Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Survival to Gastrointestinal Trait
2.3. Peptidase Activities
2.4. Hydrolysis of Immunogenic Epitopes
2.5. Gluten Degradation under Simulated Gastrointestinal Conditions
2.6. Immunogenicity Estimation of Gluten Digests Using Duodenal Explants from Celiac Disease Patients
2.7. Statistical Analyses
3. Results
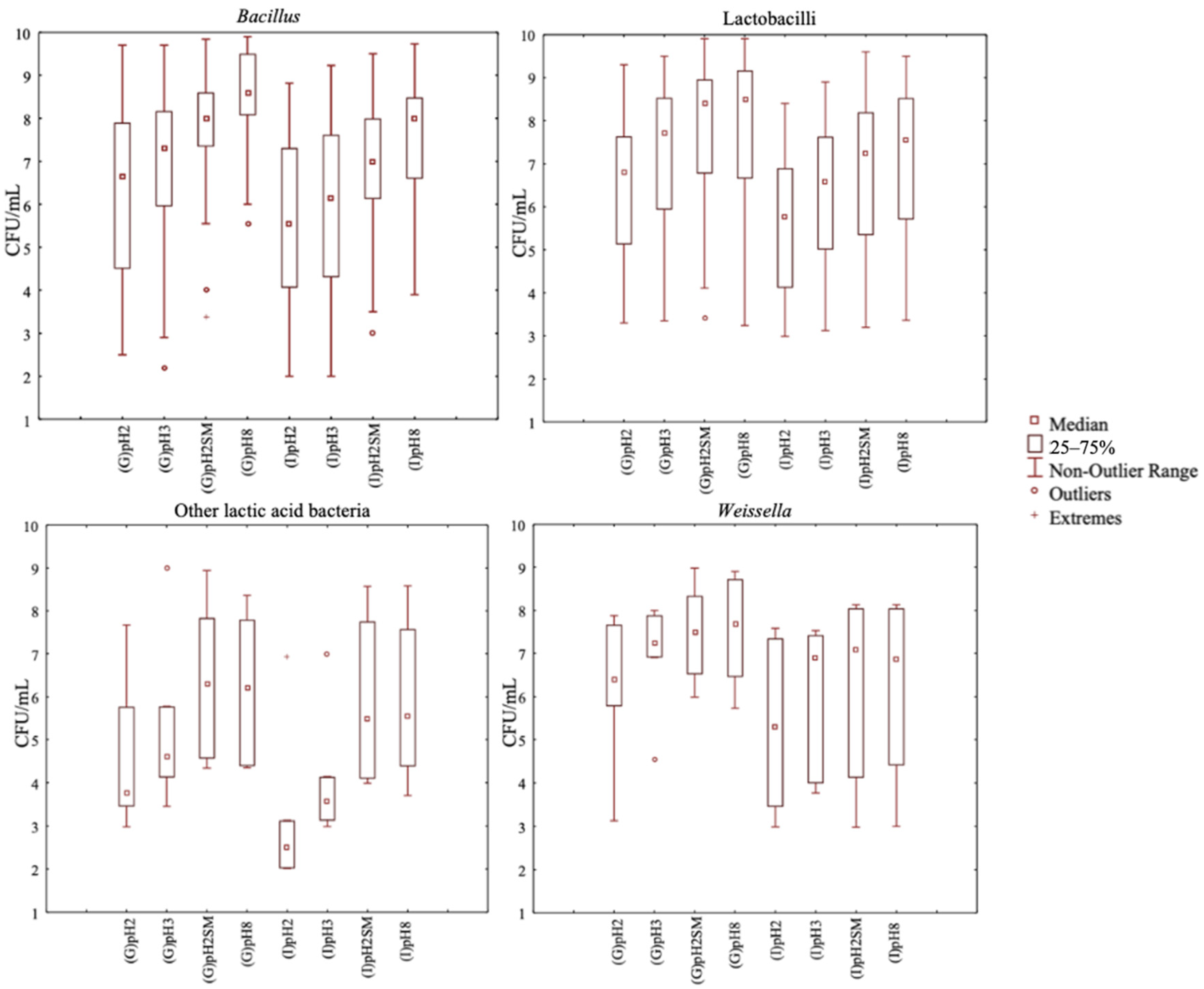
3.1. Selection of Bacterial Strains Resistant to Gastrointestinal Conditions
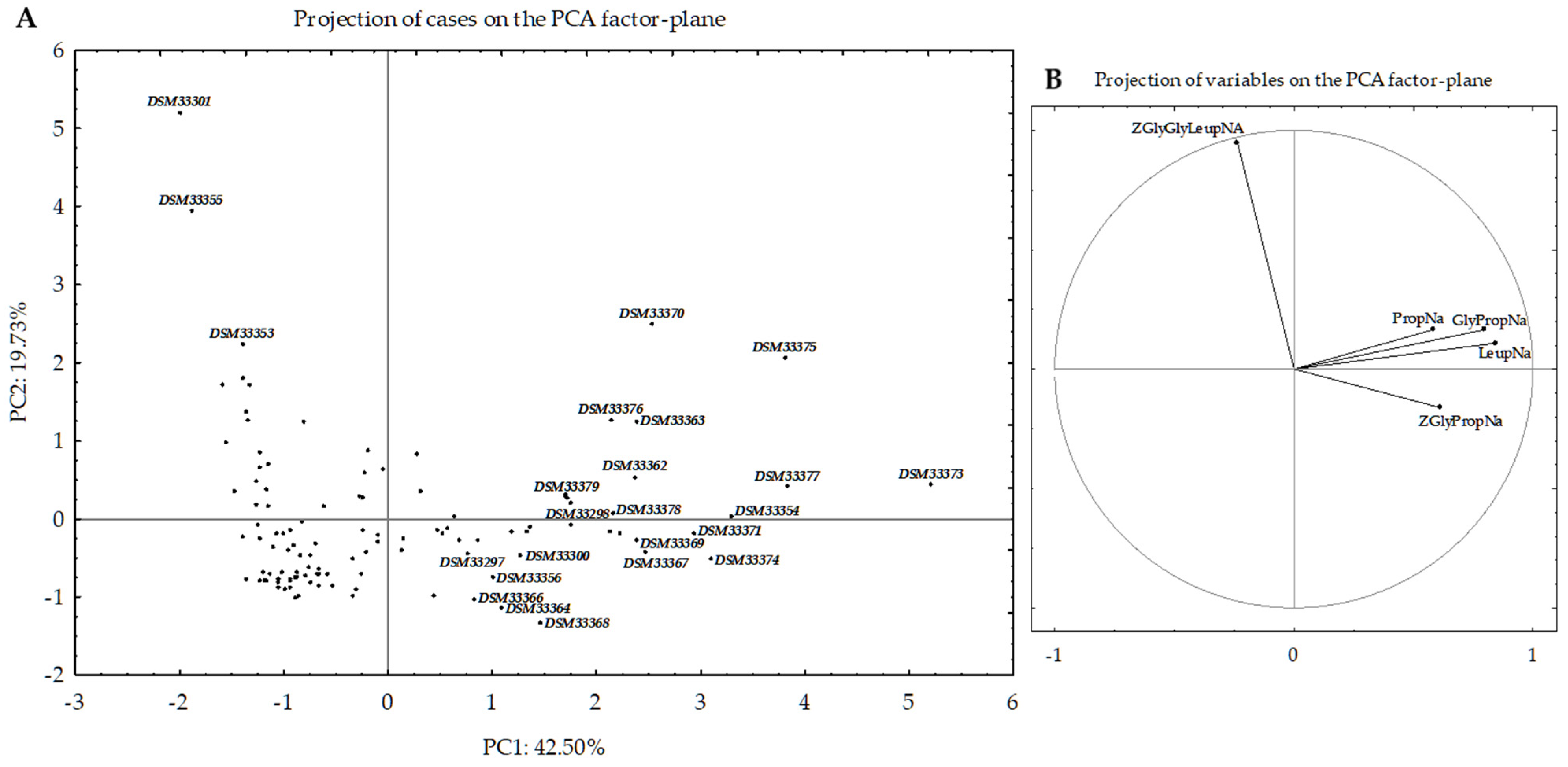
3.2. Selection of Bacterial Strains Based on Peptidase Activities
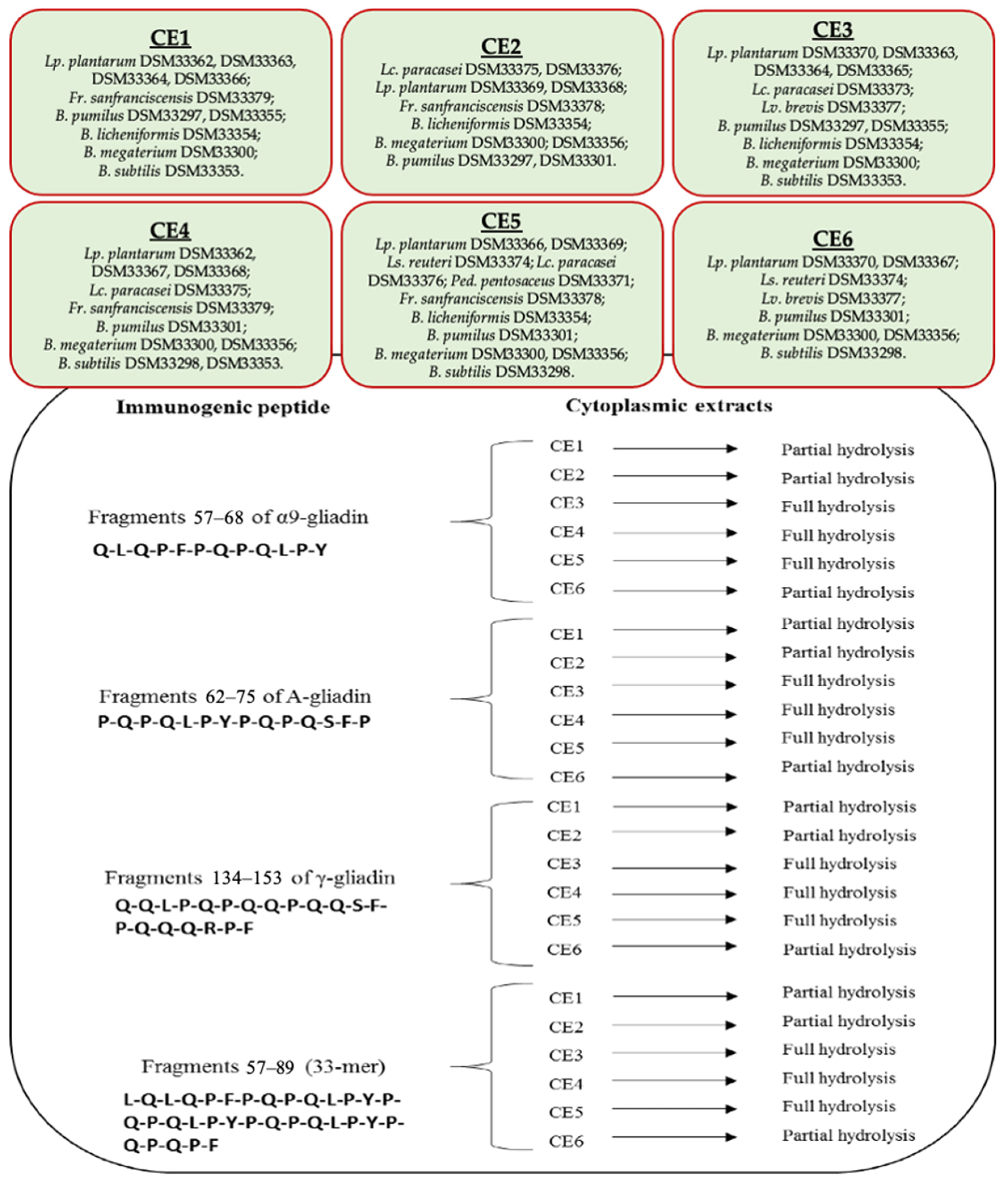
3.3. Hydrolysis of Gluten Immunogenic Epitopes
3.4. Gluten Degradation
- C3 consortium: Lp. plantarum DSM33370, DSM33363, and DSM33364; Lc. paracasei DSM33373; Lv. brevis DSM33377; B. pumilus DSM33297 and DSM33355; B. licheniformis DSM33354; B. megaterium DSM33300; and B. subtilis DSM33353.
