Valproate and Short-Chain Fatty Acids Activate Transcription of the Human Vitamin D Receptor Gene through a Proximal GC-Rich DNA Region Containing Two Putative Sp1 Binding Sites
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment of HepG2 Cells
2.2. RNA Extraction and Expression Analysis
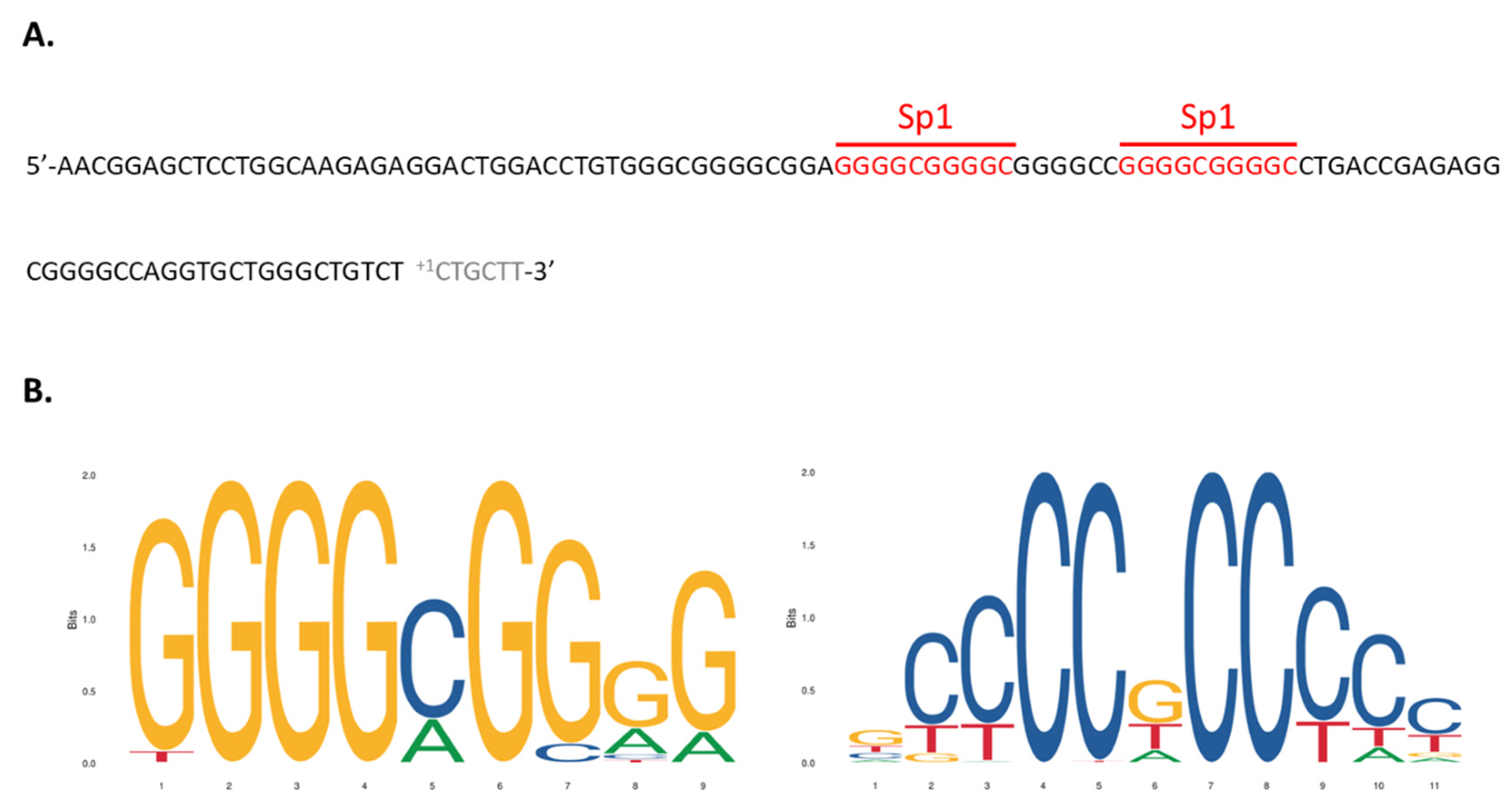
2.3. PCR Amplification of Two Promoter Regions of the hVDR Gene
2.4. Construction of Luciferase Reporter Plasmids and Transient Transfections
2.5. Statistics
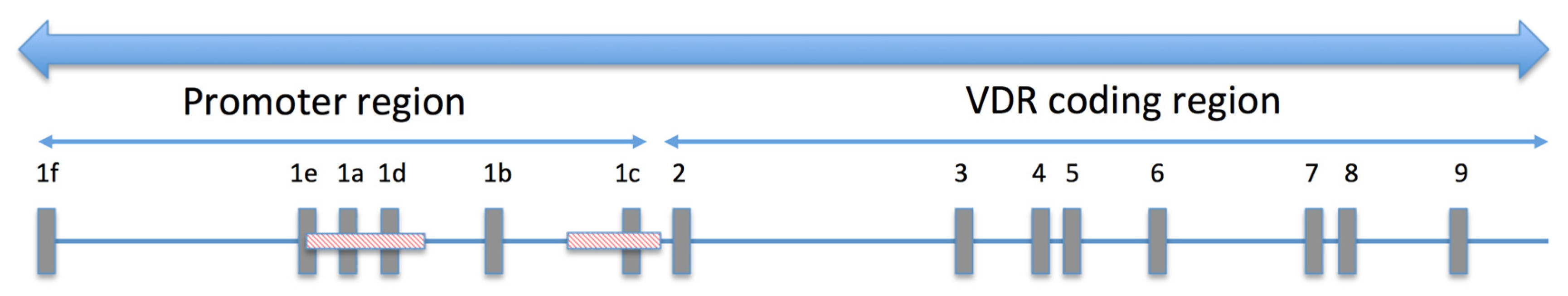
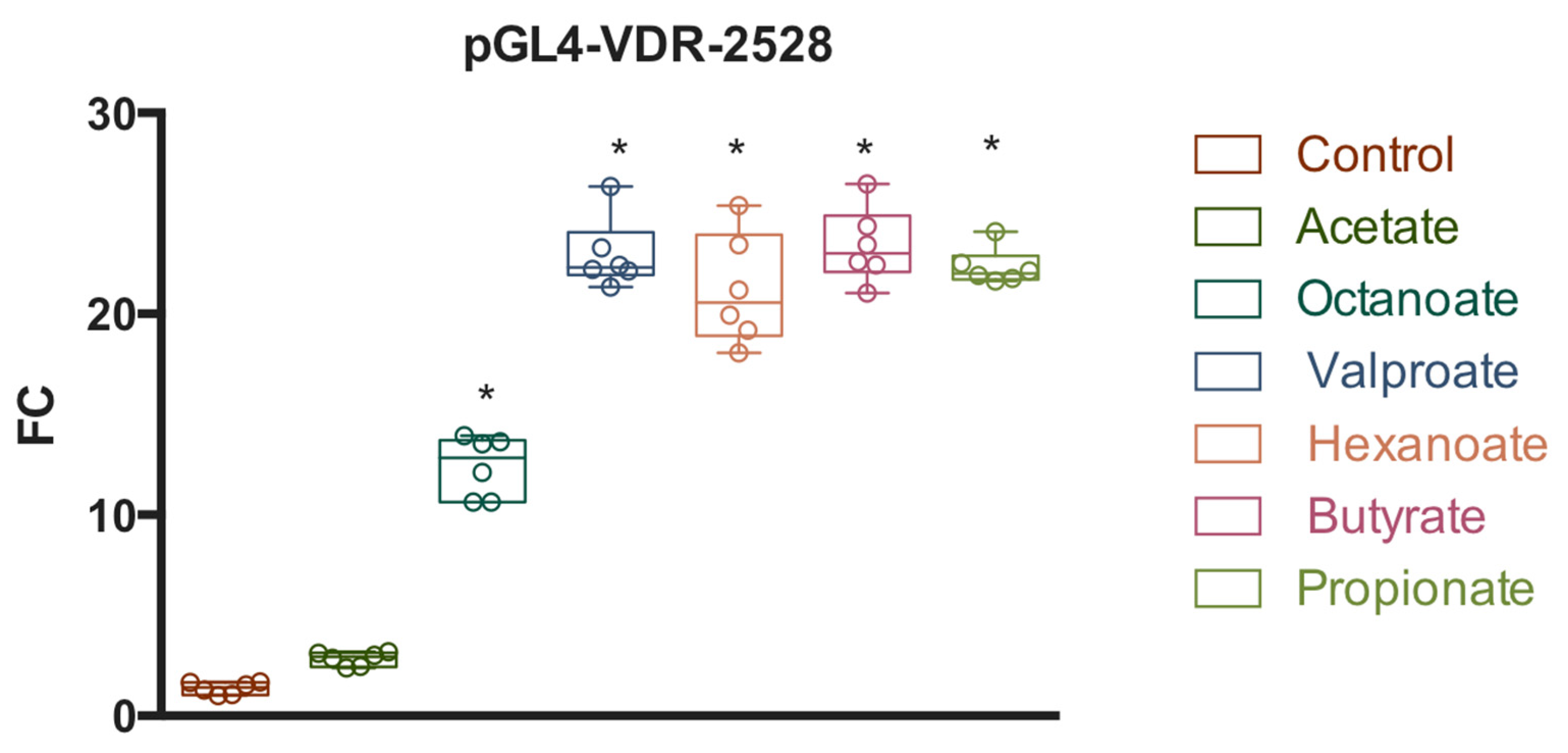
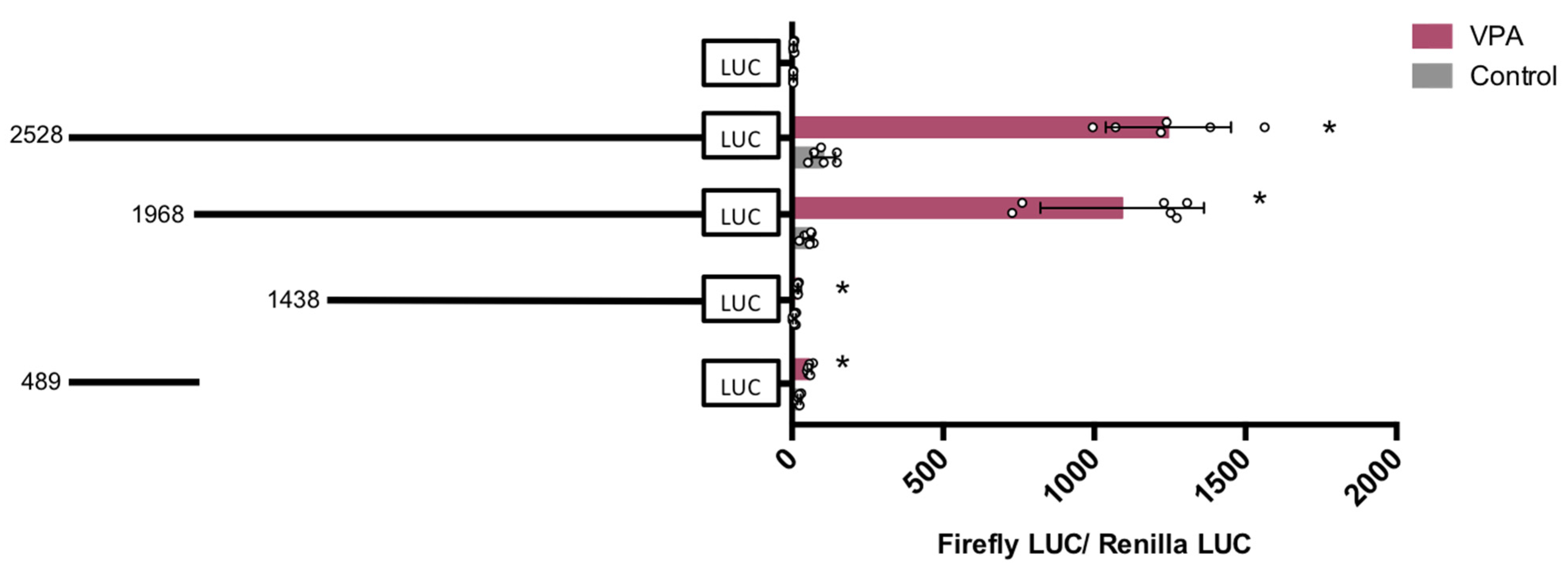
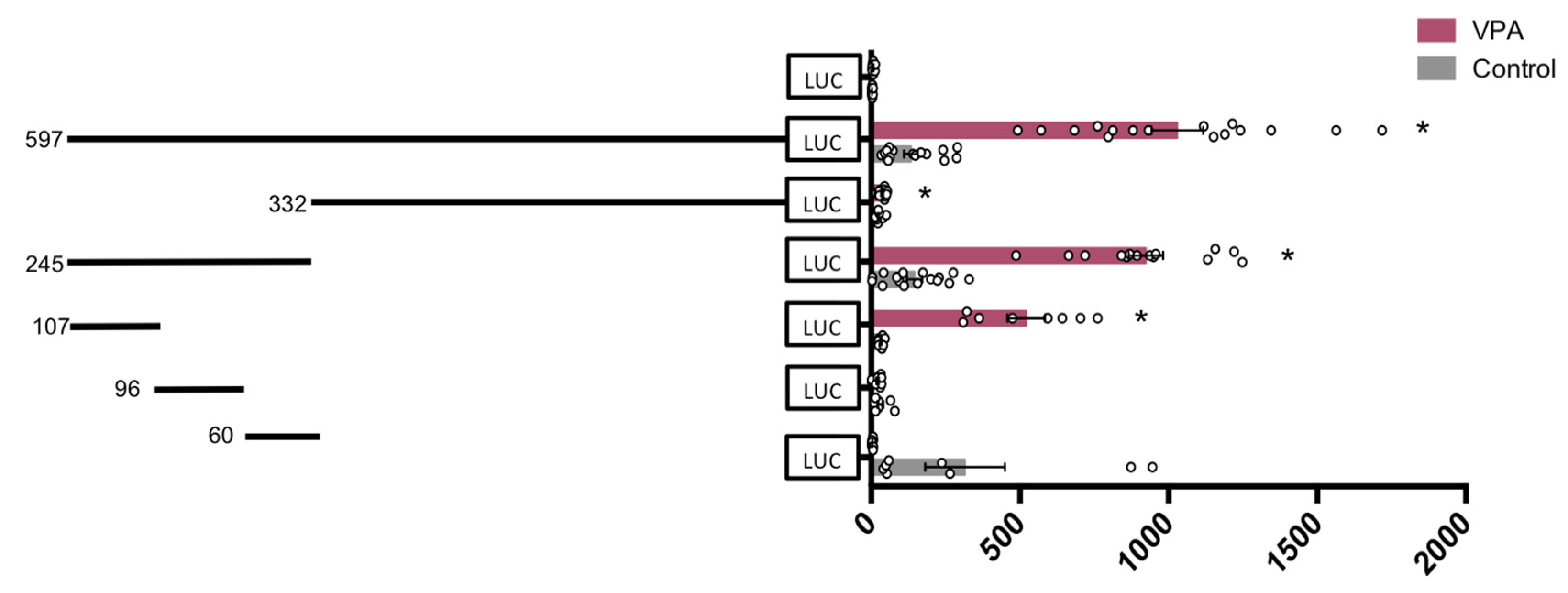
3. Results
3.1. VAP and SCFA (Butyrate) Induce VDR mRNA in HepG2 Cells and Human Hepatocytes
3.2. Functional Analysis of the hVDR Promoter
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Encío, I.J.; Detera-Wadleigh, S.D. The genomic structure of the human glucocorticoid receptor. J. Biol. Chem. 1991, 266, 7182–7188. [Google Scholar] [CrossRef]
- Huckaby, C.S.; Conneely, O.M.; Beattie, W.G.; Dobson, A.D.; Tsai, M.J.; O’Malley, B.W. Structure of the chromosomal chicken progesterone receptor gene. Proc. Natl. Acad. Sci. USA 1987, 84, 8380–8384. [Google Scholar] [CrossRef]
- Zahraoui, A.; Cuny, G. Nucleotide sequence of the chicken proto-oncogene c-erbA corresponding to domain 1 of v-erbA. Eur. J. Biochem. 1987, 166, 63–69. [Google Scholar] [CrossRef]
- Strähle, U.; Schmidt, A.; Kelsey, G.; Stewart, A.F.; Cole, T.J.; Schmid, W.; Schütz, G. At least three promoters direct expression of the mouse glucocorticoid receptor gene. Proc. Natl. Acad. Sci. USA 1992, 89, 6731–6735. [Google Scholar] [CrossRef]
- Crofts, L.A.; Hancock, M.S.; Morrison, N.A.; Eisman, J.A. Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc. Natl. Acad. Sci. USA 1998, 95, 10529–10534. [Google Scholar] [CrossRef]
- Misrahi, M.; Loosfelt, H.; Atger, M.; Mériel, C.; Zerah, V.; Dessen, P.; Milgrom, E. Organisation of the entire rabbit progesterone receptor mRNA and of the promoter and 5’ flanking region of the gene. Nucleic Acids Res. 1988, 16, 5459–5472. [Google Scholar] [CrossRef]
- Lazar, M.A. Thyroid hormone receptors: Multiple forms, multiple possibilities. Endocr. Rev. 1993, 14, 184–193. [Google Scholar]
- Haussler, M.R.; Norman, A.W. Chromosomal receptor for a vitamin D metabolite. Proc. Natl. Acad. Sci. USA 1969, 62, 155–162. [Google Scholar] [CrossRef]
