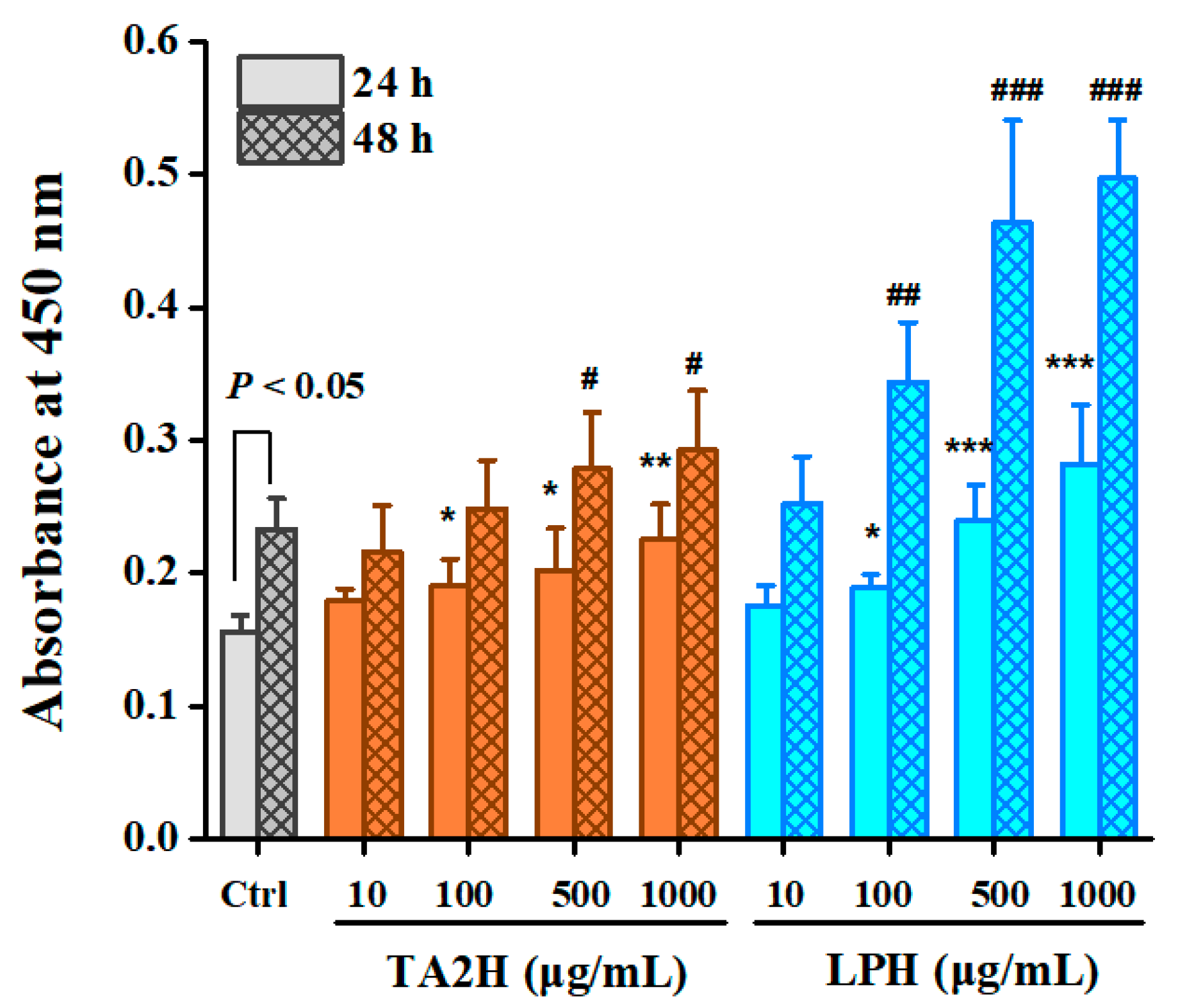
3.2. Whey Protein Hydrolysates Promote Cell Proliferation in MC3T3-E1 Osteoblasts
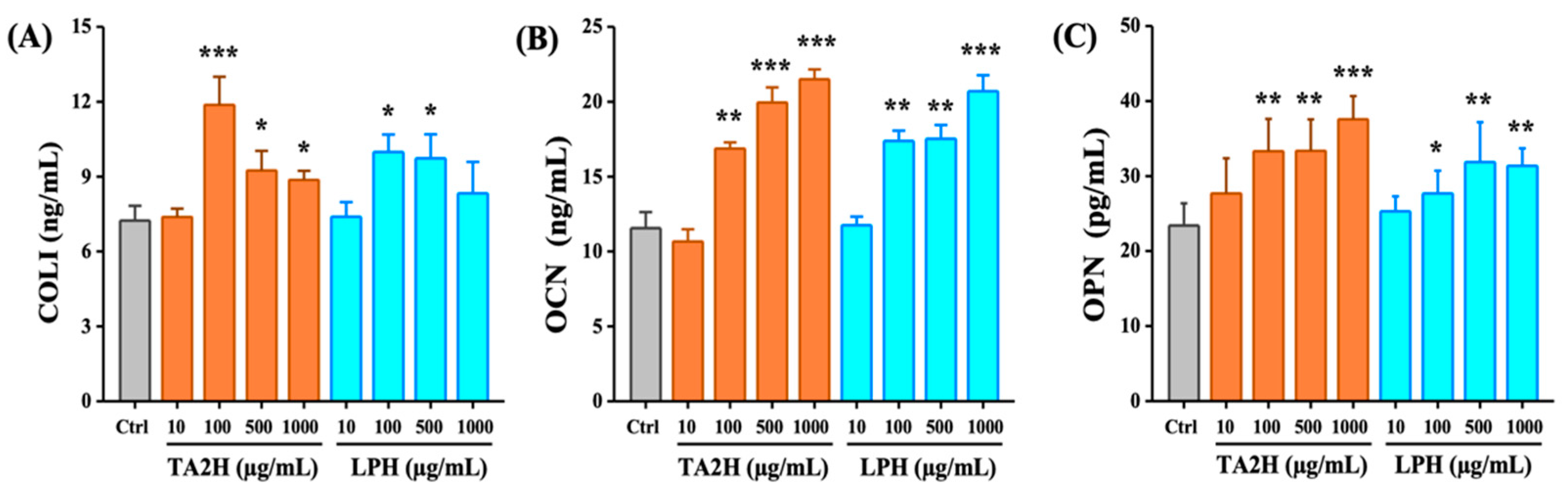
3.3. Whey Protein Hydrolysates Promote Cell Differentiation in MC3T3-E1 Osteoblasts
3.4. Whey Protein Hydrolysates Promote the Mineralization of Osteoblasts
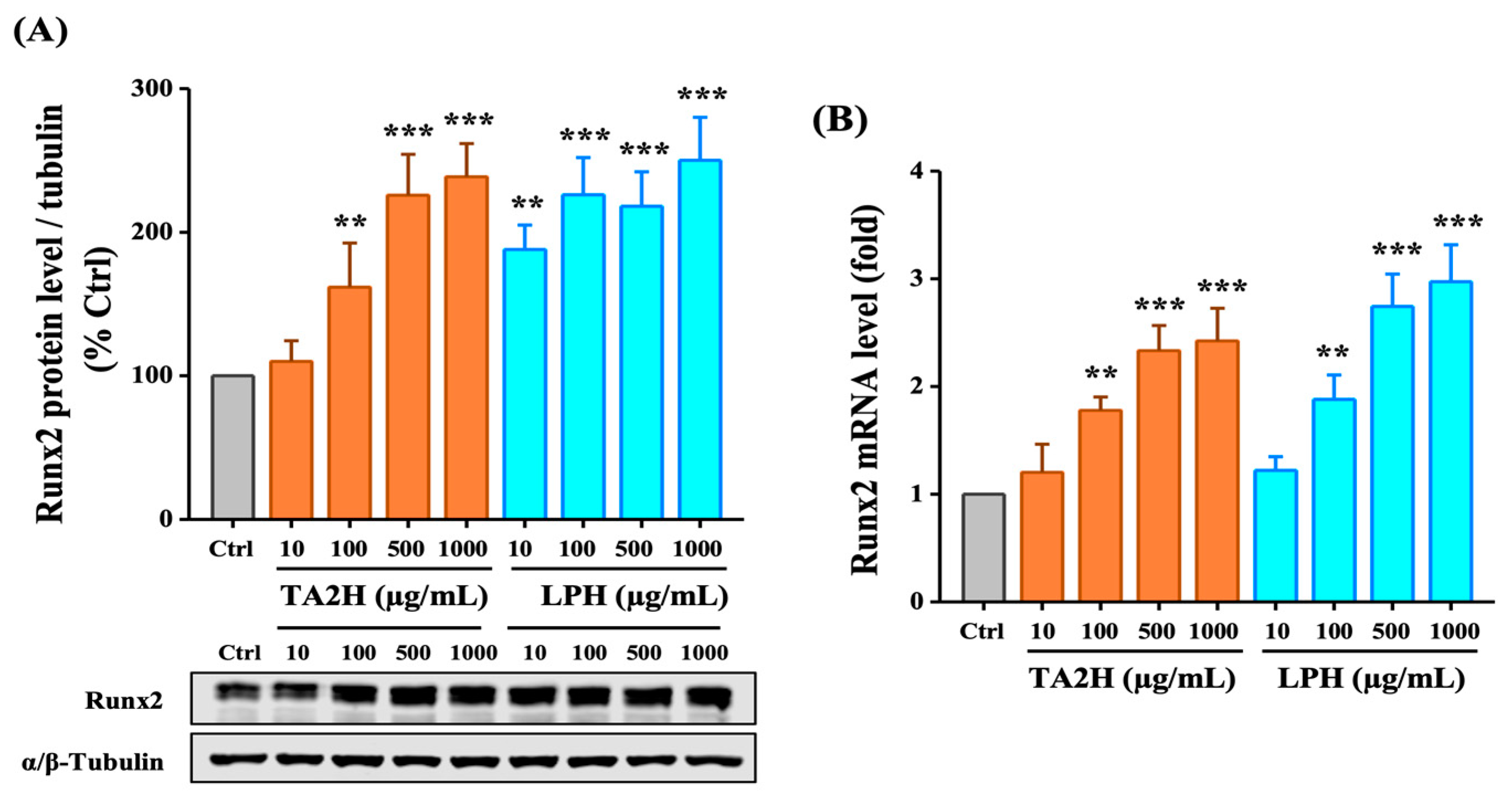
3.5. Whey Protein Hydrolysates Induce the Expression of Runx2 in Osteoblasts
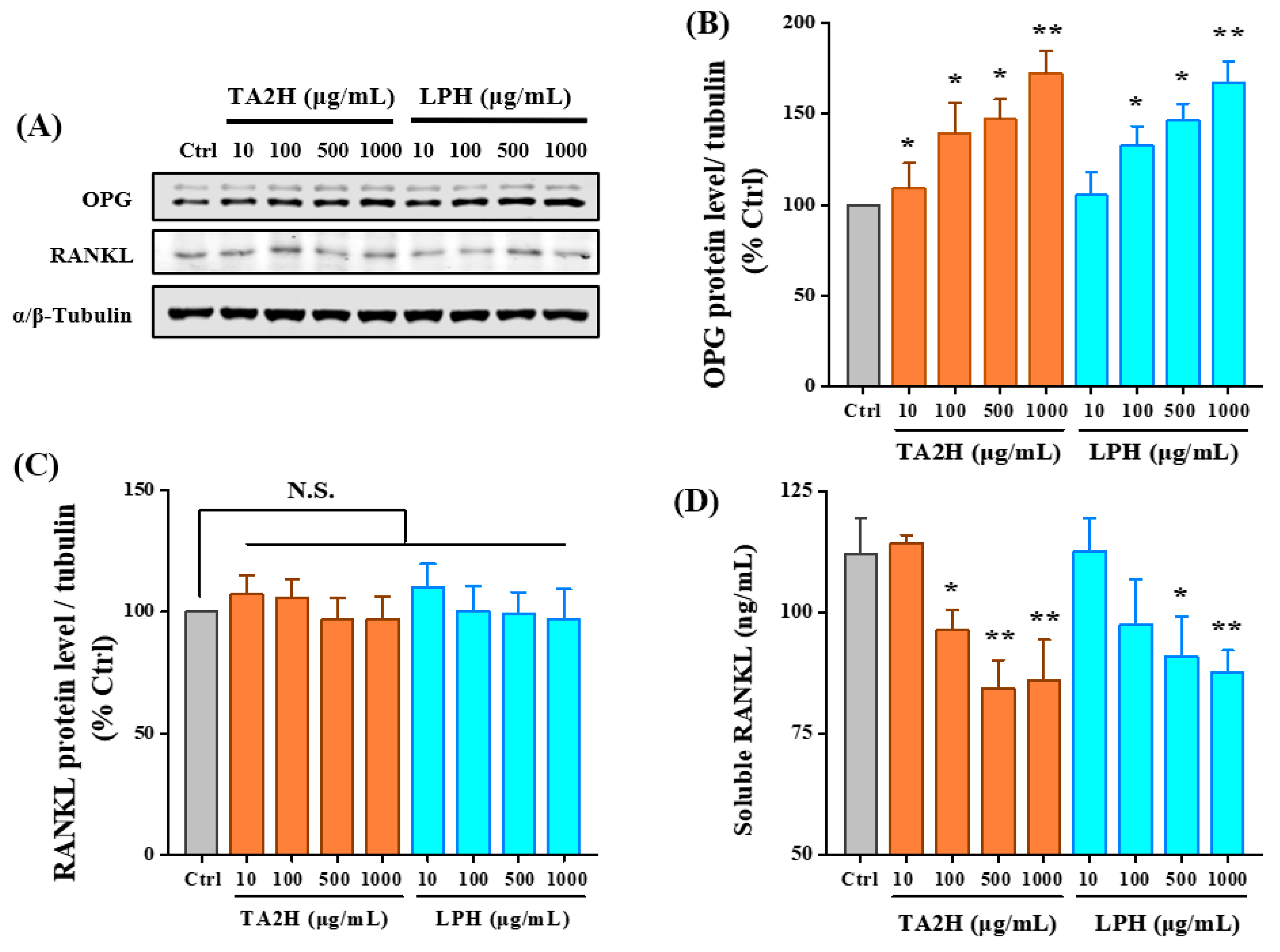
3.6. Whey Protein Hydrolysates Promote the Ratio of OPG/RANKL in Osteoblasts
3.7. LPH Activates MAPK and AKT Pathways in Osteoblasts
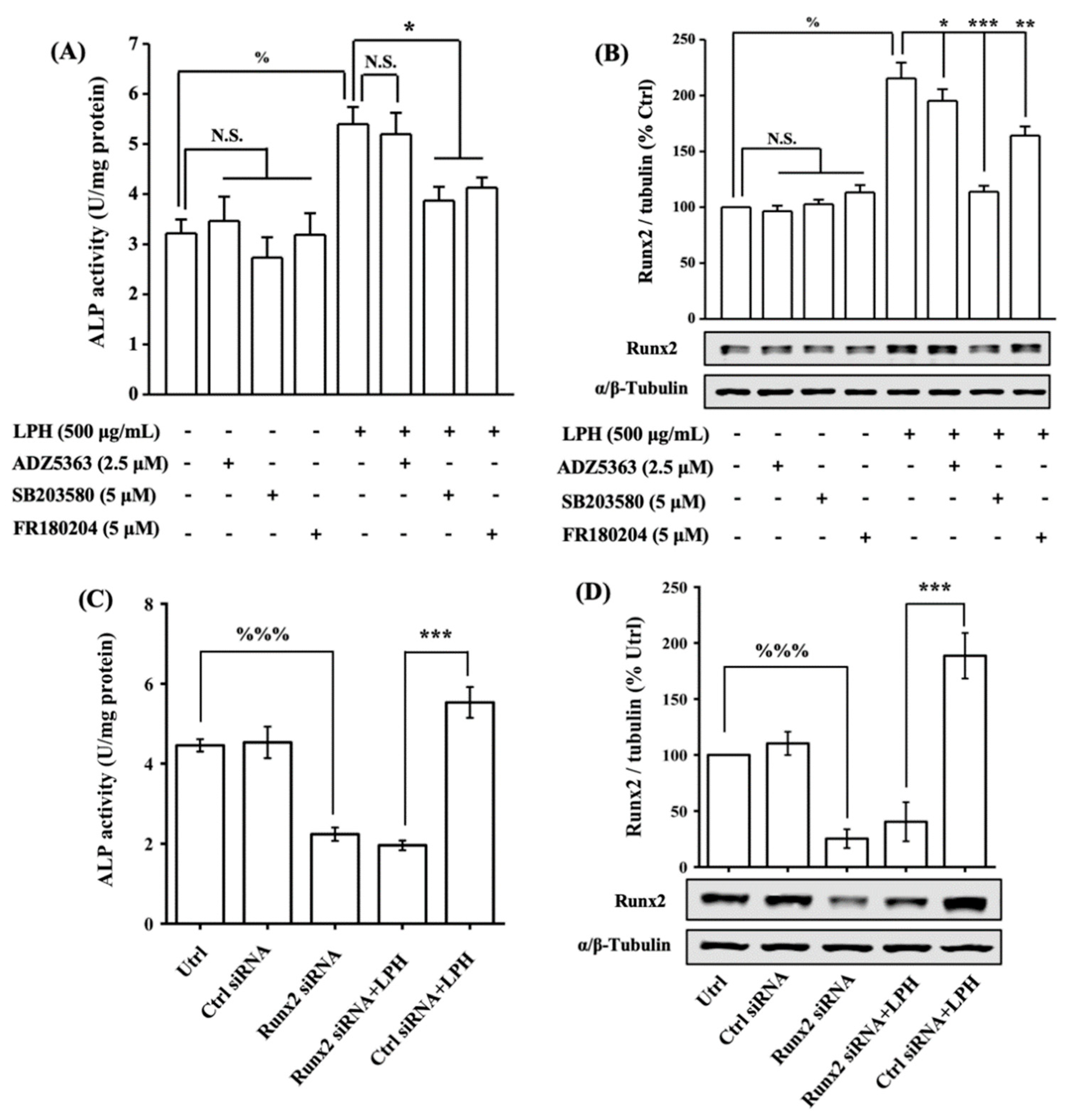
3.8. LPH Stimulates Osteoblast Differentiation through p38/Runx2 Pathway
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| BMD | Bone mineral density |
| BSP | Bone sialoprotein |
| COLI | Type-I collagen |
| ELISA | Enzyme-linked immunosorbent assay |
| ERK | Extracellular signal regulated kinase 1/2 |
| FBS | fetal bovine serum |
| JNK | Jun N-amino-terminal kinase |
| LF | Lactoferrin |
| MAPK | Mitogen-activated protein kinase |
| α-MEM | α-Minimum essential medium |
| OCN | Osteocalcin |
| OPG | Osteoprotegerin |
| OPN | Osteopontin |
| Phe | Phenylalanine |
| PI3K/Akt | Phosphatidylinositol 3-kinase/protein kinase B |
| PKU | Phenylketonuria |
| RANK | Receptor activator of nuclear factor-κB |
| RANKL | Receptor activator of nuclear factor-κB ligand |
| Runx2 | Runt-related transcription factor 2 |
| WPC | Whey protein concentrate |
Appendix A
| Amino-Acid Composition (g/100 g) | TA2H | LPH |
|---|---|---|
| Alanine | 4.32 ± 0.06 | 4.41 ± 0.07 |
| Aspartic acid | 7.70 ± 0.24 | 5.96 ± 0.12 |
| Glycine | 1.13 ± 0.06 | 0.81 ± 0.04 |
| Glutamic acid | 11.89 ± 0.51 | 6.79 ± 0.24 |
| Serine | 2.75 ± 0.04 | 2.41 ± 0.02 |