- C4 consortium: Lp. plantarum DSM33362, DSM33367, and DSM33368; Lc. paracasei DSM33375; Fr. sanfranciscensis DSM33379; B. pumilus DSM33301; B. megaterium DSM33300 and DSM33356; and B. subtilis DSM33298 and DSM33353.
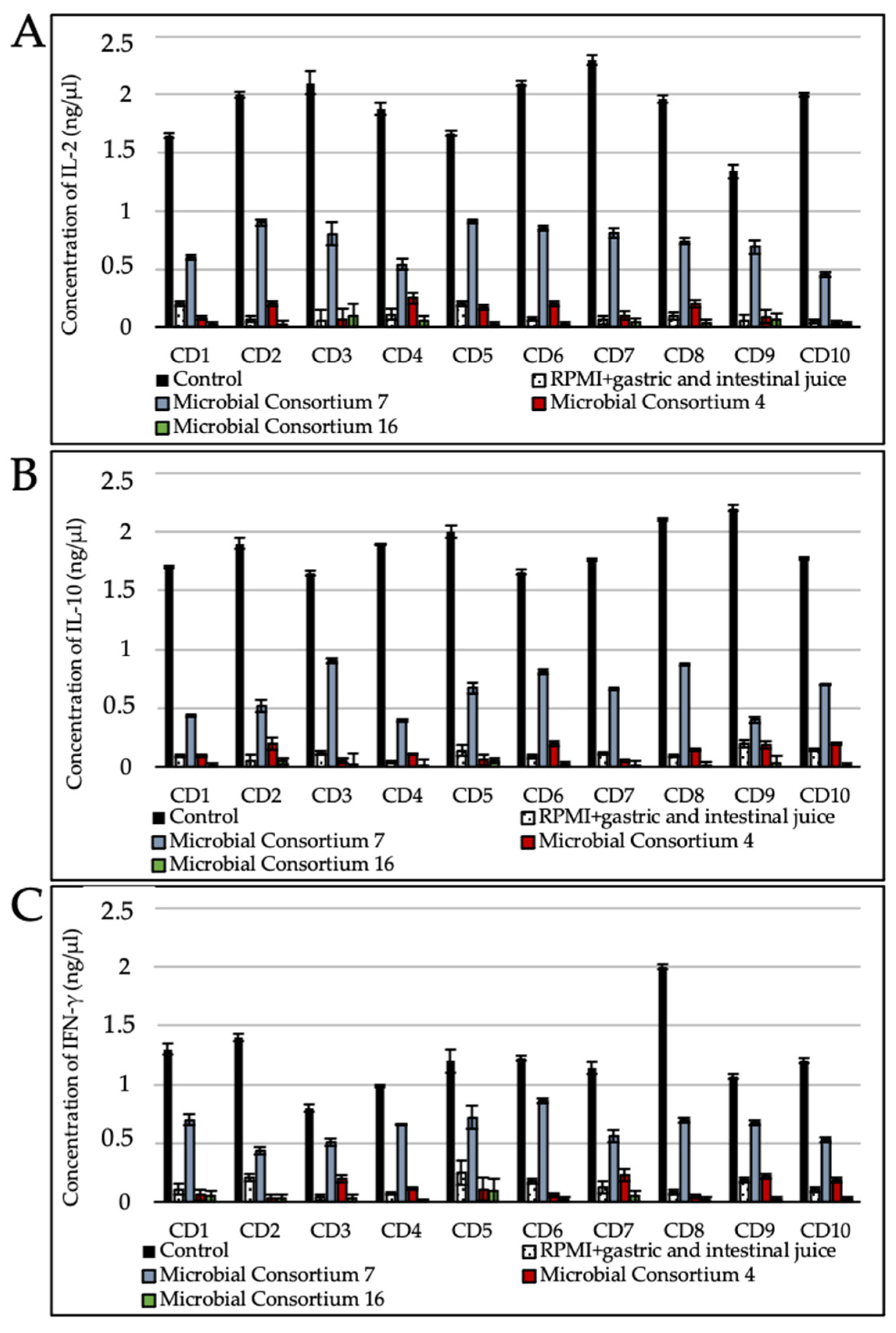
3.5. Gluten Digest Immunogenicity toward Duodenal Explants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biesiekierski, J.R. What is gluten? J. Gastroenterol. Hepatol. 2017, 32, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Khan, H. Genetic improvement for end-use quality in wheat. In Quality Breeding in Field Crops; Qureshi, A., Dar, Z., Wani, S., Eds.; Springer: Cham, Switzerland, 2019; pp. 239–253. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G.; Lafiandra, D. Genetics of wheat gluten proteins. In Advances in Genetics; Hall, J.C., Dunlap, J.C., Friedmann, T., Eds.; Academic Press: Cambridge, MA, USA, 2003; Volume 49, pp. 111–184. [Google Scholar] [CrossRef]
- Arentz–Hansen, H.; Mcadam, S.N.; Molberg, Ø.; Fleckenstein, B.; Lundin, K.E.; Jørgensen, T.J.; Jung, G.; Roepstorff, P.; Sollid, L.M. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology 2002, 123, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Barro Losada, F.; Lehisa, J.; Giménez, M.J.; García-Molina, M.D.; Ozuna, C.; Comino, I.; Sousa Martín, C.; Gil Humanes, J. Targeting of prolamins by RNAi in bread wheat: Effectiveness of seven silencing-fragment combinations for obtaining lines devoid of coeliac disease epitopes from highly immunogenic gliadins. Plant Biotechnol. J. 2016, 14, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Dewar, D.H.; Amato, M.; Ellis, H.J.; Pollock, E.L.; Gonzalez-Cinca, N.; Wieser, H.; Ciclitira, P.J. The toxicity of high molecular weight glutenin subunits of wheat to patients with coeliac disease. Eur. J. Gastroenterol. Hepatol. 2006, 18, 483–491. [Google Scholar] [CrossRef]
- Stamnaes, J.; Sollid, L.M. Celiac disease: Autoimmunity in response to food antigen. In Seminars in Immunology; Academic Press: Cambridge, MA, USA, 2015; Volume 27, pp. 343–352. [Google Scholar] [CrossRef]
- Potter, M.D.; Walker, M.M.; Keely, S.; Talley, N.J. What’s in a name? ‘Non-coeliac gluten or wheat sensitivity’: Controversies and mechanisms related to wheat and gluten causing gastrointestinal symptoms or disease. Gut 2018, 67, 2073–2077. [Google Scholar] [CrossRef]
- D’argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Del Vecchio Blanco, G.; et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 2016, 111, 879. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Laurikka, P.; Lindfors, K.; Collin, P.; Salmi, T.; Lähdeaho, M.L.; Saavalainen, O.; Mäki, M.; Mättö, J.; Kurppa, K.; et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am. J. Gastroenterol. 2014, 109, 1933–1941. [Google Scholar] [CrossRef]
