Screening and Isolation of Potential Anti-Inflammatory Compounds from Saxifraga atrata via Affinity Ultrafiltration-HPLC and Multi-Target Molecular Docking Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus and Chemicals
2.2. Preparation of the Target Fraction
2.3. Screening and Isolation of Potential Anti-Inflammatory Active Ingredients Based on Affinity Ultrafiltration-HPLC
2.3.1. Screening of the Potential Anti-Inflammatory Active Ingredient in the Target Fraction with Affinity Ultrafiltration
2.3.2. Purification of Potential COX-2 Ligand via Reversed-Phase Liquid Chromatograph
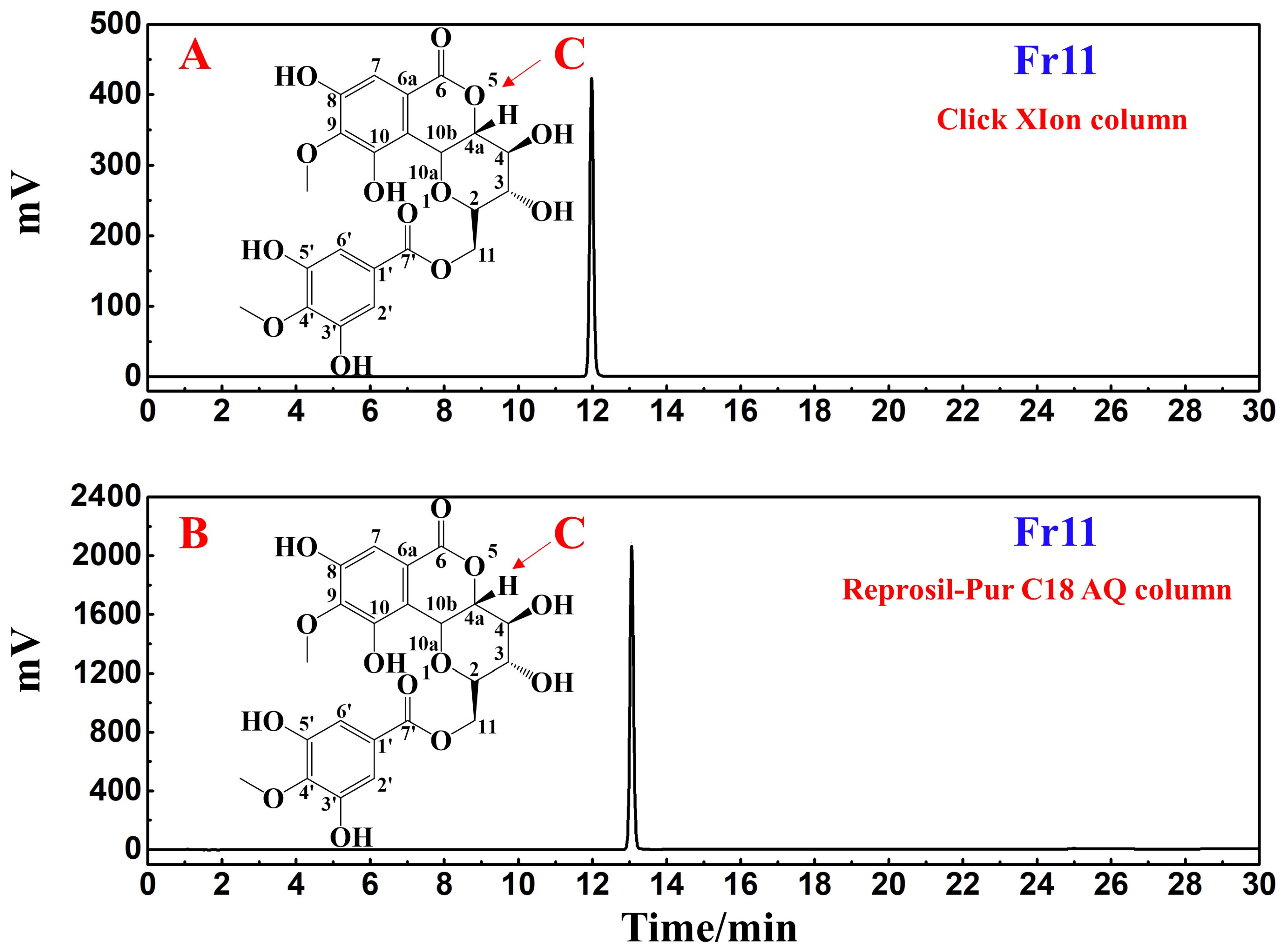
2.4. Analysis of the Purity of the Isolated Candidate COX-2 Ligand
2.5. Molecular Docking Analyses
3. Results
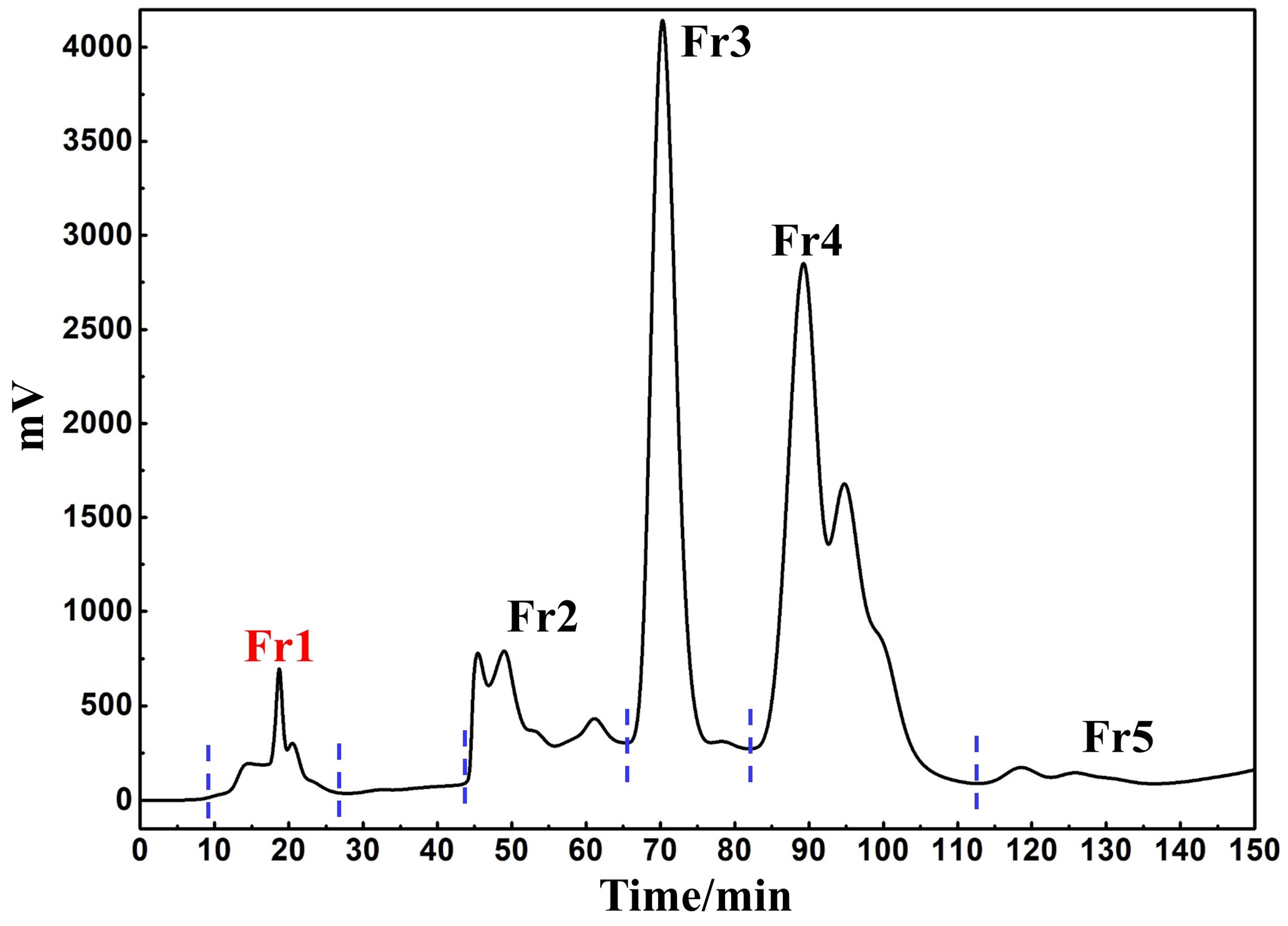
3.1. Target Fraction MCI GEL® CHP20P Medium Pressure Liquid Chromatography Sample Pretreatment
3.2. Screening and Isolation of Potential Anti-Inflammatory Active Ingredient Based on Affinity Ultrafiltration-HPLC
3.2.1. Screening of the Potential Anti-Inflammatory Active Ingredient in the Target Fraction
3.2.2. Purification of the Potential COX-2 Ligand with Reversed-Phase Liquid Chromatography
3.3. Analyses of the Purity and Structural Characterization of Potential COX-2 Ligands
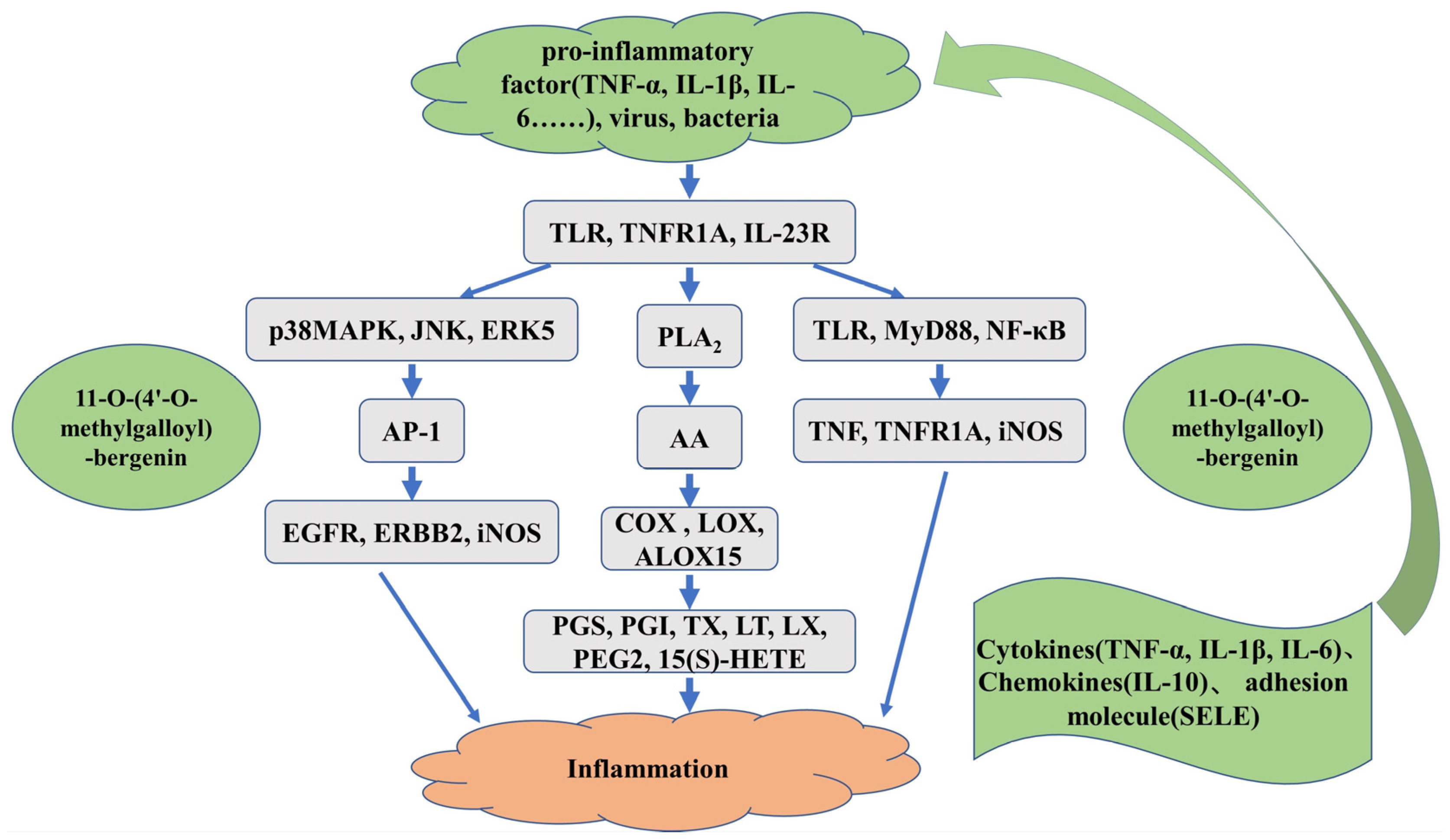
3.4. Molecular Docking Analyses of the Interactions between 11-O-(4′-O-methylgalloyl)-bergenin and Inflammation-Related Targets
3.4.1. AA Metabolic Pathway
3.4.2. MAPK Signaling Pathway
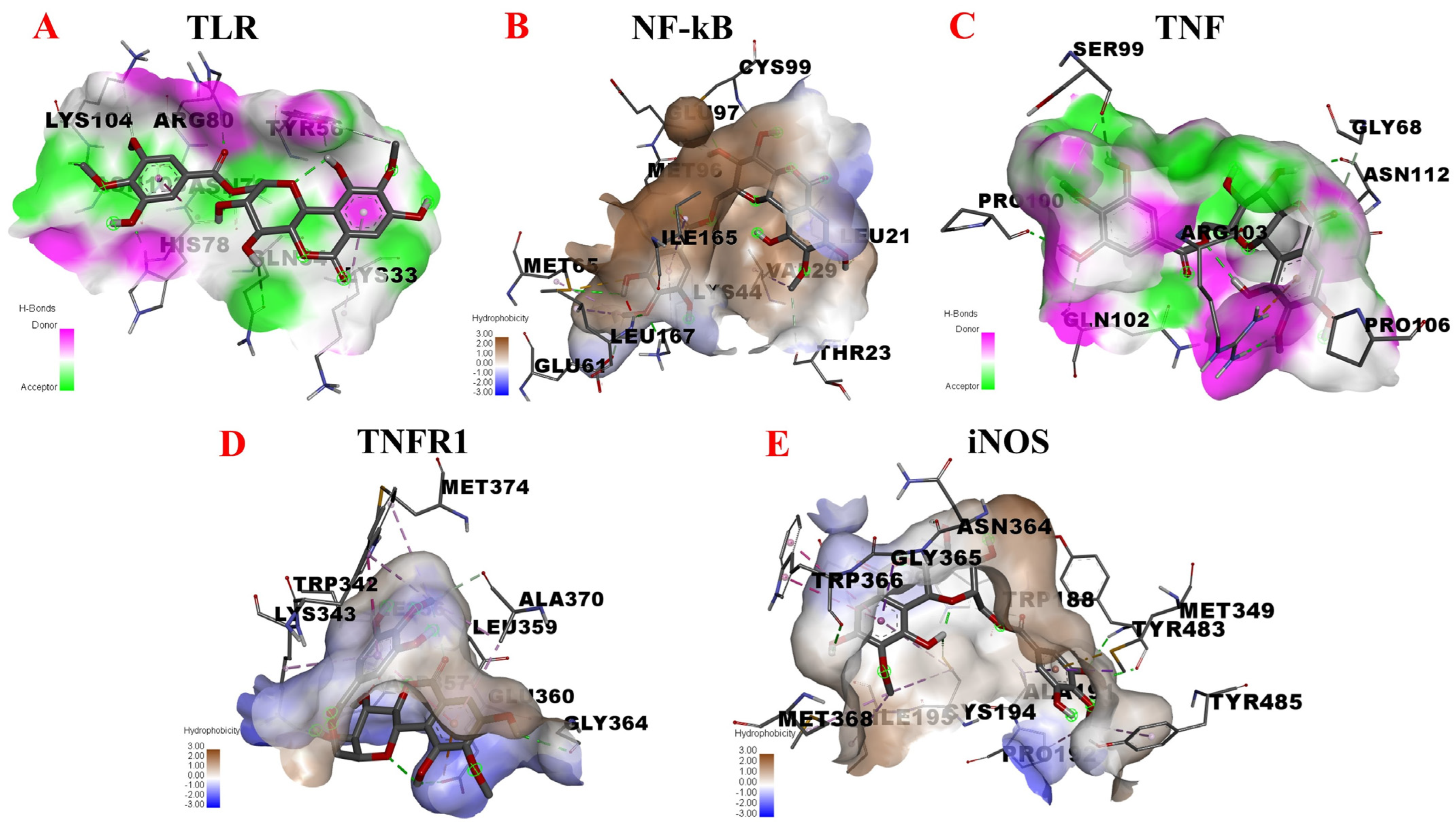
3.4.3. TLR/MyD88/NF-κB Pathway
3.4.4. Inflammation-Associated Factors and Adhesion Molecule
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef] [PubMed]
- Borsini, A.; Zunszain, P.A.; Thuret, S.; Pariante, C.M. The role of inflammatory cytokines as key modulators of neurogenesis. Trends Neurosci. 2015, 38, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yi, Y.S.; Kim, M.Y.; Cho, J.Y. Role of ginsenosides, the main active components of Panax ginseng, in inflammatory responses and diseases. J. Ginseng. Res. 2017, 41, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.F.; Zeng, Y.; Li, J.C.; Tang, C.; Zhang, Y.; Meng, X.L. The anti-arthritic activity of total glycosides from Pterocephalus hookeri, a traditional Tibetan herbal medicine. Pharm. Biol. 2017, 55, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, F.; Zheng, T.T.; Shi, L.; Zhang, Z.G.; Niu, T.M.; Wang, Q.Y.; Zhao, D.S.; Li, W.; Zhao, P. Lamiophlomis herba: A comprehensive overview of its chemical constituents, pharmacology, clinical applications, and quality control. Biomed. Pharmacother. 2021, 144, 112299. [Google Scholar] [CrossRef]
