Association Study among Comethylation Modules, Genetic Polymorphisms and Clinical Features in Mexican Teenagers with Eating Disorders: Preliminary Results
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Evaluation Instruments
2.3. DNA Extraction and Microarray Analysis
2.4. Quality Control of Genotypification Data
2.5. Quality Control of DNA Methylation Data
2.6. Comethylation Modules Construction
2.7. Enrichment Analysis of Modules
2.8. Correlation of Comethylation Modules with Clinical Features and SNPs
3. Results
3.1. Description of Comethylation Modules
3.2. Enriched Pathways on Each Module
3.3. Correlations of Modules with Clinical Features in Our Population
3.4. Correlations of SNPs with Modules
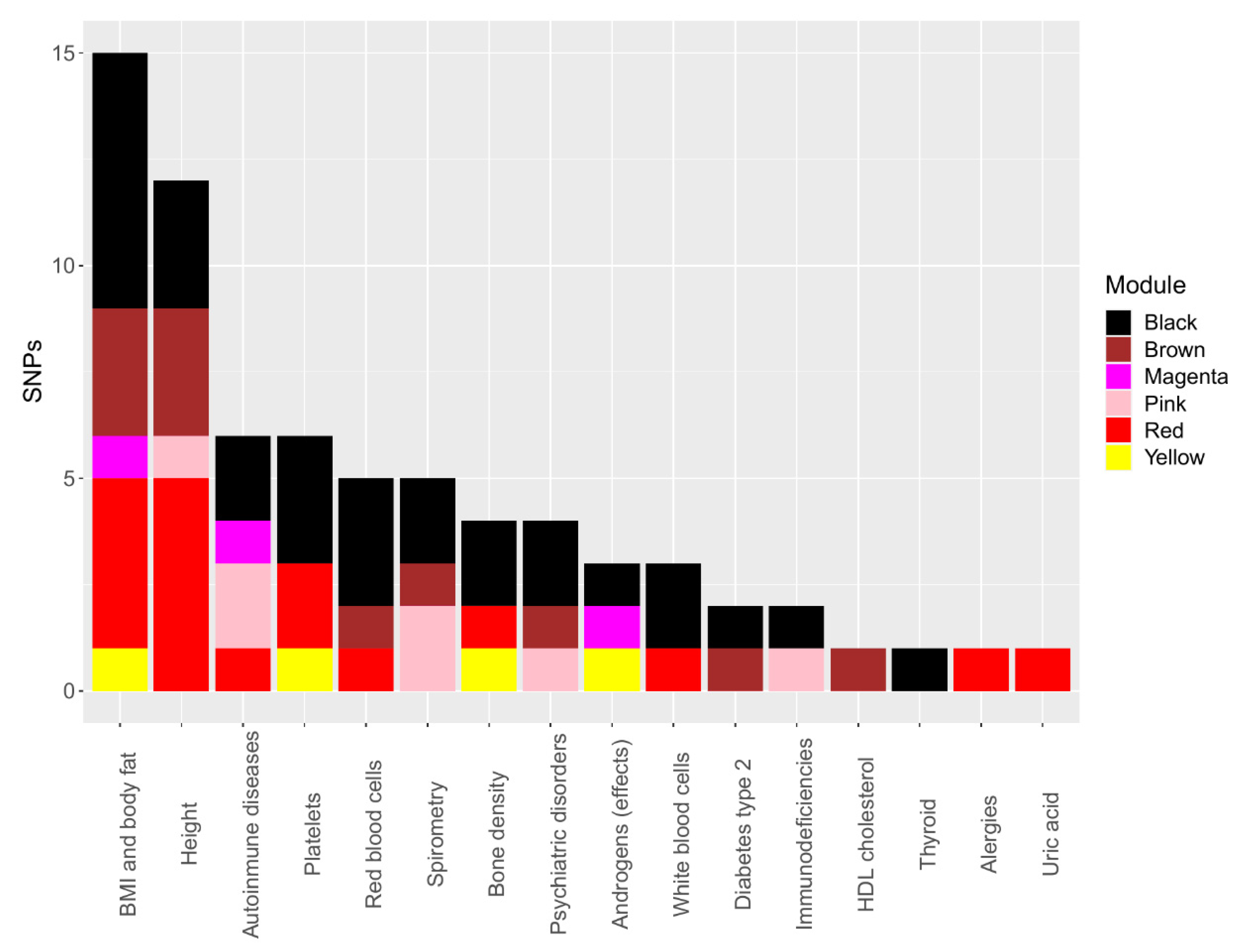
3.5. Correlated SNP PheWAS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bulik, C.M.; Blake, L.; Austin, J. Genetics of Eating Disorders: What the Clinician Needs to Know. Psychiatr. Clin. N. Am. 2019, 42, 59–73. [Google Scholar] [CrossRef]
- Steiger, H.; Booij, L. Eating Disorders, Heredity and Environmental Activation: Getting Epigenetic Concepts into Practice. J. Clin. Med. 2020, 9, 1332. [Google Scholar] [CrossRef] [PubMed]
- Duncan, L.; Yilmaz, Z.; Gaspar, H.; Walters, R.; Goldstein, J.; Anttila, V.; Bulik-Sullivan, B.; Ripke, S.; Thornton, L.; Hinney, A.; et al. Significant Locus and Metabolic Genetic Correlations Revealed in Genome-Wide Association Study of Anorexia Nervosa. Am. J. Psychiatry 2017, 174, 850–858. [Google Scholar] [CrossRef]
- Yilmaz, Z.; Hardaway, J.A.; Bulik, C.M. Genetics and Epigenetics of Eating Disorders. Adv. Genom. Genet. 2015, 5, 131–150. [Google Scholar] [CrossRef] [Green Version]
- Watson, H.J.; Yilmaz, Z.; Thornton, L.M.; Hübel, C.; Coleman, J.R.I.; Gaspar, H.A.; Bryois, J.; Hinney, A.; Leppä, V.M.; Mattheisen, M.; et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 2019, 51, 1207–1214. [Google Scholar] [CrossRef] [Green Version]
- Munn-Chernoff, M.A.; Johnson, E.C.; Chou, Y.-L.; Coleman, J.R.I.; Thornton, L.M.; Walters, R.K.; Yilmaz, Z.; Baker, J.H.; Hübel, C.; Gordon, S.; et al. Shared genetic risk between eating disorder- and substance-use-related phenotypes: Evidence from genome-wide association studies. Addict. Biol. 2021, 26, e12880. [Google Scholar] [CrossRef] [Green Version]