| Histidine | 2.07 ± 0.13 | 2.15 ± 0.10 |
| Arginine | 2.78 ± 0.11 | 2.50 ± 0.13 |
| Threonine | 4.86± 0.05 | 4.42 ± 0.20 |
| Proline | 5.16 ± 0.08 | 1.96 ± 0.11 |
| Tyrosine | 2.43 ± 0.03 | 0.78 ± 0.02 |
| valine | 3.86 ± 0.11 | 5.98 ± 0.23 |
| Methionine | 2.36 ± 0.05 | 1.87 ± 0.24 |
| Cystine | 0.44 ± 0.03 | 0.35 ± 0.02 |
| Isoleucine | 4.79 ± 0.22 | 5.80 ± 0.15 |
| Leucine | 7.62 ± 0.44 | 9.05 ± 0.37 |
| Phe | 2.33 ± 0.11 | 0.07 ± 0.02 |
| Protein content 1 (g/100 g) | 74.48 ± 1.05 | 63.34 ± 1.67 |
| Phe content 2 (mg/g protein equivalent) | 31.25 ± 0.67 | 1.17 ± 0.06 |
References
- Vockley, J.; Andersson, H.C.; Antshel, K.M.; Braverman, N.E.; Burton, B.K.; Frazier, D.M.; Mitchell, J.; Smith, W.E.; Thompson, B.H.; Berry, S.A.; et al. Phenylalanine hydroxylase deficiency: Diagnosis and management guideline. Genet. Med. 2014, 16, 188–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, R.; Hanley, W.; Levy, H.; Matalon, K.; Matalon, R.; Rouse, B.; Trefz, F.; Guttler, F.; Azen, C.; Platt, L.; et al. The maternal phenylketonuria international study: 1984–2002. Pediatrics 2003, 112, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Camp, K.M.; Lloyd-Puryear, M.A.; Huntington, K.L. Nutritional treatment for inborn errors of metabolism: Indications, regulations, and availability of medical foods and dietary supplements using phenylketonuria as an example. Mol. Genet. Metab. 2012, 107, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannini, M.; Verduci, E.; Salvatici, E.; Paci, S.; Riva, E. Phenylketonuria: Nutritional advances and challenges. Nutr. Metab. 2012, 9, 7. [Google Scholar] [CrossRef] [Green Version]
- Demirkol, M.; Giżewsk, M.; Giovannini, M.; Walter, J. Follow up of phenylketonuria patients. Mol. Genet. Metab. 2011, 104, S31–S39. [Google Scholar] [CrossRef]
