The Effects of Fatty Acids on Inflammatory Bowel Disease: A Two-Sample Mendelian Randomization Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source and Open-GWAS Statistics
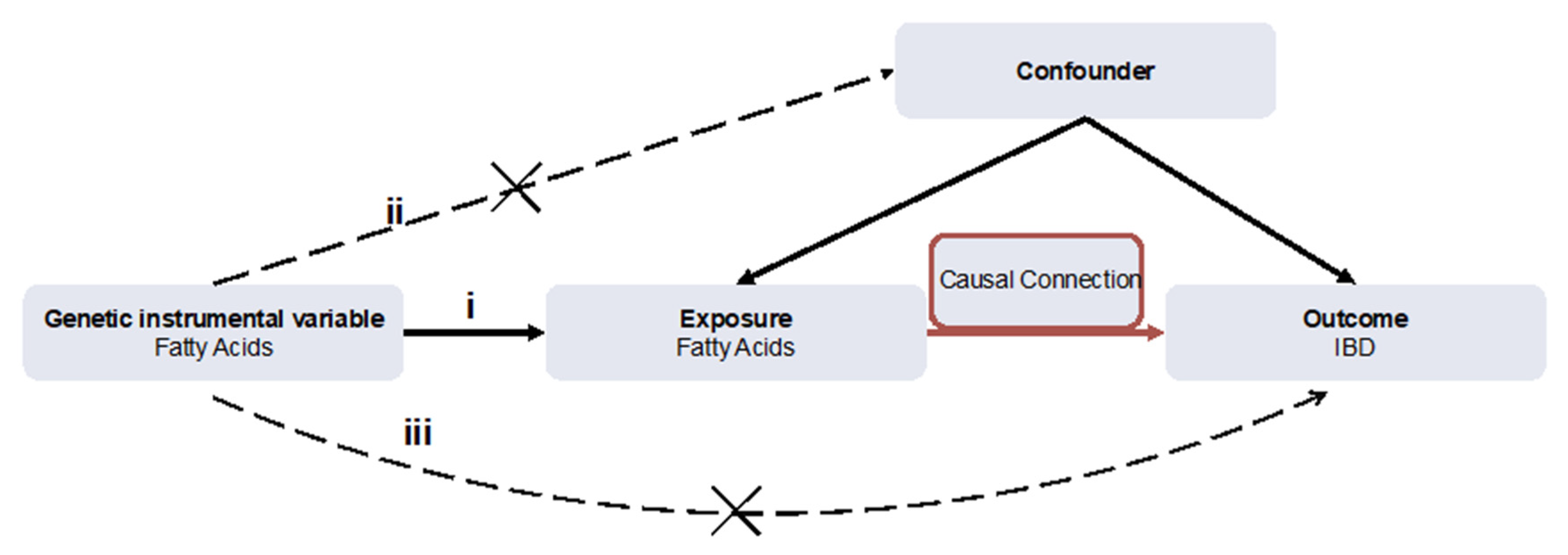
2.2. SNPs Selection and Assumption
2.3. Statistical Analysis of Primary MR
2.4. Supplementary and Sensitivity Analysis
3. Results
3.1. IIBDGC GWASs of FAs
3.2. Primary MR Analysis
3.3. Supplementary and Sensitivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torres, J.; Mehandru, S.; Colombel, J.F.; Peyrin-Biroulet, L. Crohn’s disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.F. Ulcerative colitis. Lancet 2017, 389, 1756–1770. [Google Scholar] [CrossRef]
- Gajendran, M.; Loganathan, P.; Catinella, A.P.; Hashash, J.G. A comprehensive review and update on Crohn’s disease. Disease-A-Month 2018, 64, 20–57. [Google Scholar] [CrossRef] [PubMed]
- Wawrzyniak, P.; Noureddine, N.; Wawrzyniak, M.; Lucchinetti, E.; Krämer, S.D.; Rogler, G.; Zaugg, M.; Hersberger, M. Nutritional Lipids and Mucosal Inflammation. Mol. Nutr. Food Res. 2021, 65, e1901269. [Google Scholar] [CrossRef]
- Costea, I.; Mack, D.R.; Lemaitre, R.N.; Israel, D.; Marcil, V.; Ahmad, A.; Amre, D.K. Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn’s disease. Gastroenterology 2014, 146, 929–931. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.K.; Abraham, B.; El-Serag, H. Dietary intake and risk of developing inflammatory bowel disease: A systematic review of the literature. Am. J. Gastroenterol. 2011, 106, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Abreu, M.T. Diet as a Trigger or Therapy for Inflammatory Bowel Diseases. Gastroenterology 2017, 152, 398–414.e6. [Google Scholar] [CrossRef]
- Scoville, E.A.; Allaman, M.M.; Adams, D.W.; Motley, A.K.; Peyton, S.C.; Ferguson, S.L.; Horst, S.N.; Williams, C.S.; Beaulieu, D.B.; Schwartz, D.A.; et al. Serum Polyunsaturated Fatty Acids Correlate with Serum Cytokines and Clinical Disease Activity in Crohn’s Disease. Sci. Rep. 2019, 9, 2882. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Khalili, H.; Konijeti, G.G.; Higuchi, L.M.; de Silva, P.; Fuchs, C.S.; Willett, W.C.; Richter, J.M.; Chan, A.T. Long-term intake of dietary fat and risk of ulcerative colitis and Crohn’s disease. Gut 2014, 63, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.; Thompson, S.G. Mendelian Randomization: Methods for Causal Inference Using Genetic Variants; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Zhuang, Z.; Gao, M.; Yang, R.; Liu, Z.; Cao, W.; Huang, T. Causal relationships between gut metabolites and Alzheimer’s disease: A bidirectional Mendelian randomization study. Neurobiol. Aging 2021, 100, 119.e115–119.e118. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sazonovs, A.; Stevens, C.R.; Venkataraman, G.R.; Yuan, K.; Avila, B.; Abreu, M.T.; Ahmad, T.; Allez, M.; Atzmon, G.; Baras, A. Sequencing of over 100,000 individuals identifies multiple genes and rare variants associated with Crohns disease susceptibility. medRxiv 2021. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef]
- Wooldridge, J. Instrumental variables estimation and two stage least squares. In Introductory Econometrics: A Modern Approach; South-Western: Nashville, TN, USA, 2009. [Google Scholar]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milligan, B.G. Maximum-likelihood estimation of relatedness. Genetics 2003, 163, 1153–1167. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Bowden, J.; Haycock, P.; Zheng, J.; Davis, O.; Flach, P.; Gaunt, T.; Smith, G.D. Automating Mendelian randomization through machine learning to construct a putative causal map of the human phenome. bioRxiv 2017, 173682. [Google Scholar] [CrossRef] [Green Version]
- Burgess, S.; Bowden, J.; Dudbridge, F.; Thompson, S.G. Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. arXiv 2016, arXiv:1606.03729. [Google Scholar]
- Sakamoto, N.; Kono, S.; Wakai, K.; Fukuda, Y.; Satomi, M.; Shimoyama, T.; Inaba, Y.; Miyake, Y.; Sasaki, S.; Okamoto, K.; et al. Dietary risk factors for inflammatory bowel disease: A multicenter case-control study in Japan. Inflamm. Bowel Dis. 2005, 11, 154–163. [Google Scholar] [CrossRef]
- Piotrowska, M.; Binienda, A.; Fichna, J. The role of fatty acids in Crohn’s disease pathophysiology—An overview. Mol. Cell. Endocrinol. 2021, 538, 111448. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.; Cook, N.R.; Alexander, E.K.; Friedman, S.; Walter, J.; Bubes, V.; Kotler, G.; Lee, I.M.; Manson, J.E.; Costenbader, K.H. Vitamin D and marine omega 3 fatty acid supplementation and incident autoimmune disease: VITAL randomized controlled trial. BMJ 2022, 376, e066452. [Google Scholar] [CrossRef]
- Luu, M.; Pautz, S.; Kohl, V.; Singh, R.; Romero, R.; Lucas, S.; Hofmann, J.; Raifer, H.; Vachharajani, N.; Carrascosa, L.C.; et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 2019, 10, 760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cluxton, D.; Petrasca, A.; Moran, B.; Fletcher, J.M. Differential Regulation of Human Treg and Th17 Cells by Fatty Acid Synthesis and Glycolysis. Front. Immunol. 2019, 10, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zárate, R.; El Jaber-Vazdekis, N.; Tejera, N.; Pérez, J.A.; Rodríguez, C. Significance of long chain polyunsaturated fatty acids in human health. Clin. Transl. Med. 2017, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochim. Biophys. Acta 2015, 1851, 469–484. [Google Scholar] [CrossRef]