- Hübel, C.; Marzi, S.J.; Breen, G.; Bulik, C.M. Epigenetics in eating disorders: A systematic review. Mol. Psychiatry 2019, 24, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, M.L.; Martínez-Magaña, J.J.; Ruiz-Ramos, D.; García, A.R.; Gonzalez, L.; Tovilla-Zarate, C.A.; Sarmiento, E.; Juárez-Rojop, I.E.; Nicolini, H.; Gonzalez-Castro, T.B.; et al. Individuals Diagnosed with Binge-Eating Disorder Have DNA Hypomethylated Sites in Genes of the Metabolic System: A Pilot Study. Nutrients 2021, 13, 1413. [Google Scholar] [CrossRef]
- Zhao, W.; Langfelder, P.; Fuller, T.; Dong, J.; Li, A.; Hovarth, S. Weighted Gene Coexpression Network Analysis: State of the Art. J. Biopharm. Stat. 2010, 20, 281–300. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Barton, S.; Holbrook, J.D. How to make DNA methylome wide association studies more powerful. Epigenomics 2016, 8, 1117–1129. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- APA. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Arlington, VA, USA, 2013. [Google Scholar]
- Yanovski, S.Z.; Marcus, M.D.; Wadden, T.A.; Walsh, B.T. The Questionnaire on Eating and Weight Patterns-5: An updated screening instrument for binge eating disorder. Int. J. Eat. Disord. 2015, 48, 259–261. [Google Scholar] [CrossRef] [Green Version]
- Garner, D.M.; Olmsted, M.P.; Bohr, Y.; Garfinkel, P.E. The Eating Attitudes Test: Psychometric features and clinical correlates. Psychol. Med. 1982, 12, 871–878. [Google Scholar] [CrossRef]
- Sheehan, D.V.; Sheehan, K.H.; Shytle, R.D.; Janavs, J.; Bannon, Y.; Rogers, J.E.; Milo, K.M.; Stock, S.L.; Wilkinson, B. Reliability and validity of the Mini International Neuropsychiatric Interview for Children and Adolescents (MINI-KID). J. Clin. Psychiatry 2010, 71, 313–326. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Morris, T.J.; Butcher, L.M.; Feber, A.; Teschendorff, A.E.; Chakravarthy, A.R.; Wojdacz, T.K.; Beck, S. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics 2014, 30, 428–430. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.D. IlluminaHumanMethylationEPICanno.ilm10b4.hg19: Annotation for Illumina’s EPIC Methylation Arrays, R Package Version 0.6.0. 2017.