- Hillman, L.; Schlotzhauer, C.; Lee, D.; Grasela, J.; Witter, S.; Allen, S.; Hillman, R. Decreased bone mineralization in children with phenylketonuria under treatment. Eur. J. Pediatr. 1996, 155, S148–S152. [Google Scholar] [CrossRef]
- Nagasaka, H.; Tsukahara, H.; Takatani, T.; Sanayama, Y.; Takayanagi, M.; Ohura, T.; Sakamoto, O.; Ito, T.; Wada, M.; Yoshino, M.; et al. Cross-sectional study of bone metabolism with nutrition in adult classical phenylketonuric patients diagnosed by neonatal screening. J. Bone Miner. Metab. 2011, 29, 737–743. [Google Scholar] [CrossRef]
- Wang, K.D.; Shen, M.; Li, H.L.; Li, X.W.; He, C. Reduced bone mineral density in Chinese children with phenylketonuria. J. Pediatr. Endocr. Met. 2017, 30, 651–656. [Google Scholar] [CrossRef]
- de Castro, M.J.; de Lamas, C.; Sanchez-Pintos, P.; Gonzalez-Lamuno, D.; Couce, M.L. Bone Status in Patients with Phenylketonuria: A Systematic Review. Nutrients 2020, 12, 2154. [Google Scholar] [CrossRef]
- Demirdas, S.; Coakley, K.E.; Bisschop, P.H.; Hollak, C.E.M.; Bosch, A.M.; Singh, R.H. Bone health in phenylketonuria: A systematic review and meta-analysis. Mol. Genet. Metab. 2015, 114, 17. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.E.; Ney, D. A systematic review of bone mineral density and fractures in phenylketonuria. J. Inherit. Metab. Dis. 2014, 37, 875–880. [Google Scholar] [CrossRef] [Green Version]
- Roato, I.; Porta, F.; Mussa, A.; D’Amico, L.; Fiore, L.; Garelli, D.; Spada, M.; Ferracini, R. Bone Impairment in Phenylketonuria Is Characterized by Circulating Osteoclast Precursors and Activated T Cell Increase. PLoS ONE 2010, 5, e14167. [Google Scholar] [CrossRef]
- Porta, F.; Roato, I.; Mussa, A.; Repici, M.; Gorassini, E.; Spada, M.; Ferracini, R. Increased spontaneous osteoclastogenesis from peripheral blood mononuclear cells in phenylketonuria. J. Inherit. Metab. Dis. 2008, 31, S339–S342. [Google Scholar] [CrossRef]