- Faber, J.; Berkhout, M.; Vos, A.P.; Sijben, J.W.; Calder, P.C.; Garssen, J.; van Helvoort, A. Supplementation with a fish oil-enriched, high-protein medical food leads to rapid incorporation of EPA into white blood cells and modulates immune responses within one week in healthy men and women. J. Nutr. 2011, 141, 964–970. [Google Scholar] [CrossRef] [Green Version]
- Browning, L.M.; Walker, C.G.; Mander, A.P.; West, A.L.; Madden, J.; Gambell, J.M.; Young, S.; Wang, L.; Jebb, S.A.; Calder, P.C. Incorporation of eicosapentaenoic and docosahexaenoic acids into lipid pools when given as supplements providing doses equivalent to typical intakes of oily fish. Am. J. Clin. Nutr. 2012, 96, 748–758. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Yen, J.H.; Vassiliou, E.; Adhikary, S.; Toscano, M.G.; Ganea, D. Docosahexaenoic acid prevents dendritic cell maturation and in vitro and in vivo expression of the IL-12 cytokine family. Lipids Health Dis. 2010, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.W.; Kwon, M.J.; Choi, A.M.; Kim, H.P.; Nakahira, K.; Hwang, D.H. Fatty acids modulate Toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J. Biol. Chem. 2009, 284, 27384–27392. [Google Scholar] [CrossRef] [Green Version]
- Szanto, A.; Nagy, L. The many faces of PPARgamma: Anti-inflammatory by any means? Immunobiology 2008, 213, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Smith, G.D.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Holmes, M.V.; Minelli, C.; Relton, C.L. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019, 4, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Exposures | GWAS ID | Consortium | Sample Size | No. of Strongly Related SNPs | Adjustment | Population |
|---|---|---|---|---|---|---|
| Total FA | met-d-Total_FA | UK biobank | 114,999 | 64 | Depression, Worrier, Cholesterol, Smoking, Contraceptives and sclerosing cholangitis | European |
| Saturated FA | met-d-SFA | UK biobank | 114,999 | 55 | ||
| Monounsaturated FA | met-d-MUFA | UK biobank | 114,999 | 66 | ||
| Polyunsaturated FA | met-d-PUFA | UK biobank | 114,999 | 66 | ||
| Omega-3 FA | met-d-Omega_3 | UK biobank | 114,999 | 52 | ||
| Omega-6 FA | met-d-Omega_6 | UK biobank | 114,999 | 63 | ||
| Outcomes | GWAS ID | Consortium | Cases | Control | Population | |