- Zhang, M.R.; Jiang, K.; Yang, J.L.; Shi, Y.P. Flavonoids as key bioactive components of Oxytropis falcata bunge, a traditional anti-inflammatory and analgesic Tibetan medicine. Nat. Prod. Res. 2020, 34, 3335–3352. [Google Scholar] [CrossRef]
- Gerschwitz-Eidt, M.A.; Kadereit, J.W. Species composition of Saxifraga sect. Saxifraga subsect. Arachnoideae (Saxifragaceae) based on DNA sequence evidence. Willdenowia 2020, 50, 225–233. [Google Scholar] [CrossRef]
- Durovic, S.Z.; Kuzmanovic, N.; Tomovic, G.; Niketic, M. Nomenclatural notes and typification of the names related to Silene saxifraga group (Caryophyllaceae). Phytotaxa 2018, 344, 201–214. [Google Scholar] [CrossRef]
- Yang, Y.; He, T.; Lu, S.; Huang, R.; Wang, Z. Records of Tibetan Medicine; Qinghai Publishing House: Qinghai, China, 1991; pp. 450–451. [Google Scholar]
- Zhao, X.M.; Li, R.J.; Zhou, C.; Zhang, J.; He, C.H.; Zheng, Y.T.; Wu, W.D.; Zhang, H.B. Separation and purification of deoxynivalenol (DON) mycotoxin from wheat culture using a simple two-step silica gel column chromatography. J. Integr. Agric. 2016, 15, 694–701. [Google Scholar] [CrossRef]
- Huang, M.X.; Pineau, A.; Bouaziz, O.; Vu, T.D. Recrystallization induced plasticity in austenite and ferrite. Mater. Sci. Eng. A. 2012, 541, 196–198. [Google Scholar] [CrossRef]
- Jandera, P.; Janas, P. Recent advances in stationary phases and understanding of retention in hydrophilic interaction chromatography. A review. Anal. Chim. Acta 2017, 967, 12–32. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Ju, Z.C.; Yang, Y.B.; Zhang, Y.H.; Yang, L.; Wang, Z.T. Phytochemical analysis of Panax species: A review. J. Ginseng. Res. 2021, 45, 1–21. [Google Scholar] [CrossRef]
- Gong, Y.; Huang, X.Y.; Pei, D.; Duan, W.D.; Zhang, X.; Sun, X.; Di, D.L. The applicability of high-speed counter current chromatography to the separation of natural antioxidants. J. Chromatogr. A 2020, 1623, 461150. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, S.S.; Zhang, X.K.; He, F.; Duan, C.Q. An effective method for the semi-preparative isolation of high-purity anthocyanin monomers from grape pomace. Food Chem. 2020, 310, 125830. [Google Scholar] [CrossRef]
- Sun, G.Y.; Abuduaini, M.; Adili, G.; Zhao, Y.X.; Aisa, H.A. Dual-tautomerism separation method based on asymmetric transformation: Gram-scale preparation of high-purity punicalagin from pomegranate peel wastes. J. Chromatogr. A 2021, 1651, 462281. [Google Scholar] [CrossRef]