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Watanabe, K.; Stringer, S.; Frei, O.; Umićević Mirkov, M.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef]
- Genis-Mendoza, A.D.; Martínez-Magaña, J.J.; Ruiz-Ramos, D.; Gonzalez-Covarrubias, V.; Tovilla-Zarate, C.A.; Narvaez, M.L.L.; Castro, T.B.G.; Juárez-Rojop, I.E.; Nicolini, H. Interaction of FTO rs9939609 and the native American-origin ABCA1 p.Arg230Cys with circulating leptin levels in Mexican adolescents diagnosed with eating disorders: Preliminary results. Psychiatry Res. 2020, 291, 113270. [Google Scholar] [CrossRef] [PubMed]
- Hay, P. Current approach to eating disorders: A clinical update. Intern. Med. J. 2020, 50, 24–29. [Google Scholar] [CrossRef]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaioannou, V.E. T-box genes in development: From hydra to humans. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 2001; Volume 207, pp. 1–70. [Google Scholar]
- Singh, M.K.; Petry, M.; Haenig, B.; Lescher, B.; Leitges, M.; Kispert, A. The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev. 2005, 122, 131–144. [Google Scholar] [CrossRef]
- Lee, K.Y.; Singh, M.K.; Ussar, S.; Wetzel, P.; Hirshman, M.F.; Goodyear, L.J.; Kispert, A.; Kahn, C.R. Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat. Commun. 2015, 6, 8054. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Sharma, R.; Gase, G.; Ussar, S.; Li, Y.; Welch, L.; Berryman, D.E.; Kispert, A.; Bluher, M.; Kahn, C.R. Tbx15 Defines a Glycolytic Subpopulation and White Adipocyte Heterogeneity. Diabetes 2017, 66, 2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Zhang, D.; Huang, H.; Yu, Y.; Yang, Z.; Niu, Y.; Liu, J. MicroRNA-1225-5p acts as a tumor-suppressor in laryngeal cancer via targeting CDC14B. Biol. Chem. 2019, 400, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Levy, J.D.; Zhang, Y.; Frontini, A.; Kolodin, D.P.; Svensson, K.J.; Lo, J.C.; Zeng, X.; Ye, L.; Khandekar, M.J.; et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell 2014, 156, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Arensdorf, A.M.; Dillard, M.E.; Menke, J.M.; Frank, M.W.; Rock, C.O.; Ogden, S.K. Sonic Hedgehog Activates Phospholipase A2 to Enhance Smoothened Ciliary Translocation. Cell Rep. 2017, 19, 2074–2087. [Google Scholar] [CrossRef] [Green Version]
- Iyer, A.; Lim, J.; Poudyal, H.; Reid, R.C.; Suen, J.Y.; Webster, J.; Prins, J.B.; Whitehead, J.P.; Fairlie, D.P.; Brown, L. An Inhibitor of Phospholipase A2; Group IIA Modulates Adipocyte Signaling and Protects Against Diet-Induced Metabolic Syndrome in Rats. Diabetes 2012, 61, 2320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuefner, M.S.; Deng, X.; Stephenson, E.J.; Pham, K.; Park, E.A. Secretory phospholipase A2 group IIA enhances the metabolic rate and increases glucose utilization in response to thyroid hormone. FASEB J. 2019, 33, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Taketomi, Y.; Ushida, A.; Isogai, Y.; Kojima, T.; Hirabayashi, T.; Miki, Y.; Yamamoto, K.; Nishito, Y.; Kobayashi, T.; et al. The Adipocyte-Inducible Secreted Phospholipases PLA2G5 and PLA2G2E Play Distinct Roles in Obesity. Cell Metab. 