- Zenata, O.; Vrzal, R. Fine tuning of vitamin D receptor (VDR) activity by post-transcriptional and post-translational modifications. Oncotarget 2017, 8, 35390–35402. [Google Scholar] [CrossRef]
- Jehan, F.; d’Alésio, A.; Garabédian, M. Exons and functional regions of the human vitamin D receptor gene around and within the main 1a promoter are well conserved among mammals. J. Steroid Biochem. Mol. Biol. 2007, 103, 361–367. [Google Scholar] [CrossRef]
- Byrne, I.M.; Flanagan, L.; Tenniswood, M.P.; Welsh, J. Identification of a hormone-responsive promoter immediately upstream of exon 1c in the human vitamin D receptor gene. Endocrinology 2000, 141, 2829–2836. [Google Scholar] [CrossRef]
- Dusso, A.S.; Brown, A.J.; Slatopolsky, E. Vitamin D. Am. J. Physiol. Renal Physiol. 2005, 289, F8–F28. [Google Scholar] [CrossRef]
- Simboli-Campbell, M.; Narvaez, C.J.; Tenniswood, M.; Welsh, J. 1,25-Dihydroxyvitamin D3 induces morphological and biochemical markers of apoptosis in MCF-7 breast cancer cells. J. Steroid Biochem. Mol. Biol. 1996, 58, 367–376. [Google Scholar] [CrossRef]
- VanWeelden, K.; Flanagan, L.; Binderup, L.; Tenniswood, M.; Welsh, J. Apoptotic regression of MCF-7 xenografts in nude mice treated with the vitamin D3 analog, EB1089. Endocrinology 1998, 139, 2102–2110. [Google Scholar] [CrossRef]
- Makishima, M.; Lu, T.T.; Xie, W.; Whitfield, G.K.; Domoto, H.; Evans, R.M.; Haussler, M.R.; Mangelsdorf, D.J. Vitamin D receptor as an intestinal bile acid sensor. Science 2002, 296, 1313–1316. [Google Scholar] [CrossRef]
- Costa, E.M.; Hirst, M.A.; Feldman, D. Regulation of 1,25-dihydroxyvitamin D3 receptors by vitamin D analogs in cultured mammalian cells. Endocrinology 1985, 117, 2203–2210. [Google Scholar] [CrossRef]
- Pan, L.C.; Price, P.A. Ligand-dependent regulation of the 1,25-dihydroxyvitamin D3 receptor in rat osteosarcoma cells. J. Biol. Chem. 1987, 262, 4670–4675. [Google Scholar] [CrossRef]
- Krishnan, A.V.; Feldman, D. Cyclic adenosine 3′,5′-monophosphate up-regulates 1,25-dihydroxyvitamin D3 receptor gene expression and enhances hormone action. Mol. Endocrinol. 1992, 6, 198–206. [Google Scholar]
- Krishnan, A.V.; Feldman, D. Activation of protein kinase-C inhibits vitamin D receptor gene expression. Mol. Endocrinol. 1991, 5, 605–612. [Google Scholar] [CrossRef]