- Fan, C.; Li, N.; Cao, X.L.; Wen, L.J. Ionic liquid-modified countercurrent chromatographic isolation of high-purity delphinidin-3-rutinoside from eggplant peel. J. Food Sci. 2020, 85, 1132–1139. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, T.T.; Yan, X.; Xue, S.; Chen, J.W.; Wu, Z.; Qiu, Y.K. Gradual gradient two-dimensional preparative liquid chromatography system for preparative separation of complex natural products. Chromatographia 2019, 82, 543–552. [Google Scholar] [CrossRef]
- Dawa, Y.Z.; Du, Y.R.; Wang, Q.; Chen, C.B.; Zou, D.L.; Qi, D.S.; Ma, J.B.; Dang, J. Targeted isolation of 1,1-diphenyl-2-picrylhydrazyl inhibitors from Saxifraga atrata using medium- and high- pressure liquid chromatography combined with online high performance liquid chromatography-1,1-diphenyl-2-picrylhydrazyl detection. J. Chromatogr. A 2021, 1635, 461690. [Google Scholar] [CrossRef]
- Ma, Z.H.; Wei, X.Y.; Zhou, M.Y.; Liu, G.H.; Liu, F.J.; Liu, Z.Q.; Yu, X.Y.; Zong, Z.M. Isolation and purification of carbazole contained in anthracene slag by extraction combined with medium pressure liquid chromatography. Chin. J. Chem. Eng. 2019, 27, 2925–2929. [Google Scholar] [CrossRef]
- Dang, J.; Wang, Q.; Wang, Q.L.; Yuan, C.; Li, G.; Ji, T.F. Preparative isolation of antioxidative gallic acid derivatives from Saxifraga tangutica using a class separation method based on medium-pressure liquid chromatography and reversed-phase liquid chromatography. J. Sep. Sci. 2021, 44, 3734–3746. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Choi, S.I.; Han, S.H.; Lee, Y.J.; Choi, J.G.; Lee, Y.S.; Choi, J.H.; Lee, S.E.; Kim, G.S. Potential of Pseudoshikonin I Isolated from Lithospermi Radix as Inhibitors of MMPs in IL-1 beta-Induced SW1353 Cells. Int. J. Mol. Sci. 2016, 17, 1350. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Ma, J.B.; Dawa, Y.Z.; Liu, C.; Ji, T.F.; Wang, Q.L. Preparative separation of 1,1-diphenyl-2-picrylhydrazyl inhibitors originating from Saxifraga sinomontana employing medium-pressure liquid chromatography in combination with reversed-phase liquid chromatography. Rsc. Adv. 2021, 11, 38739–38749. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.Q.; Shen, J.W.; Ma, Y.H.; He, Y.F.; Bao, Y.; Li, R.R.; Wang, S.S.; Wang, Q.; Lin, P.C.; Dang, J. Preparative separation of isoquinoline alkaloids from Corydalis impatiens using a middle-pressure chromatogram isolated gel column coupled with two-dimensional liquid chromatography. J. Sep. Sci. 2019, 42, 3182–3190. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.X.; Chen, G.L.; Sun, B.Q.; Wu, J.L.; Li, N.; Sarker, S.D.; Nahar, L.; Guo, M.Q. Screening for natural inhibitors of human topoisomerases from medicinal plants with bio-affinity ultrafiltration and LC-MS. Phytochem. Rev. 2020, 19, 1231–1261. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, G.L.; Yang, J.P.; Yang, C.L.; Guo, M.Q. Screening and characterisation of potential antioxidant, hypoglycemic and hypolipidemic components revealed in Portulaca oleracea via multi-target affinity ultrafiltration LC-MS and molecular docking. Phytochem. Anal. 2022, 33, 272–285. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Salman, M.M.; Al-Obaidi, Z.; Kitchen, P.; Loreto, A.; Bill, R.M.; Wade-Martins, R. Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 4688. [Google Scholar] [CrossRef]
- Cheng, F.J.; Huynh, T.K.; Yang, C.S.; Hu, D.W.; Shen, Y.C.; Tu, C.Y.; Wu, Y.C.; Tang, C.H.; Huang, W.C.; Chen, Y.; et al. Hesperidin Is a Potential Inhibitor against SARS-CoV-2 Infection. Nutrients 2021, 13, 2800. [Google Scholar] [CrossRef]
- Cinelli, M.A.; Do, H.T.; Miley, G.P.; Silverman, R.B. Inducible nitric oxide synthase: Regulation, structure, and inhibition. Med. Res. Rev. 2020, 40, 158–189. [Google Scholar] [CrossRef]
- Dang, J.; Ma, J.B.; Du, Y.R.; Dawa, Y.; Wang, Q.; Chen, C.B.; Wang, Q.L.; Tao, Y.D.; Ji, T.F. Large-scale preparative isolation of bergenin standard substance from Saxifraga atrata using polyamide coupled with MCI GEL® CHP20P as stationary phases in medium pressure chromatography. J. Chromatogr. B 2021, 1170, 122617. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Galan, C.; Barbosa, O.; Hernandez, K.; dos Santos, J.C.S.; Rodrigues, R.C.; Fernandez-Lafuente, R. Evaluation of Styrene-Divinylbenzene Beads as a Support to Immobilize Lipases. Molecules 2014, 19, 7629–7645. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lv, Y.P. Purification, characterization, and DNA damage protection of active components from tartary buckwheat (Fagopyrum tataricum) hull. Food Sci. Biotechnol. 2015, 24, 1959–1966. [Google Scholar] [CrossRef]
- Zielinska, D.; Zielinski, H.; Laparra-Llopis, J.M.; Szawara-Nowak, D.; Honke, J.; Gimenez-Bastida, J.A. Caffeic Acid Modulates Processes Associated with Intestinal Inflammation. Nutrients 2021, 13, 554. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Jang, D.S.; Jin, J.L.; Yun-Choi, H.S. Anti-platelet aggregating and anti-oxidative activities of 11-O-(4′-O-methylgalloyl)-bergenin, a new compound isolated from Crassula cv. ‘Himaturi’. Planta Med. 2005, 71, 776–777. [Google Scholar] [CrossRef]
- Zarrin, A.A.; Bao, K.; Lupardus, P.; Vucic, D. Kinase inhibition in autoimmunity and inflammation. Nat. Rev. Drug Discov. 2021, 20, 39–63. [Google Scholar] [CrossRef]
- Bai, J.R.; Zhang, Y.S.; Tang, C.; Hou, Y.; Ai, X.P.; Chen, X.R.; Zhang, Y.; Wang, X.B.; Meng, X.L. Gallic acid: Pharmacological activities and molecular mechanisms involved in inflammation-related diseases. Biomed. Pharmacother. 2021, 133, 110985. [Google Scholar] [CrossRef]