2014, 20, 119–132. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.-Y.; Fauman, E.B.; Petersen, A.-K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.-P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Coan, P.M.; Hummel, O.; Garcia Diaz, A.; Barrier, M.; Alfazema, N.; Norsworthy, P.J.; Pravenec, M.; Petretto, E.; Hübner, N.; Aitman, T.J. Genetic, physiological and comparative genomic studies of hypertension and insulin resistance in the spontaneously hypertensive rat. Dis. Models Mech. 2017, 10, 297–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M.; et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Belanger, K.; Nutter, C.A.; Li, J.; Tasnim, S.; Liu, P.; Yu, P.; Kuyumcu-Martinez, M.N. CELF1 contributes to aberrant alternative splicing patterns in the type 1 diabetic heart. Biochem. Biophys. Res. Commun. 2018, 503, 3205–3211. [Google Scholar] [CrossRef]
- Chang, K.-T.; Cheng, C.-F.; King, P.-C.; Liu, S.-Y.; Wang, G.-S. CELF1 Mediates Connexin 43 mRNA Degradation in Dilated Cardiomyopathy. Circ. Res. 2017, 121, 1140–1152. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Tao, Y.; Zhou, H.; Lai, H. Promoting role of circ-Jarid2/miR-129-5p/Celf1 axis in cardiac hypertrophy. Gene Ther. 2020, 27, 1–11. [Google Scholar] [CrossRef]
- Sciarretta, S.; Forte, M.; Frati, G.; Sadoshima, J. New Insights into the Role of mTOR Signaling in the Cardiovascular System. Circ. Res. 2018, 122, 489–505. [Google Scholar] [CrossRef]
- Sciarretta, S.; Volpe, M.; Sadoshima, J. Mammalian Target of Rapamycin Signaling in Cardiac Physiology and Disease. Circ. Res. 2014, 114, 549–564. [Google Scholar] [CrossRef] [Green Version]
- Fayssoil, A.; Melchior, J.C.; Hanachi, M. Heart and anorexia nervosa. Heart Fail. Rev. 2021, 26, 65–70. [Google Scholar] [CrossRef]
- Westmoreland, P.; Krantz, M.J.; Mehler, P.S. Medical Complications of Anorexia Nervosa and Bulimia. Am. J. Med. 2016, 129, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouidrat, Y.; Amad, A.; Lalau, J.-D.; Loas, G. Eating Disorders in Schizophrenia: Implications for Research and Management. Schizophr. Res. Treat. 2014, 2014, 791573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Cheng, D.; Zhang, C.; Li, Y.; Zhang, Z.; Wang, J.; Shi, Y. Association of PDE4B Polymorphisms with Susceptibility to Schizophrenia: A Meta-Analysis of Case-Control Studies. PLoS ONE 2016, 11, e0147092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardiñas, A.F.; Holmans, P.; Pocklington, A.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.; et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018, 50, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Astle, W.J.; Elding, H.; Jiang, T.; Allen, D.; Ruklisa, D.; Mann, A.L.; Mead, D.; Bouman, H.; Riveros-Mckay, F.; Kostadima, M.A.; et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 2016, 167, 1415–1429.e1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.-G.; Juran, B.D.; Mucha, S.; Folseraas, T.; Jostins, L.; Melum, E.; Kumasaka, N.; Atkinson, E.J.; Schlicht, E.M.; Liu, J.Z.; et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat. Genet. 