- Kular, J.; Tickner, J.; Chim, S.M.; Xu, J.K. An overview of the regulation of bone remodelling at the cellular level. Clin. Biochem. 2012, 45, 863–873. [Google Scholar] [CrossRef]
- Long, F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2011, 13, 27–38. [Google Scholar] [CrossRef]
- Bu, T.T.; Zheng, J.X.; Liu, L.; Li, S.S.; Wu, J.P. Milk proteins and their derived peptides on bone health: Biological functions, mechanisms, and prospects. Compr. Rev. Food Sci. F 2021, 20, 2234–2262. [Google Scholar] [CrossRef]
- Jang, J.H.; Kim, S.; Lee, H.J.; Suh, H.J.; Jo, K. Stimulating effect of whey protein hydrolysate on bone growth in MC3T3-E1 cells and a rat model. Food Funct. 2021, 12, 5109–5117. [Google Scholar] [CrossRef]
- Cornish, J.; Naot, D. Lactoferrin as an effector molecule in the skeleton. Biometals 2010, 23, 425–430. [Google Scholar] [CrossRef]
- Pandey, M.; Kapila, R.; Kapila, S. Osteoanabolic activity of whey-derived anti-oxidative (MHIRL and YVEEL) and angiotensin-converting enzyme inhibitory (YLLF, ALPMHIR, IPA and WLAHK) bioactive peptides. Peptides 2018, 99, 1–7. [Google Scholar] [CrossRef]
- Pandey, M.; Kapila, S.; Kapila, R.; Trivedi, R.; Karvande, A. Evaluation of the osteoprotective potential of whey derived- antioxidative ( YVEEL) and angiotensin- converting enzyme inhibitory ( YLLF) bioactive peptides in ovariectomised rats. Food Funct. 2018, 9, 4791–4801. [Google Scholar] [CrossRef]
- Rodriguez-Carballo, E.; Gamez, B.; Ventura, F. p38 MAPK Signaling in Osteoblast Differentiation. Front. Cell Dev. Biol. 2016, 4, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S.; Arumugam, B.; Miranda, P.J.; Selvamurugan, N. Runx2: Structure, function, and phosphorylation in osteoblast differentiation. Int. J. Biol. Macromol. 