- Chen, J.; Ye, C.; Wan, C.; Li, G.; Peng, L.C.; Peng, Y.Y.; Fang, R.D. The Roles of c-Jun N-Terminal Kinase (JNK) in Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 9640. [Google Scholar] [CrossRef]
- Lu, N.; Malemud, C.J. Extracellular Signal-Regulated Kinase: A Regulator of Cell Growth, Inflammation, Chondrocyte and Bone Cell Receptor-Mediated Gene Expression. Int. J. Mol. Sci. 2019, 20, 3792. [Google Scholar] [CrossRef]
- Maisel, S.A.; Broka, D.; Atwell, B.; Bunch, T.; Kupp, R.; Singh, S.K.; Mehta, S.; Schroeder, J. Stapled EGFR peptide reduces inflammatory breast cancer and inhibits additional HER-driven models of cancer. J. Transl. Med. 2019, 17, 201. [Google Scholar] [CrossRef]
- Akdis, M.; Burgler, S.; Crameri, R.; Eiwegger, T.; Fujita, H.; Gomez, E.; Klunker, S.; Meyer, N.; O’Mahony, L.; Palomares, O.; et al. Interleukins, from 1 to 37, and interferon-gamma: Receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2011, 127, 701–721. [Google Scholar] [CrossRef] [PubMed]
- Grancieri, M.; Martino, H.S.D.; de Mejia, E.G. Protein Digests and Pure Peptides from Chia Seed Prevented Adipogenesis and Inflammation by Inhibiting PPAR gamma and NF-kappa B Pathways in 3T3L-1 Adipocytes. Nutrients 2021, 13, 176. [Google Scholar] [CrossRef] [PubMed]
- Stachon, T.; Latta, L.; Kolev, K.; Seitz, B.; Langenbucher, A.; Szentmáry, N. Increased NF-κB and iNOS Expression in Keratoconus Keratocytes—Hints for an Inflammatory Component? Klin. Monbl. Augenheilkd. 2021, 238, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Lohberger, B.; Bernhart, E.; Stuendl, N.; Glaenzer, D.; Leithner, A.; Rinner, B.; Bauer, R.; Kretschmer, N. Periplocin mediates TRAIL-induced apoptosis and cell cycle arrest in human myxofibrosarcoma cells via the ERK/p38/JNK pathway. Phytomedicine 2020, 76, 153262. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.R.; Ramakrishnan, P. Regulation of Nuclear Factor-kappaB Function by O-GlcNAcylation in Inflammation and Cancer. Front. Cell. Dev. Biol. 2021, 9, 751761. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, L.J.; Chen, J.; Dong, L.L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct. Target. Ther. 2021, 26, 94. [Google Scholar] [CrossRef]
- Du, B.; Lin, C.Y.; Bian, Z.X.; Xu, B.J. An insight into anti-inflammatory effects of fungal beta-glucans. Trends Food Sci. Technol. 2015, 41, 49–59. [Google Scholar] [CrossRef]
- Huang, X.F.; Cheng, W.B.; Jiang, Y.; Liu, Q.; Liu, X.H.; Xu, W.F.; Huang, H.T. A network pharmacology-based strategy for predicting anti-inflammatory targets of ephedra in treating asthma. Int. Immunopharmacol. 2020, 83, 106423. [Google Scholar] [CrossRef]