- Niland, B.; Cash, B.D. Health benefits and adverse effects of a gluten-free diet in non–celiac disease patients. Gastroenterol. Hepatol. 2018, 14, 82. [Google Scholar]
- Larretxi, I.; Simon, E.; Benjumea, L.; Miranda, J.; Bustamante, M.A.; Lasa, A.; Churruca, I. Gluten-free-rendered products contribute to imbalanced diets in children and adolescents with celiac disease. Eur. J. Nutr. 2018, 58, 775–783. [Google Scholar] [CrossRef]
- Wild, D.; Robins, G.G.; Burley, V.J.; Howdle, P.D. Evidence of high sugar intake, and low fibre and mineral intake, in the gluten-free diet. Aliment. Pharmacol. Ther. 2010, 32, 573–581. [Google Scholar] [CrossRef]
- Herrán, A.R.; Pérez-Andrés, J.; Caminero, A.; Nistal, E.; Vivas, S.; de Morales, J.M.R.; Casqueiro, J. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res. Microbiol. 2017, 168, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Pontonio, E.; Filannino, P.; Rizzello, C.G.; De Angelis, M.; Di Cagno, R. How to improve the gluten-free diet: The state of the art from a food science perspective. Food Res. Int. 2018, 110, 22–32. [Google Scholar] [CrossRef]
- Ribeiro, M.; Nunes, F.M.; Rodriguez-Quijano, M.; Carrillo, J.M.; Branlard, G.; Igrejas, G. Next-generation therapies for celiac disease: The gluten-targeted approaches. Trends Food Sci. Technol. 2018, 75, 56–71. [Google Scholar] [CrossRef]
- Francavilla, R.; Piccolo, M.; Francavilla, A.; Polimeno, L.; Semeraro, F.; Cristofori, F.; Castellaneta, S.; Barone, M.; Indrio, F.; Gobbetti, M.; et al. Clinical and microbiological effect of a multispecies probiotic supplementation in celiac patients with persistent IBS-type symptoms: A randomized, double-blind, placebo-controlled, multicenter trial. J. Clin. Gastroenterol. 2019, 53, e117. [Google Scholar] [CrossRef]
- Cavaletti, L.; Taravella, A.; Carrano, L.; Carenzi, G.; Sigurtà, A.; Solinas, N.; De Caro, S.; Di Stasio, L.; Picascia, S.; Laezza, M.; et al. E40, a novel microbial protease efficiently detoxifying gluten proteins, for the dietary management of gluten intolerance. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Serena, G.; Kelly, C.P.; Fasano, A. Nondietary therapies for celiac disease. Gastroenterol. Clin. N. Am. 2019, 48, 145–163. [Google Scholar] [CrossRef]
- Caputo, I.; Lepretti, M.; Martucciello, S.; Esposito, C. Enzymatic strategies to detoxify gluten: Implications for celiac disease. Enzym. Res. 2010, 2010, 174354. [Google Scholar] [CrossRef]
- de Sousa Moraes, L.F.; Grzeskowiak, L.M.; de Sales Teixeira, T.F.; Peluzio, M.D.C.G. Intestinal microbiota and probiotics in celiac disease. Clin. Microbiol. Rev. 2014, 27, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Plugis, N.M.; Khosla, C. Therapeutic approaches for celiac disease. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 503–521. [Google Scholar] [CrossRef]