2015, 78, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Arora, H.; Shang, N.; Bhullar, K.S.; Wu, J.P. Pea protein-derived tripeptide LRW shows osteoblastic activity on MC3T3-E1 cells via the activation of the Akt/Runx2 pathway. Food Funct. 2020, 11, 7197–7207. [Google Scholar] [CrossRef]
- Zhang, W.; Guo, H.Y.; Jing, H.; Li, Y.X.; Wang, X.Y.; Zhang, H.; Jiang, L.; Ren, F.Z. Lactoferrin Stimulates Osteoblast Differentiation Through PKA and p38 Pathways Independent of Lactoferrin’s Receptor LRP1. J. Bone Miner. Res. 2014, 29, 1232–1243. [Google Scholar] [CrossRef] [Green Version]
- Bu, T.T.; Zhou, M.J.; Zheng, J.X.; Yang, P.; Song, H.L.; Li, S.S.; Wu, J.P. Preparation and characterization of a low-phenylalanine whey hydrolysate using two-step enzymatic hydrolysis and macroporous resin adsorption. Lwt-Food Sci. Technol. 2020, 132, 109753. [Google Scholar] [CrossRef]
- Bu, T.T.; Zhang, L.; Liu, L.; Yu, S.F.; Zheng, J.X.; Wu, J.P.; Yang, K. Evaluation of the anti-osteoporotic effect of a low-phenylalanine whey protein hydrolysate in an ovariectomized mice model. Food Funct. 2022, 13, 3957–3967. [Google Scholar] [CrossRef]
- Grey, A.; Banovic, T.; Zhu, Q.; Watson, M.; Callon, K.; Palmano, K.; Ross, J.; Naot, D.; Reid, I.R.; Cornish, J. The low-density lipoprotein receptor-related protein 1 is a mitogenic receptor for lactoferrin in osteoblastic cells. Mol. Endocrinol. 2004, 18, 2268–2278. [Google Scholar] [CrossRef] [Green Version]
- Shang, N.; Wu, J.P. Egg White Ovotransferrin Shows Osteogenic Activity in Osteoblast Cells. J. Agric. Food Chem. 2018, 66, 2775–2782. [Google Scholar] [CrossRef]
- Zhang, J.L.; Han, X.; Shan, Y.J.; Zhang, L.W.; Du, M.; Liu, M.; Yi, H.X.; Ma, Y. Effect of bovine lactoferrin and human lactoferrin on the proliferative activity of the osteoblast cell line MC3T3-E1 in vitro. J. Dairy Sci. 2018, 101, 1827–1833. [Google Scholar] [CrossRef] [Green Version]