2017, 49, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Andersen, G.; Yorgov, D.; Ferrara, T.M.; Ben, S.; Brownson, K.M.; Holland, P.J.; Birlea, S.A.; Siebert, J.; Hartmann, A.; et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 2016, 48, 1418–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronson, P.G.; Chang, D.; Bhangale, T.; Seldin, M.F.; Ortmann, W.; Ferreira, R.C.; Urcelay, E.; Pereira, L.F.; Martin, J.; Plebani, A.; et al. Common variants at PVT1, ATG13–AMBRA1, AHI1 and CLEC16A are associated with selective IgA deficiency. Nat. Genet. 2016, 48, 1425–1429. [Google Scholar] [CrossRef] [Green Version]
- Julià, A.; López-Longo, F.J.; Pérez Venegas, J.J.; Bonàs-Guarch, S.; Olivé, À.; Andreu, J.L.; Aguirre-Zamorano, M.Á.; Vela, P.; Nolla, J.M.; de la Fuente, J.L.M.; et al. Genome-wide association study meta-analysis identifies five new loci for systemic lupus erythematosus. Arthritis Res. Ther. 2018, 20, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raevuori, A.; Haukka, J.; Vaarala, O.; Suvisaari, J.M.; Gissler, M.; Grainger, M.; Linna, M.S.; Suokas, J.T. The Increased Risk for Autoimmune Diseases in Patients with Eating Disorders. PLoS ONE 2014, 9, e104845. [Google Scholar] [CrossRef]
- Zerwas, S.; Larsen, J.T.; Petersen, L.; Thornton, L.M.; Quaranta, M.; Koch, S.V.; Pisetsky, D.; Mortensen, P.B.; Bulik, C.M. Eating Disorders, Autoimmune, and Autoinflammatory Disease. Pediatrics 2017, 140, e20162089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almstrup, K.; Lindhardt Johansen, M.; Busch, A.S.; Hagen, C.P.; Nielsen, J.E.; Petersen, J.H.; Juul, A. Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Sci. Rep. 2016, 6, 28657. [Google Scholar] [CrossRef]
- Abreu, A.P.; Kaiser, U.B. Pubertal development and regulation. Lancet Diabetes Endocrinol. 2016, 4, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Herbison, A.E. Control of puberty onset and fertility by gonadotropin-releasing hormone neurons. Nat. Rev. Endocrinol. 2016, 12, 452–466. [Google Scholar] [CrossRef]
- Livadas, S.; Chrousos, G.P. Control of the onset of puberty. Curr. Opin. Pediatrics 2016, 28, 551–558. [Google Scholar] [CrossRef]
- Fang, H.; Judd, R.L. Adiponectin Regulation and Function. Compr. Physiol. 2018, 8, 1031–1063. [Google Scholar] [CrossRef]
- Wang, Z.V.; Scherer, P.E. Adiponectin, the past two decades. J. Mol. Cell Biol. 2016, 8, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Sitticharoon, C.; Sukharomana, M.; Likitmaskul, S.; Churintaraphan, M.; Maikaew, P. Increased high molecular weight adiponectin, but decreased total adiponectin and kisspeptin, in central precocious puberty compared with aged-matched prepubertal girls. Reprod. Fertil. Dev. 2017, 29, 2466–2478. [Google Scholar] [CrossRef]
- Woo, J.G.; Dolan, L.M.; Daniels, S.R.; Goodman, E.; Martin, L.J. Adolescent Sex Differences in Adiponectin Are Conditional on Pubertal Development and Adiposity. Obes. Res. 2005, 13, 2095–2101. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA Package FAQ. Available online: https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/faq.html (accessed on 24 August 2021).