- Francavilla, R.; Cristofori, F.; Vacca, M.; Barone, M.; De Angelis, M. Advances in understanding the potential therapeutic applications of gut microbiota and probiotic mediated therapies in celiac disease. Expert Rev. Gastroenterol. Hepatol. 2020, 14, 323–333. [Google Scholar] [CrossRef]
- Mickowska, B.; Socha, P.; Urminská, D. Immunochemical evaluation of proteolysis of cereal proteins causing celiac disease by microbial proteases. Food Agric. Immunol. 2016, 27, 743–757. [Google Scholar] [CrossRef]
- Krishnareddy, S.; Stier, K.; Recanati, M.; Lebwohl, B.; Green, P.H. Commercially available glutenases: A potential hazard in coeliac disease. Ther. Adv. Gastroenterol. 2017, 10, 473–481. [Google Scholar] [CrossRef]
- Francavilla, R.; De Angelis, M.; Rizzello, C.G.; Cavallo, N.; Dal Bello, F.; Gobbetti, M. Selected probiotic lactobacilli have the capacity to hydrolyze gluten peptides during simulated gastrointestinal digestion. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.F.; Boris, S.; Barbes, C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 2003, 94, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Zárate, G.; Chaia, A.P.; González, S.; Oliver, G. Viability and beta-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 2000, 63, 1214–1221. [Google Scholar] [CrossRef]
- De Angelis, M.; Siragusa, S.; Berloco, M.; Caputo, L.; Settanni, L.; Alfonsi, G.; Amerio, M.; Grandi, A.; Ragni, A.; Gobbetti, M. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 2006, 157, 792–801. [Google Scholar] [CrossRef]
- De Angelis, M.; Cassone, A.; Rizzello, C.G.; Gagliardi, F.; Minervini, F.; Calasso, M.; Di Cagno, R.; Francavilla, R.; Gobbetti, M. Mechanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl. Environ. Microbiol. 2010, 76, 508–518. [Google Scholar] [CrossRef]
- Walker-Smith, J.A.G.S. Revised criteria for diagnosis of celiac disease. Arch. Dis. Child. 1990, 65, 909–911. [Google Scholar]
- Browning, T.H.; Trier, J.S. Organ culture of mucosal biopsies of human small intestine. J. Clin. Investig. 1969, 48, 1423–1432. [Google Scholar] [CrossRef]
- De Angelis, M.; Rizzello, C.G.; Fasano, A.; Clemente, M.G.; De Simone, C.; Silano, M.; De Vincenzi, M.; Losito, I.; Gobbetti, M. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for celiac sprue probiotics and gluten intolerance. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2006, 1762, 80–93. [Google Scholar] [CrossRef]
- Rizzello, C.G.; De Angelis, M.; Di Cagno, R.; Camarca, A.; Silano, M.; Losito, I.; De Vincenzi, M.; De Bari, M.D.; Palmisano, F.; Maurano, F.; et al. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: New perspectives for celiac disease. Appl. Environ. Microbiol. 2007, 73, 4499–4507. [Google Scholar] [CrossRef]