- Takarada, T.; Hinoi, E.; Nakazato, R.; Ochi, H.; Xu, C.; Tsuchikane, A.; Takeda, S.; Karsenty, G.; Abe, T.; Kiyonari, H.; et al. An Analysis of Skeletal Development in Osteoblast-Specific and Chondrocyte-Specific Runt- Related Transcription Factor-2 ( Runx2) Knockout Mice. J. Bone Miner. Res. 2013, 28, 2064–2069. [Google Scholar] [CrossRef]
- Chen, H.Y.; Ghori-Javed, F.Y.; Rashid, H.; Adhami, M.D.; Serra, R.; Gutierrez, S.E.; Javed, A. Runx2 Regulates Endochondral Ossification Through Control of Chondrocyte Proliferation and Differentiation. J. Bone Miner. Res. 2014, 29, 2653–2665. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.Q.; Deng, C.X.; Li, Y.P. TGF-beta and BMP Signaling in Osteoblast Differentiation and Bone Formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.C.; Lee, Y.H. Phosphorylation, acetylation and ubiquitination: The molecular basis of RUNX regulation. Gene 2006, 366, 58–66. [Google Scholar] [CrossRef]
- Xiao, G.Z.; Jiang, D.; Gopalakrishnan, R.; Franceschi, R.T. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J. Biol. Chem. 2002, 277, 36181–36187. [Google Scholar] [CrossRef] [Green Version]
- Wee, H.J.; Haung, G.; Shigesada, K.; Ito, Y. Serine phosphorylation of RUNX2 with novel potential functions as negative regulatory mechanisms. Embo Rep. 2002, 3, 967–974. [Google Scholar] [CrossRef] [Green Version]
- Pande, S.; Browne, G.; Padmanabhan, S.; Zaidi, S.K.; Lian, J.B.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J. Cell Physiol. 2013, 228, 1784–1792. [Google Scholar] [CrossRef] [Green Version]