| Crohn’s disease | ieu-a-30 | IIBDGC | 5956 | 14,927 | European | |
| Ulcerative colitis | ieu-a-32 | IIBDGC | 6968 | 20,464 | ||
| Fat Acids (Exposure) | MR Method | No. SNP | β | SE | OR(95%CI) | p |
|---|---|---|---|---|---|---|
| total FA | MR Egger | 14 | 0.769 | 0.595 | 2.158 (0.672, 6.926) | 0.220 |
| Weighted median | −0.053 | 0.316 | 0.948 (0.510, 1.762) | 0.867 | ||
| IVW(fixed effects) | −0.366 | 0.232 | 0.694 (0.441, 1.092) | 0.114 | ||
| Maximum likelihood | −0.359 | 0.235 | 0.697 (0.440, 1.108) | 0.128 | ||
| Penalised weighted median | 0.021 | 0.315 | 1.020 (0.550, 1.896) | 0.948 | ||
| saturated FA | MR Egger | 23 | −0.097 | 0.394 | 0.907 (0.419, 1.964) | 0.807 |
| Weighted median | −0.226 | 0.246 | 0.797 (0.491, 1.295) | 0.356 | ||
| IVW(random effects) | −0.255 | 0.217 | 0.775 (0.507, 1.184) | 0.238 | ||
| Maximum likelihood | −0.257 | 0.143 | 0.774 (0.585, 1.024) | 0.072 | ||
| Penalised weighted median | −0.104 | 0.261 | 0.901 (0.541, 1.501) | 0.689 | ||
| polyunsaturated FA | MR Egger | 19 | 0.430 | 0.355 | 1.537 (0.766, 3.085) | 0.243 |
| Weighted median | −0.039 | 0.231 | 0.961 (0.612, 1.510) | 0.864 | ||
| IVW(fixed effects) | 0.093 | 0.162 | 1.097 (0.799, 1.506) | 0.567 | ||
| Maximum likelihood | 0.094 | 0.163 | 1.098 (0.798, 1.512) | 0.565 | ||
| Penalised weighted median | −0.067 | 0.234 | 0.935 (0.591, 1.480) | 0.775 | ||
| monounsaturated FA | MR Egger | 19 | 0.641 | 0.584 | 1.898 (0.604, 5.970) | 0.288 |
| Weighted median | 0.578 | 0.263 | 1.784 (1.066, 2.987) | 0.027 | ||
| IVW(random effects) | 0.112 | 0.308 | 1.118 (0.612, 2.043) | 0.716 | ||
| Maximum likelihood | 0.118 | 0.169 | 1.125 (0.808, 1.568) | 0.485 | ||
| Penalised weighted median | 0.815 | 0.264 | 2.259 (1.346, 3.793) | 0.71 | ||
| Omega-3 FA | MR Egger | 21 | 0.188 | 0.483 | 1.207 (0.468, 3.111) | 0.701 |
| Weighted median | −0.161 | 0.281 | 0.851 (0.491, 1.476) | 0.566 | ||
| IVW(random effects) | 0.0675 | 0.238 | 1.070 (0.671, 1.705) | 0.776 | ||
| Maximum likelihood | 0.069 | 0.150 | 1.071 (0.798, 1.436) | 0.648 | ||
| Penalised weighted median | −0.457 | 0.270 | 0.633 (0.373, 1.075) | 0.0905 | ||
| Omega-6 FA | MR Egger | 14 | 0.213 | 0.388 | 1.237 (0.578, 2.648) | 0.594 |
| Weighted median | −0.011 | 0.265 | 0.989 (0.588, 1.662) | 0.966 | ||
| IVW(fixed effects) | 0.103 | 0.186 | 1.108 (0.770, 1.595) | 0.580 | ||
| Maximum likelihood | 0.107 | 0.187 | 1.112 (0.771, 1.605) | 0.569 | ||
| Penalised weighted median | −0.076 | 0.245 | 0.927 (0.574, 1.499) | 0.758 |
| Fat Acids (Exposure) | MR Method | No. SNP | β | SE | OR(95%CI) | p |