- Levy, J.; Zuili, I.; Yankowitz, N.; Shany, S. Induction of cytosolic receptors for 1 alpha, 25-dihydroxyvitamin D3 in the immature rat uterus by oestradiol. J. Endocrinol. 1984, 100, 265–269. [Google Scholar] [CrossRef]
- Mahonen, A.; Pirskanen, A.; Mäenpää, P.H. Homologous and heterologous regulation of 1,25-dihydroxyvitamin D-3 receptor mRNA levels in human osteosarcoma cells. Biochim. Biophys. Acta 1991, 1088, 111–118. [Google Scholar] [CrossRef]
- Chen, T.L.; Cone, C.M.; Morey-Holton, E.; Feldman, D. 1 alpha,25-dihydroxyvitamin D3 receptors in cultured rat osteoblast-like cells. Glucocorticoid treatment increases receptor content. J. Biol. Chem. 1983, 258, 4350–4355. [Google Scholar] [CrossRef]
- Petkovich, P.M.; Heersche, J.N.; Tinker, D.O.; Jones, G. Retinoic acid stimulates 1,25-dihydroxyvitamin D3 binding in rat osteosarcoma cells. J. Biol. Chem. 1984, 259, 8274–8280. [Google Scholar] [CrossRef]
- Chen, T.L.; Feldman, D. Retinoic acid modulation of 1,25(OH)2 vitamin D3 receptors and bioresponse in bone cells: Species differences between rat and mouse. Biochem. Biophys. Res. Commun. 1985, 132, 74–80. [Google Scholar] [CrossRef]
- Krishnan, A.V.; Feldman, D. Stimulation of 1,25-dihydroxyvitamin D3 receptor gene expression in cultured cells by serum and growth factors. J. Bone Miner. Res. 1991, 6, 1099–1107. [Google Scholar] [CrossRef]
- Zhao, X.; Feldman, D. Regulation of vitamin D receptor abundance and responsiveness during differentiation of HT-29 human colon cancer cells. Endocrinology 1993, 132, 1808–1814. [Google Scholar] [CrossRef]
- Martínez-Sena, T.; Soluyanova, P.; Guzmán, C.; Valdivielso, J.M.; Castell, J.V.; Jover, R. The Vitamin D Receptor Regulates Glycerolipid and Phospholipid Metabolism in Human Hepatocytes. Biomolecules 2020, 10, 493. [Google Scholar] [CrossRef]
- Bozic, M.; Guzmán, C.; Benet, M.; Sánchez-Campos, S.; García-Monzón, C.; Gari, E.; Gatius, S.; Valdivielso, J.M.; Jover, R. Hepatocyte vitamin D receptor regulates lipid metabolism and mediates experimental diet-induced steatosis. J. Hepatol. 2016, 65, 748–757. [Google Scholar] [CrossRef]
- López-Riera, M.; Conde, I.; Tolosa, L.; Zaragoza, A.; Castell, J.V.; Gómez-Lechón, M.J.; Jover, R. New microRNA Biomarkers for Drug-Induced Steatosis and Their Potential to Predict the Contribution of Drugs to Non-alcoholic Fatty Liver Disease. Front. Pharmacol. 2017, 8, 3. [Google Scholar] [CrossRef]