- Li, Y.; Huang, H.; Liu, B.; Zhang, Y.; Pan, X.; Yu, X.Y.; Shen, Z.; Song, Y.H. Inflammasomes as therapeutic targets in human diseases. Signal Transduct. Target. Ther. 2021, 6, 247. [Google Scholar] [CrossRef]
- Doña, I.; Pérez-Sánchez, N.; Eguiluz-Gracia, I.; Muñoz-Cano, R.; Bartra, J.; Torres, M.J.; Cornejo-García, J.A. Progress in understanding hypersensitivity reactions to nonsteroidal anti-inflammatory drugs. Allergy 2020, 75, 561–575. [Google Scholar] [CrossRef]
- Rastogi, A.; Tiwari, M.K.; Ghangrekar, M.M. A review on environmental occurrence, toxicity and microbial degradation of Non-Steroidal Anti-Inflammatory Drugs (NSAIDs). J. Environ. Manag. 2021, 300, 113694. [Google Scholar] [CrossRef] [PubMed]




| Proteins | Protein Function | Binding Energy (kcal/mol) | Binding Residues | Type |
|---|---|---|---|---|
| COX-2 | Inflammation, pain | −6.35 | LEU-153 TYR-131 PRO-154 GLU-466 GLN-462 HIS-39 GLY-136 CYS-47 CYS-36 PRO-157 ASP-158 | Pi-Alkyl Conventional Hydrogen Bond Pi-Alkyl Carbon Hydrogen Bond Conventional Hydrogen Bond van der Waals Carbon Hydrogen Bond van der Waals Carbon Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond |
| 5-LOX | Inflammation, pain | −3.66 | ASP-170 GLU-622 ARG-401 PHE-402 LEU-615 LEU-615 ALA-398 | Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Sigma Conventional Hydrogen Bond |
| ALOX15 | Inflammation, immunity, neuroprotection | −9.36 | LEU-597 HIS-545 HIS-545 LEU-549 ASP-600 MET-640 ASP-550 ASP-550 LEU-215 GLU-650 GLU-650 | Pi-Alkyl Carbon Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Anion Pi-Alkyl Conventional Hydrogen Bond Pi-Anion |
| Proteins | Protein Function | Binding Energy (kcal/mol) | Binding Residues | Type |
|---|---|---|---|---|
| p38 MAPK | Inflammation, apoptosis, proliferation, differentiation | −6.21 | ASP-168 GLU-71 GLU-71 LYS-53 ILE-84 PHE-169 LEU-75 TYR-35 TYR-35 | Conventional Hydrogen Bond Pi-Alkyl Pi-Anion Conventional Hydrogen Bond Pi-Pi Stacked Pi-Alkyl Pi-Alkyl Pi-Pi Stacked Pi-Alkyl |
| JNK | Inflammation, apoptosis, proliferation, differentiation | −6.87 | VAL-78 LYS-93 LYS-93 LEU-206 LEU-206 GLU-147 ASN-194 SER-72 SER-72 SER-193 ASN-152 | Pi-Sigma Conventional Hydrogen Bond Pi-Alkyl Pi-Sigma Pi-Alkyl Conventional Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond |
| ERK5 | proliferation, differentiation, development | −6.31 | LEU-189 ASP-143 MET-140 ASN-63 GLY-62 ASP-138 GLY-67 VAL-69 LYS-84 | Pi-Alkyl Conventional Hydrogen Bond Conventional Hydrogen Bond van der Waals Amide-Pi Stacked Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Pi-Cation |