| Features | Sample (n = 50) |
|---|---|
| Age (years) | 13.98 ± 1.74 |
| Gender | |
| Male | 13 (26) |
| Female | 37 (74) |
| Body Mass Index (BMI) zscore | 1.03 ± 0.97 |
| BMI classification | |
| Underweight | 1 (2) |
| Normal weight | 20 (40) |
| Overweight | 11 (22) |
| Obesity | 18 (36) |
| Diagnosis | |
| Binge eating disorder | 17 (34) |
| Bulimia nervosa | 22 (44) |
| Anorexia nervosa | 11 (22) |
| Comorbidities | |
| Any | 46 (92) |
| Major depressive disorder | 21 (42) |
| Suicide behavior | 16 (32) |
| Dysthymia disorder | 18 (36) |
| Attention-Deficit/Hyperactivity Disorder | 15 (30) |
| Generalized Anxiety Disorder | 10 (20) |
| Oppositional Defiant Disorder | 6 (12) |
| Conduct Disorder | 5 (10) |
| Psychotic Disorder | 5 (10) |
| Eating Attitudes | |
| Fear of gain weight | 35 (70) |
| Binge | 34 (68) |
| Restriction | 24 (48) |
| Vomit | 21 (42) |
| Other behaviors | 10 (20) |
| Module | TSS1500 | TSS200 | 5′UTR | Body | 1stExon | ExonBnd | 3′UTR | |
|---|---|---|---|---|---|---|---|---|
| Turquoise | 359 (10.23) | 97 (2.77) | 436 (12.43) | 2430 (69.27) | 33 (0.94) | 37 (1.05) | 116 (3.31) | |
| Blue | 158 (7.85) | 75 (3.72) | 309 (15.34) | 1366 (67.83) | 23 (1.14) | 16 (0.79) | 67 (3.33) | |
| Brown | 14 (10.77) | 4 (3.08) | 23 (17.69) | 78 (60.00) | 2 (1.54) | 3 (2.31) | 6 (4.62) | |
| Yellow | 13 (12.15) | 6 (5.61) | 13 (12.15) | 69 (64.49) | 2 (1.87) | 0 (0) | 4 (3.74) | |
| Green | 11 (11.22) | 3 (3.06) | 13 (13.27) | 68 (69.39) | 1 (1.02) | 1 (1.02) | 1 (1.02) | |
| Red | 20 (18.52) | 1 (0.93) | 15 (13.89) | 66 (61.11) | 2 (1.85) | 1 (0.93) | 3 (2.78) | |
| Black | 11 (11.22) | 2 (2.04) | 9 (9.18) | 66 (67.35) | 2 (2.04) | 2 (2.04) | 6 (6.12) | |
| Pink | 13 (13.00) | 2 (2.00) | 9 (9.00) | 71 (71.00) | 2 (2.00) | 1 (1.00) | 2 (2.00) | |
| Magenta | 10 (11.36) | 4 (4.55) | 12 (13.64) | 56 (63.64) | 1 (1.14) | 0 (0) | 5 (5.68) | |
| Purple | 8 (11.27) | 5 (7.04) | 8 (11.27) | 40 (56.34) | 1 (1.41) | 2 (2.82) | 7 (9.86) | |
| Module | OpenSea | Island | N Shore | S Shore | N Shelf | S Shelf | |
|---|---|---|---|---|---|---|---|
| Turquoise | 4207 (82.93) | 13 (0.26) | 247 (4.87) | 212 (4.18) | 202 (3.98) | 192 (3.78) | |
| Blue | 2476 (84.56) | 15 (0.51) | 128 (4.37) | 83 (2.83) | 104 (3.55) | 122 (4.17) | |
| Brown | 153 (79.27) | 6 (3.11) | 9 (4.66) | 17 (8.81) | 4 (2.07) | 4 (2.07) | |
| Yellow | 125 (75.30) | 1 (0.60) | 14 (8.43) | 10 (6.02) | 12 (7.23) | 4 (2.41) | |
| Green | 114 (75.50) | 5 (3.31) | 10 (6.62) | 8 (5.30) | 9 (5.96) | 5 (3.31) | |
| Red | 121 (80.67) | 1 (0.67) | 6 (4.00) | 2 (1.33) | 8 (5.33) | 12 (8.00) | |
| Black | 115 (77.70) | 1 (0.68) | 7 (4.73) | 6 (4.05) | 11 (7.43) | 8 (5.41) | |
| Pink | 114 (78.62) | 6 (4.14) | 7 (4.83) | 7 (4.83) | 8 (5.52) | 3 (2.07) | |
| Magenta | 96 (71.11) | 6 (4.44) | 9 (6.67) | 9 (6.67) | 8 (5.93) | 7 (5.19) | |
| Purple | 85 (76.58) | 17 (15.32) | 4 (3.60) | 3 (2.70) | 1 (0.90) | 1 (0.90) | |