- Guo, H.L.; Jing, X.; Sun, J.-Y.; Hu, Y.; Xu, Z.-J.; Ni, M.-M.; Chen, F.; Lu, X.-P.; Qiu, J.-C.; Wang, T. Valproic Acid and the Liver Injury in Patients with Epilepsy: An Update. Curr. Pharm. Des. 2019, 25, 343–351. [Google Scholar] [CrossRef]
- Balagura, G.; Iapadre, G.; Verrotti, A.; Striano, P. Moving beyond sodium valproate: Choosing the right anti-epileptic drug in children. Expert Opin. Pharmacother. 2019, 20, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Phiel, C.J.; Zhang, F.; Huang, E.Y.; Guenther, M.G.; Lazar, M.A.; Klein, P.S. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001, 276, 36734–36741. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Hong, I.; Kim, M.; Lee, B.H.; Kim, J.H.; Kang, K.S.; Kim, H.L.; Yoon, B.I.; Chung, H.; Kong, G.; et al. Gene expression profiles of murine fatty liver induced by the administration of valproic acid. Toxicol. Appl. Pharmacol. 2007, 220, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Vrzal, R.; Doricakova, A.; Novotna, A.; Bachleda, P.; Bitman, M.; Pavek, P.; Dvorak, Z. Valproic acid augments vitamin D receptor-mediated induction of CYP24 by vitamin D3: A possible cause of valproic acid-induced osteomalacia? Toxicol. Lett. 2011, 200, 146–153. [Google Scholar] [CrossRef]
- Schaefer, M.; Schänzle, G.; Bischoff, D.; Süssmuth, R.D. Upcyte Human Hepatocytes: A Potent In Vitro Tool for the Prediction of Hepatic Clearance of Metabolically Stable Compounds. Drug Metab. Dispos. 2016, 44, 435–444. [Google Scholar] [CrossRef]
- Tolosa, L.; Gómez-Lechón, M.J.; López, S.; Guzmán, C.; Castell, J.V.; Donato, M.T.; Jover, R. Human Upcyte Hepatocytes: Characterization of the Hepatic Phenotype and Evaluation for Acute and Long-Term Hepatotoxicity Routine Testing. Toxicol. Sci. 2016, 152, 214–229. [Google Scholar] [CrossRef]
- Petrov, P.D.; Fernández-Murga, L.; Conde, I.; Martínez-Sena, T.; Guzmán, C.; Castell, J.V.; Jover, R. Epistane, an anabolic steroid used for recreational purposes, causes cholestasis with elevated levels of cholic acid conjugates, by upregulating bile acid synthesis (CYP8B1) and cross-talking with nuclear receptors in human hepatocytes. Arch. Toxicol. 2020, 94, 589–607. [Google Scholar] [CrossRef]
- Kel, A.E.; Gössling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef]