- Greenblatt, M.B.; Shim, J.H.; Zou, W.G.; Sitara, D.; Schweitzer, M.; Hu, D.; Lotinun, S.; Sano, Y.; Baron, R.; Park, J.M.; et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J. Clin. Investig. 2010, 120, 2457–2473. [Google Scholar] [CrossRef] [Green Version]
- Ulsamer, A.; Ortuno, M.J.; Ruiz, S.; Susperregui, A.R.G.; Osses, N.; Rosa, J.L.; Ventura, F. BMP-2 induces osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J. Biol. Chem. 2008, 283, 3816–3826. [Google Scholar] [CrossRef] [Green Version]
- Ortuno, M.J.; Ruiz-Gaspa, S.; Rodriguez-Carballo, E.; Susperregui, A.R.G.; Bartrons, R.; Rosa, J.L.; Ventura, F. p38 Regulates Expression of Osteoblast-specific Genes by Phosphorylation of Osterix. J. Biol. Chem. 2010, 285, 31985–31994. [Google Scholar] [CrossRef] [Green Version]
- Artigas, N.; Urena, C.; Rodriguez-Carballo, E.; Rosa, J.L.; Ventura, F. Mitogen-activated Protein Kinase (MAPK)-regulated Interactions between Osterix and Runx2 Are Critical for the Transcriptional Osteogenic Program. J. Biol. Chem. 2014, 289, 27105–27117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, A.; Guicheux, J.; Palmer, G.; Miura, Y.; Oiso, Y.; Bonjour, J.P.; Caverzasio, J. Evidence for a role of p38 MAP kinase in expression of alkaline phosphatase during osteoblastic cell differentiation. Bone 2002, 30, 91–98. [Google Scholar] [CrossRef]
- Reddi, S.; Kumar, N.; Vij, R.; Mada, S.B.; Kapila, S.; Kapila, R. Akt drives buffalo casein-derived novel peptide-mediated osteoblast differentiation. J. Nutr. Biochem. 2016, 38, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Hikita, A.; Yana, I.; Wakeyama, H.; Nakamura, M.; Kadono, Y.; Oshima, Y.; Nakamura, K.; Seiki, M.; Tanaka, S. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappa B ligand. J. Biol. Chem. 2006, 281, 36846–36855. [Google Scholar] [CrossRef] [Green Version]