| Total | 7606 (82.67) | 71 (0.77) | 441 (4.79) | 357 (3.88) | 367 (3.99) | 358 (3.89) | |
| Brown | Green | Yellow | Magenta | Red | Black | Pink | Total | |
|---|---|---|---|---|---|---|---|---|
| 3′UTR | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (5.88) | 1 (1.89) | 0 (0.00) | 0 (0.00) | 2 (0.89) |
| Downstream | 0 (0.00) | 2 (6.25) | 1 (3.70) | 0 (0.00) | 4 (7.55) | 1 (2.44) | 1 (5.00) | 9 (4) |
| Intergenic | 7 (17.95) | 9 (28.13) | 4 (14.81) | 5 (29.41) | 13 (24.53) | 13 (31.71) | 4 (20.00) | 55 (24.4) |
| Intron | 19 (48.72) | 7 (21.88) | 15 (55.56) | 7 (41.18) | 18 (33.96) | 14 (34.15) | 7 (35.00) | 87 (38.67) |
| Missense | 3 (7.69) | 1 (3.13) | 2 (7.41) | 1 (5.88) | 4 (7.55) | 4 (9.76) | 2 (10.00) | 17 (7.56) |
| Non coding transcript | 3 (7.69) | 8 (28.13) | 2 (7.41) | 1 (5.88) | 6 (13.21) | 5 (14.64) | 3 (20.00) | 28 (12.44) |
| Regulatory | 1 (2.56) | 3 (9.38) | 0 (0.00) | 2 (11.76) | 2 (3.77) | 1 (2.44) | 1 (5.00) | 10 (4.44) |
| Synonymous | 2 (5.13) | 0 (0.00) | 2 (7.41) | 0 (0.00) | 2 (3.77) | 0 (0.00) | 0 (0.00) | 6 (2.67) |
| Upstream | 4 (10.26) | 1 (3.13) | 1 (3.70) | 0 (0.00) | 2 (3.77) | 2 (4.88) | 1 (5.00) | 11 (4.89) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nolasco-Rosales, G.A.; Martínez-Magaña, J.J.; Juárez-Rojop, I.E.; González-Castro, T.B.; Tovilla-Zarate, C.A.; García, A.R.; Sarmiento, E.; Ruiz-Ramos, D.; Genis-Mendoza, A.D.; Nicolini, H. Association Study among Comethylation Modules, Genetic Polymorphisms and Clinical Features in Mexican Teenagers with Eating Disorders: Preliminary Results. Nutrients 2021, 13, 3210. https://doi.org/10.3390/nu13093210
Nolasco-Rosales GA, Martínez-Magaña JJ, Juárez-Rojop IE, González-Castro TB, Tovilla-Zarate CA, García AR, Sarmiento E, Ruiz-Ramos D, Genis-Mendoza AD, Nicolini H. Association Study among Comethylation Modules, Genetic Polymorphisms and Clinical Features in Mexican Teenagers with Eating Disorders: Preliminary Results. Nutrients. 2021; 13(9):3210. https://doi.org/10.3390/nu13093210
Chicago/Turabian StyleNolasco-Rosales, Germán Alberto, José Jaime Martínez-Magaña, Isela Esther Juárez-Rojop, Thelma Beatriz González-Castro, Carlos Alfonso Tovilla-Zarate, Ana Rosa García, Emmanuel Sarmiento, David Ruiz-Ramos, Alma Delia Genis-Mendoza, and Humberto Nicolini. 2021. "Association Study among Comethylation Modules, Genetic Polymorphisms and Clinical Features in Mexican Teenagers with Eating Disorders: Preliminary Results" Nutrients 13, no. 9: 3210. https://doi.org/10.3390/nu13093210
APA StyleNolasco-Rosales, G. A., Martínez-Magaña, J. J., Juárez-Rojop, I. E., González-Castro, T. B., Tovilla-Zarate, C. A., García, A. R., Sarmiento, E., Ruiz-Ramos, D., Genis-Mendoza, A. D., & Nicolini, H. (2021). Association Study among Comethylation Modules, Genetic Polymorphisms and Clinical Features in Mexican Teenagers with Eating Disorders: Preliminary Results. Nutrients, 13(9), 3210. https://doi.org/10.3390/nu13093210