- Andrés, V.; Ureña, J.; Poch, E.; Chen, D.; Goukassian, D. Role of Sp1 in the induction of p27 gene expression in vascular smooth muscle cells in vitro and after balloon angioplasty. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 342–347. [Google Scholar] [CrossRef][Green Version]
- Wierstra, I. Sp1: Emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 2008, 372, 1–13. [Google Scholar] [CrossRef]
- Krämer, O.H.; Zhu, P.; Ostendorff, H.P.; Golebiewski, M.; Tiefenbach, J.; Peters, M.A.; Brill, B.; Groner, B.; Bach, I.; Heinzel, T.; et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. Embo J. 2003, 22, 3411–3420. [Google Scholar] [CrossRef] [PubMed]
- Gurvich, N.; Tsygankova, O.M.; Meinkoth, J.L.; Klein, P.S. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004, 64, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Tripathi, S.; Pandey, K.N. Histone deacetylase inhibitors modulate the transcriptional regulation of guanylyl cyclase/natriuretic peptide receptor-a gene: Interactive roles of modified histones, histone acetyltransferase, p300, AND Sp1. J. Biol. Chem. 2014, 289, 6991–7002. [Google Scholar] [CrossRef] [PubMed]
- Doetzlhofer, A.; Rotheneder, H.; Lagger, G.; Koranda, M.; Kurtev, V.; Brosch, G.; Wintersberger, E.; Seiser, C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 1999, 19, 5504–5511. [Google Scholar] [CrossRef]
- Karlseder, J.; Rotheneder, H.; Wintersberger, E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 1996, 16, 1659–1667. [Google Scholar] [CrossRef]
- Lin, S.Y.; Black, A.R.; Kostic, D.; Pajovic, S.; Hoover, C.N.; Azizkhan, J.C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol. Cell. Biol. 1996, 16, 1668–1675. [Google Scholar] [CrossRef]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef]
- Swingler, T.E.; Kevorkian, L.; Culley, K.L.; Illman, S.A.; Young, D.A.; Parker, A.E.; Lohi, J.; Clark, I.M. MMP28 gene expression is regulated by Sp1 transcription factor acetylation. Biochem. J. 2010, 427, 391–400. [Google Scholar] [CrossRef]
- Tan, N.Y.; Khachigian, L.M. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 2009, 29, 2483–2488. [Google Scholar] [CrossRef]
- Suzuki, T.; Kimura, A.; Nagai, R.; Horikoshi, M. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 2000, 5, 29–41. [Google Scholar] [CrossRef]