- Proell, V.; Xu, H.Q.; Schuler, C.; Weber, K.; Hofbauer, L.C.; Erben, R.G. Orchiectomy upregulates free soluble RANKL in bone marrow of aged rats. Bone 2009, 45, 677–681. [Google Scholar] [CrossRef]



| Gene Name | Primer Sequence (5′ to 3′) |
|---|---|
| Runx2 | Forward: CCTTCAAGGTTGTAGCCCTC |
| Reverse: GGAGTAGTTCTCATCATTCC | |
| COLI | Forward: CAAGATGTGCCACTCTGACT |
| Reverse: TCTGACCTGTCTCCATGTTG | |
| OCN | Forward: AGACTCCGGCGCTACCTTGG |
| Reverse: CGGTCTTCAAGCCATACTGG | |
| OPN | Forward: TCAGGCATGTCCCTCGGTAT |
| Reverse: TGGCAGGTAGGTATGGTAGT | |
| β-Actin | Forward: TTGCTGACAGGATGCAGAAG |
| Reverse: ACATCTGCTGGAAGGTGGAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, T.; Ren, Y.; Yu, S.; Zheng, J.; Liu, L.; Sun, P.; Wu, J.; Yang, K. A Low-Phenylalanine-Containing Whey Protein Hydrolysate Stimulates Osteogenic Activity through the Activation of p38/Runx2 Signaling in Osteoblast Cells. Nutrients 2022, 14, 3135. https://doi.org/10.3390/nu14153135
Bu T, Ren Y, Yu S, Zheng J, Liu L, Sun P, Wu J, Yang K. A Low-Phenylalanine-Containing Whey Protein Hydrolysate Stimulates Osteogenic Activity through the Activation of p38/Runx2 Signaling in Osteoblast Cells. Nutrients. 2022; 14(15):3135. https://doi.org/10.3390/nu14153135
Chicago/Turabian StyleBu, Tingting, Yuting Ren, Songfeng Yu, Jiexia Zheng, Ling Liu, Peilong Sun, Jianping Wu, and Kai Yang. 2022. "A Low-Phenylalanine-Containing Whey Protein Hydrolysate Stimulates Osteogenic Activity through the Activation of p38/Runx2 Signaling in Osteoblast Cells" Nutrients 14, no. 15: 3135. https://doi.org/10.3390/nu14153135
APA StyleBu, T., Ren, Y., Yu, S., Zheng, J., Liu, L., Sun, P., Wu, J., & Yang, K. (2022). A Low-Phenylalanine-Containing Whey Protein Hydrolysate Stimulates Osteogenic Activity through the Activation of p38/Runx2 Signaling in Osteoblast Cells. Nutrients, 14(15), 3135. https://doi.org/10.3390/nu14153135