|---|---|---|---|---|---|---|
| total FA | MR Egger | 14 | −0.051 | 0.602 | 0.951 (0.292, 3.091) | 0.934 |
| Weighted median | −0.196 | 0.294 | 0.822 (0.462, 1.462) | 0.505 | ||
| IVW(fixed effects) | −0.353 | 0.215 | 0.703 (0.461, 1.071) | 0.101 | ||
| Maximum likelihood | −0.355 | 0.218 | 0.701 (0.457, 1.075) | 0.103 | ||
| Penalised weighted median | −0.189 | 0.306 | 0.827 (0.455, 1.506) | 0.535 | ||
| saturated FA | MR Egger | 23 | −0.105 | 0.339 | 1.111 (0.572, 2.157) | 0.759 |
| Weighted median | 0.108 | 0.200 | 1.114 (0.758, 1.637) | 0.590 | ||
| IVW(random effects) | −0.236 | 0.191 | 0.790 (0.544, 1.148) | 0.216 | ||
| Maximum likelihood | −0.234 | 0.132 | 0.791 (0.611, 1.025) | 0.076 | ||
| Penalised weighted median | 0.109 | 0.192 | 1.115 (0.749, 1.659) | 0.517 | ||
| polyunsaturated FA | MR Egger | 19 | −0.589 | 0.472 | 0.555 (0.220, 1.401) | 0.230 |
| Weighted median | −0.073 | 0.240 | 0.930 (0.581, 1.488) | 0.761 | ||
| IVW(random effects) | −0.193 | 0.234 | 0.825 (0.521, 1.305) | 0.411 | ||
| Maximum likelihood | −0.194 | 0.151 | 0.823 (0.613, 1.107) | 0.199 | ||
| Penalised weighted median | −0.017 | 0.242 | 0.983 (0.611, 1.582) | 0.944 | ||
| monounsaturated FA | MR Egger | 19 | −0.091 | 0.425 | 0.913 (0.397, 2.098) | 0.832 |
| Weighted median | 0.088 | 0.232 | 1.092 (0.692, 1.723) | 0.705 | ||
| IVW(random effects) | −0.045 | 0.216 | 0.956 (0.626, 1.460) | 0.835 | ||
| Maximum likelihood | −0.046 | 0.156 | 0.955 (0.703, 1.295) | 0.765 | ||
| Penalised weighted median | 0.134 | 0.239 | 1.144 (0.717, 1.826) | 0.573 | ||
| Omega-3 FA | MR Egger | 21 | −0.863 | 0.307 | 0.422 (0.231, 0.770) | 0.0111 |
| Weighted median | −0.723 | 0.215 | 0.485 (0.318, 0.740) | 7.706 × 10−4 | ||
| IVW(fixed effects) | −0.509 | 0.136 | 0.601 (0.461, 0.784) | 1.766 × 10−4 | ||
| Maximum likelihood | −0.493 | 0.137 | 0.610 (0.466, 0.799) | 3.262 × 10−4 | ||
| Penalised weighted median | −0.787 | 0.209 | 0.455 (0.302, 0.685) | 1.628 × 10−4 | ||
| Omega-6 FA | MR Egger | 14 | −0.874 | 0.456 | 0.417 (0.171, 1.021) | 0.079 |
| Weighted median | −0.303 | 0.249 | 0.738 (0.454, 1.202) | 0.223 | ||
| IVW(random effects) | −0.285 | 0.246 | 0.752 (0.464, 1.217) | 0.246 | ||
| Maximum likelihood | −0.278 | 0.173 | 0.757 (0.539, 1.063) | 0.108 | ||
| Penalised weighted median | −0.280 | 0.263 | 0.756 (0.451, 1.265) | 0.286 |
| Traits (Outcome) | Fat Acids (Exposure) | Methods | Q | Q-dif | p |
|---|---|---|---|---|---|
| CD | total FA | MR Egger | 13.249 | 12 | 0.351 |
| IVW | 18.072 | 13 | 0.155 | ||
| saturated FA | MR Egger | 51.622 | 21 | 2.160 × 10−4 | |