- Chen, C.J.; Chang, W.C.; Chen, B.K. Attenuation of c-Jun and Sp1 expression and p300 recruitment to gene promoter confers the trichostatin A-induced inhibition of 12(S)-lipoxygenase expression in EGF-treated A431 cells. Eur. J. Pharmacol. 2008, 591, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kosaras, B.; Aleyasin, H.; Han, J.A.; Park, D.S.; Ratan, R.R.; Kowall, N.W.; Ferrante, R.J.; Lee, S.W.; Ryu, H. Role of cyclooxygenase-2 induction by transcription factor Sp1 and Sp3 in neuronal oxidative and DNA damage response. Faseb J. 2006, 20, 2375–2377. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhao, S.; Ammanamanchi, S.; Brattain, M.; Venkatasubbarao, K.; Freeman, J.W. Trichostatin A induces transforming growth factor beta type II receptor promoter activity and acetylation of Sp1 by recruitment of PCAF/p300 to a Sp1.NF-Y complex. J. Biol. Chem. 2005, 280, 10047–10054. [Google Scholar] [CrossRef] [PubMed]
- Song, C.Z.; Keller, K.; Chen, Y.; Stamatoyannopoulos, G. Functional interplay between CBP and PCAF in acetylation and regulation of transcription factor KLF13 activity. J. Mol. Biol. 2003, 329, 207–215. [Google Scholar] [CrossRef]
- Ryu, H.; Lee, J.; Olofsson, B.A.; Mwidau, A.; Dedeoglu, A.; Escudero, M.; Flemington, E.; Azizkhan-Clifford, J.; Ferrante, R.J.; Ratan, R.R. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl. Acad. Sci. USA 2003, 100, 4281–4286. [Google Scholar] [CrossRef]
- Dempsey, N.J.; Ojalvo, L.S.; Wu, D.W.; Little, J.A. Induction of an embryonic globin gene promoter by short-chain fatty acids. Blood 2003, 102, 4214–4222. [Google Scholar] [CrossRef]
- Qin, X.; Wang, X. Role of vitamin D receptor in the regulation of CYP3A gene expression. Acta Pharm. Sin. B 2019, 9, 1087–1098. [Google Scholar] [CrossRef]
- Thompson, P.D.; Jurutka, P.W.; Whitfield, G.K.; Myskowski, S.M.; Eichhorst, K.R.; Dominguez, C.E.; Haussler, C.A.; Haussler, M.R. Liganded VDR induces CYP3A4 in small intestinal and colon cancer cells via DR3 and ER6 vitamin D responsive elements. Biochem. Biophys. Res. Commun. 2002, 299, 730–738. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, K.N.; Chen, C. The role of CYP3A4 in the biotransformation of bile acids and therapeutic implication for cholestasis. Ann. Transl. Med. 2014, 2, 7. [Google Scholar]
- Jahn, D.; Dorbath, D.; Schilling, A.K.; Gildein, L.; Meier, C.; Vuille-Dit-Bille, R.N.; Schmitt, J.; Kraus, D.; Fleet, J.C.; Hermanns, H.M.; et al. Intestinal vitamin D receptor modulates lipid metabolism, adipose tissue inflammation and liver steatosis in obese mice. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1567–1578. [Google Scholar] [CrossRef]