| EGFR | Inflammation, proliferation, differentiation | −6.51 | ARG-841 ARG-841 ASN-842 THR-854 GLU-762 LEU-718 LYS-745 VAL-726 MET-766 MET-793 LEU-844 ALA-743 | Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Alkyl Sulfur-X Conventional Hydrogen Bond Pi-Alkyl Pi-Alkyl |
| ERBB2 | Inflammation, proliferation | −8.35 | ALA-751 LEU-852 LEU-726 MET-801 MET-801 ARG-849 GLN-799 ASN-850 THR-862 GLY-729 LYS-753 MET-774 PHE-864 PHE-864 LEU-785 | Pi-Alkyl Pi-Sigma Pi-Sigma Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Carbon Hydrogen Bond Pi-Alkyl Pi-Alkyl Pi-Alkyl Pi-Pi T-Shaped Pi-Alkyl |
| Proteins | Protein Function | Binding Energy (kcal/mol) | Binding Residues | Type |
|---|---|---|---|---|
| TLR | Inflammation, immunity, survival, proliferation | −5.42 | ASN-103 LYS-104 HIS-78 ASN-79 ARG-80 GLN-54 TYR-56 LYS-33 | Carbon Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Amide-Pi Stacked Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl |
| NF-κB | Inflammation, immunity | −7.33 | LEU-21 LEU-21 CYS-99 VAL-29 THR-23 MET-65 MET-65 MET-65 GLU-97 ILE-165 LYS-44 LYS-44 MET-96 GLU-61 | Conventional Hydrogen Bond Pi-Sigma Conventional Hydrogen Bond Pi-Alkyl Carbon Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Pi-Sulfur Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Alkyl Pi-Alkyl Carbon Hydrogen Bond |
| TNF | Fever, proliferation, differentiation, inflammation, cytotoxicity | −5.62 | PRO-100 GLN-102 SER-99 SER-99 ARG-103 ARG-103 PRO-106 ASN-112 GLY-68 | Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Pi-Cation Carbon Hydrogen Bond Conventional Hydrogen Bond Carbon Hydrogen Bond |
| TNFR1A | Inflammation, apoptosis | −5.87 | GLY-364 ASP-357 GLU-360 GLU-360 LYS-343 LYS-343 LEU-359 MET-374 TRP-342 TRP-342 ALA-370 | Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Anion Conventional Hydrogen Bond Pi-Alkyl Pi-Alkyl Pi-Alkyl Pi-Alkyl Pi-Pi Stacked Carbon Hydrogen Bond |
| iNOS | Inflammation, apoptosis | −9.30 | MET-368 ILE-195 GLY-365 GLN-199 ASN-364 TRP-366 TRP-366 CYS-194 CYS-194 TYR-483 TRP-188 ALA-191 MET-349 PRO-192 TYR-485 | Pi-Alkyl Pi-Alkyl Pi-Sigma Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Sulfur Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Carbon Hydrogen Bond Pi-Sigma Conventional Hydrogen Bond Pi-Alkyl Pi-Alkyl |
| Proteins | Protein Function | Binding Energy (kcal/mol) | Binding Residues | Type |
|---|---|---|---|---|
| IL-6 | Inflammation, immunity, hematopoiesis | −5.62 | PHE-136 PHE-136 ARG-154 ASN-135 PRO-157 PRO-157 GLU-133 THR-130 GLY-127 GLU-129 | Conventional Hydrogen Bond Pi-Alkyl Pi-Sigma Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Carbon Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Pi-Anion |