| IVW | 52.196 | 22 | 2.937 × 10−3 | ||
| polyunsaturated FA | MR Egger | 20.667 | 17 | 0.242 | |
| IVW | 22.130 | 18 | 0.226 | ||
| monounsaturated FA | MR Egger | 58.748 | 17 | 1.688 × 10−6 | |
| IVW | 62.658 | 18 | 7.558 × 10−7 | ||
| Omega-3 FA | MR Egger | 51.832 | 19 | 7.012 × 10−5 | |
| IVW | 52.060 | 20 | 1.116 × 10−4 | ||
| Omega-6 FA | MR Egger | 14.321 | 12 | 0.281 | |
| IVW | 14.452 | 13 | 0.343 | ||
| UC | total FA | MR Egger | 15.198 | 12 | 0.231 |
| IVW | 15.579 | 13 | 0.273 | ||
| saturated FA | MR Egger | 44.057 | 21 | 2.299 × 10−3 | |
| IVW | 47.138 | 22 | 1.408 × 10−3 | ||
| polyunsaturated FA | MR Egger | 42.533 | 17 | 5.614 × 10−4 | |
| IVW | 44.869 | 18 | 4.328 × 10−4 | ||
| monounsaturated FA | MR Egger | 35.630 | 17 | 5.13 × 10−3 | |
| IVW | 35.670 | 18 | 7.78 × 10−3 | ||
| Omega-3 FA | MR Egger | 24.271 | 19 | 0.186 | |
| IVW | 26.541 | 20 | 0.149 | ||
| Omega-6 FA | MR Egger | 22.678 | 12 | 0.031 | |
| IVW | 26.945 | 13 | 0.013 |
| Traits (Outcome) | Fat Acids (Exposure) | Egger_Intercept | se | p |
|---|---|---|---|---|
| CD | total FA | −0.041 | 0.020 | 0.059 |
| saturated FA | −8.271 × 103 | 0.017 | 0.634 | |
| polyunsaturated FA | −0.018 | 0.016 | 0.288 | |
| monounsaturated FA | −0.026 | 0.025 | 0.302 | |
| Omega-3 FA | −6.115 × 103 | 0.021 | 0.775 | |
| Omega-6 FA | −5.612 × 103 | 0.017 | 0.746 | |
| UC | total FA | −0.011 | 0.020 | 0.594 |
| saturated FA | −0.018 | 0.015 | 0.239 | |
| polyunsaturated FA | 0.020 | 0.021 | 0.347 | |
| monounsaturated FA | 2.28 × 103 | 0.018 | 0.899 | |
| Omega-3 FA | 0.018 | 0.013 | 0.198 | |
| Omega-6 FA | 0.030 | 0.020 | 0.159 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Luo, X.; Xin, H.; Lai, Q.; Zhou, Y.; Bai, Y. The Effects of Fatty Acids on Inflammatory Bowel Disease: A Two-Sample Mendelian Randomization Study. Nutrients 2022, 14, 2883. https://doi.org/10.3390/nu14142883
He J, Luo X, Xin H, Lai Q, Zhou Y, Bai Y. The Effects of Fatty Acids on Inflammatory Bowel Disease: A Two-Sample Mendelian Randomization Study. Nutrients. 2022; 14(14):2883. https://doi.org/10.3390/nu14142883
Chicago/Turabian StyleHe, Jian, Xiaobei Luo, Hongjie Xin, Qianwei Lai, Yuanping Zhou, and Yang Bai. 2022. "The Effects of Fatty Acids on Inflammatory Bowel Disease: A Two-Sample Mendelian Randomization Study" Nutrients 14, no. 14: 2883. https://doi.org/10.3390/nu14142883
APA StyleHe, J., Luo, X., Xin, H., Lai, Q., Zhou, Y., & Bai, Y. (2022). The Effects of Fatty Acids on Inflammatory Bowel Disease: A Two-Sample Mendelian Randomization Study. Nutrients, 14(14), 2883. https://doi.org/10.3390/nu14142883