- Farinelli, E.; Giampaoli, D.; Cenciarini, A.; Cercado, E.; Verrotti, A. Valproic acid and nonalcoholic fatty liver disease: A possible association? World J. Hepatol. 2015, 7, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.K.; Darwish, H.M.; Moss, V.E.; DeLuca, H.F. Vitamin D-influenced gene expression via a ligand-independent, receptor-DNA complex intermediate. Proc. Natl. Acad. Sci. USA 1993, 90, 9257–9260. [Google Scholar] [CrossRef] [PubMed]
- Christakos, S.; Raval-Pandya, M.; Wernyj, R.P.; Yang, W. Genomic mechanisms involved in the pleiotropic actions of 1,25-dihydroxyvitamin D3. Biochem. J. 1996, 316 Pt 2, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.A.; Richon, V.M.; Breslow, R.; Rifkind, R.A. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 2001, 13, 477–483. [Google Scholar] [CrossRef]
- Krämer, O.H.; Göttlicher, M.; Heinzel, T. Histone deacetylase as a therapeutic target. Trends Endocrinol. Metab. 2001, 12, 294–300. [Google Scholar] [CrossRef]
- Gore, S.D.; Carducci, M.A. Modifying histones to tame cancer: Clinical development of sodium phenylbutyrate and other histone deacetylase inhibitors. Expert Opin. Investig. Drugs 2000, 9, 2923–2934. [Google Scholar] [CrossRef]
- Cheson, B.D.; Zwiebel, J.A.; Dancey, J.; Murgo, A. Novel therapeutic agents for the treatment of myelodysplastic syndromes. Semin. Oncol. 2000, 27, 560–577. [Google Scholar]
- Göttlicher, M.; Minucci, S.; Zhu, P.; Krämer, O.H.; Schimpf, A.; Giavara, S.; Sleeman, J.P.; Lo Coco, F.; Nervi, C.; Pelicci, P.G.; et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. Embo J. 2001, 20, 6969–6978. [Google Scholar] [CrossRef]
- Cinatl, J., Jr.; Cinatl, J.; Scholz, M.; Driever, P.H.; Henrich, D.; Kabickova, H.; Vogel, J.U.; Doerr, H.W.; Kornhuber, B. Antitumor activity of sodium valproate in cultures of human neuroblastoma cells. Anticancer Drugs 1996, 7, 766–773. [Google Scholar] [CrossRef]
- Blaheta, R.A.; Cinatl, J., Jr. Anti-tumor mechanisms of valproate: A novel role for an old drug. Med. Res. Rev. 2002, 22, 492–511. [Google Scholar] [CrossRef]
- Yang, W.M.; Yao, Y.L.; Sun, J.M.; Davie, J.R.; Seto, E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997, 272, 28001–28007. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]