| IL-23R | Inflammation, immunity | −6.20 | LYS-104 SER-203 PHE-106 LEU-107 LEU-107 LEU-107 TYR-201 ASN-200 THR-202 TRP-90 MET-189 | Conventional Hydrogen Bond Carbon Hydrogen Bond Pi-Pi T-Shaped Conventional Hydrogen Bond Pi-Alkyl Pi-Lone Pair Carbon Hydrogen Bond Conventional Hydrogen Bond Carbon Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl |
| IL-1β | Inflammation, immunity, proliferation, differentiation | −5.52 | GLU-64 LYS-63 VAL-40 MET-20 MET-20 GLN-38 VAL-41 LYS-65 GLU-37 GLN-39 MET-36 | Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Carbon Hydrogen Bond |
| IL-10 | Inflammation, immunity, apoptosis | −4.02 | PHE-143 VAL-124 GLU-142 ALA-139 LYS-138 LYS-138 | Carbon Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Sigma Amide-Pi Stacked Pi-Alkyl |
| SELE | Inflammation | −7.93 | CYS-325 THR-327 ASN-269 ALA-326 ASP-272 ASP-272 LEU-273 LYS-270 LYS-270 GLY-45 ARG-178 ARG-178 GLU-43 THR-48 VAL-179 SER-47 THR-177 LYS-51 | Sulfur-X Pi-Sigma Conventional Hydrogen Bond Pi-Alkyl Conventional Hydrogen Bond Pi-Alkyl Pi-Alkyl Conventional Hydrogen Bond Pi-Sigma Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Lone Pair Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Conventional Hydrogen Bond Pi-Lone Pair Conventional Hydrogen Bond |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Fang, Y.; Ma, Y.; Dawa, Y.; Wang, Q.; Gan, J.; Dang, J. Screening and Isolation of Potential Anti-Inflammatory Compounds from Saxifraga atrata via Affinity Ultrafiltration-HPLC and Multi-Target Molecular Docking Analyses. Nutrients 2022, 14, 2405. https://doi.org/10.3390/nu14122405
Li G, Fang Y, Ma Y, Dawa Y, Wang Q, Gan J, Dang J. Screening and Isolation of Potential Anti-Inflammatory Compounds from Saxifraga atrata via Affinity Ultrafiltration-HPLC and Multi-Target Molecular Docking Analyses. Nutrients. 2022; 14(12):2405. https://doi.org/10.3390/nu14122405
Chicago/Turabian StyleLi, Gang, Yan Fang, Yonggui Ma, Yangzom Dawa, Qilan Wang, Jing Gan, and Jun Dang. 2022. "Screening and Isolation of Potential Anti-Inflammatory Compounds from Saxifraga atrata via Affinity Ultrafiltration-HPLC and Multi-Target Molecular Docking Analyses" Nutrients 14, no. 12: 2405. https://doi.org/10.3390/nu14122405
APA StyleLi, G., Fang, Y., Ma, Y., Dawa, Y., Wang, Q., Gan, J., & Dang, J. (2022). Screening and Isolation of Potential Anti-Inflammatory Compounds from Saxifraga atrata via Affinity Ultrafiltration-HPLC and Multi-Target Molecular Docking Analyses. Nutrients, 14(12), 2405. https://doi.org/10.3390/nu14122405