| Promoter | Name | Sequence |
|---|---|---|
| PBGD-FW | PBGD-FW | CGGAAGAAAACAGCCCAAAGA |
| PBGD-RV | PBGD-RV | TGAAGCCAGGAGGAAGCACAGT |
| VDR-FW | VDR-FW | CACCCCTGGGCTCCACTTACC |
| VDR-RV | VDR-RV | CCGCCACAGGCTGTCCTAGTC |
| 3838-FW | VDR-UP3094-KpnI | CTCGGTACCTCAGTTGTACAATGGAACGGT |
| 3838-RV | VDR-DN6813-HindIII | TGAAAGCTTCTGCCGAAGAGGAGTAAAGG |
| 2514-FW | VDR_UP25869-KpnI | GCTGGTACCGGACTTGGGCAGTAGGAGC |
| 2514-RV | VDR-DN28360-HindIII | GATAAGCTTGGGGAGGGGTGTGCATTATAT |
| 271-FW | 508-VDR:161U26 | AGCGGTACCGGAGCTCCTGGCAAGAG |
| 271-RV | 508-VDR:421L27 | ATGCTCGAGGCCGGGCGCTCAGGCCCC |
| 355-FW | 508-VDR:433u26 | CCCGGTACCGAGCATTAGAGTCTAAG |
| 355-RV | 508-VDR:672L27 | CAACTCGAGCATCACAGGTGACCATAC |
| 99(1)-FW | VDR-287_1-Sac-UP | AACGGAGCTCCTGGCAAGAGAGGAC |
| 99(1)-RV | VDR-287_1-Hind-DN | TGACAAGCTTAGACAGCCCAGCACC |
| 99(2)-FW | VDR-287_2-Sac-UP | CCAGGAGCTCGGCTGTCTCTGCTTG |
| 99(2)-RV | VDR-287_2-Hind-DN | CCCGAAGCTTCCGGGTTCGCACCTG |
| 63-FW | VDR-287_3-Sac-UP | GTGCGAGCTCGGGAGCAGCGGGAAA |
| 63-RV | VDR-287_3-Hind-DN | GCTCAAGCTTCGGTATCCCAGACGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Torres, M.; Guzmán, C.; Petrov, P.D.; Jover, R. Valproate and Short-Chain Fatty Acids Activate Transcription of the Human Vitamin D Receptor Gene through a Proximal GC-Rich DNA Region Containing Two Putative Sp1 Binding Sites. Nutrients 2022, 14, 2673. https://doi.org/10.3390/nu14132673
Moreno-Torres M, Guzmán C, Petrov PD, Jover R. Valproate and Short-Chain Fatty Acids Activate Transcription of the Human Vitamin D Receptor Gene through a Proximal GC-Rich DNA Region Containing Two Putative Sp1 Binding Sites. Nutrients. 2022; 14(13):2673. https://doi.org/10.3390/nu14132673
Chicago/Turabian StyleMoreno-Torres, Marta, Carla Guzmán, Petar D. Petrov, and Ramiro Jover. 2022. "Valproate and Short-Chain Fatty Acids Activate Transcription of the Human Vitamin D Receptor Gene through a Proximal GC-Rich DNA Region Containing Two Putative Sp1 Binding Sites" Nutrients 14, no. 13: 2673. https://doi.org/10.3390/nu14132673
APA StyleMoreno-Torres, M., Guzmán, C., Petrov, P. D., & Jover, R. (2022). Valproate and Short-Chain Fatty Acids Activate Transcription of the Human Vitamin D Receptor Gene through a Proximal GC-Rich DNA Region Containing Two Putative Sp1 Binding Sites. Nutrients, 14(13), 2673. https://doi.org/10.3390/nu14132673