- Smecuol, E.; Hwang, H.J.; Sugai, E.; Corso, L.; Chernavsky, A.C.; Bellavite, F.P.; González, A.; Vodánovich, F.; Moreno, M.L.; Vázquez, H.; et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J. Clin. Gastroenterol. 2013, 47, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Sarno, M.; Lania, G.; Cuomo, M.; Nigro, F.; Passannanti, F.; Budelli, A.; Fasano, F.; Troncone, R.; Auricchio, S.; Barone, M.V.; et al. Lactobacillus paracasei CBA L74 interferes with gliadin peptides entrance in Caco-2 cells. Int. J. Food Sci. Nutr. 2014, 65, 953–959. [Google Scholar] [CrossRef]
- Giorgi, A.; Cerrone, R.; Capobianco, D.; Filardo, S.; Mancini, P.; Zanni, F.; Fanelli, S.; Mastromarino, P.; Mosca, L. A probiotic preparation hydrolyzes gliadin and protects intestinal cells from the toxicity of pro-inflammatory peptides. Nutrients 2020, 12, 495. [Google Scholar] [CrossRef]
- Laparra, J.M.; Sanz, Y. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J. Cell Biochem. 2010, 109, 801–807. [Google Scholar] [CrossRef]
- Lindfors, K.; Blomqvist, T.; Juuti-Uusitalo, K.; Stenman, S.; Venäläinen, J.; Mäki, M.; Kaukinen, K. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin. Exp. Immunol. 2008, 152, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; Herrán, A.R.; Nistal, E.; Pérez-Andrés, J.; Vaquero, L.; Vivas, S.; Ruiz de Morales, J.M.G.; Albillos, S.M.; Casqueiro, J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: Isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol. Ecol. 2014, 88, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Caminero, A.; Galipeau, H.J.; McCarville, J.L.; Johnston, C.W.; Bernier, S.P.; Russell, A.K.; Jury, J.; Herran, A.R.; Casqueiro, J.; Tye-Din, J.A.; et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 2016, 151, 670–683. [Google Scholar] [CrossRef]
- Socha, P.; Mickowska, B.; Urminská, D.; Kačmárová, K. The use of different proteases to hydrolyze gliadins. J. Microbiol. Biotechnol. Food Sci. 2020, 9, 101–104. [Google Scholar] [CrossRef]
- Shan, L.; Marti, T.; Sollid, L.M.; Gray, G.M.; Khosla, C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. Biochem. J. 2004, 383, 311–318. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Calasso, M.; Archetti, G.; Rizzello, C.G. Novel insights on the functional/nutritional features of the sourdough fermentation. Int. J. Food Microbiol. 2019, 302, 103–113. [Google Scholar] [CrossRef]
- Kontakou, M.; Przemioslo, R.T.; Sturgess, R.P.; Limb, G.A.; Ellis, H.J.; Day, P.; Ciclitira, P.J. Cytokine mRNA expression in the mucosa of treated coeliac patients after wheat peptide challenge. Gut 1995, 37, 52–57. [Google Scholar] [CrossRef]
- Goel, G.; Tye-Din, J.A.; Qiao, S.W.; Russell, A.K.; Mayassi, T.; Ciszewski, C.; Sarna, V.K.; Wang, S.; Goldstein, K.E.; Dzuris, J.L.; et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci. Adv. 2019, 5, eaaw7756. [Google Scholar] [CrossRef]
- Makharia, G.K.D. Current and emerging therapy for celiac disease. Front. Med. 2014, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Freitag, T.L.; Rietdijk, S.; Junker, Y.; Popov, Y.; Bhan, A.K.; Kelly, C.P.; Terhorst, C.; Schuppan, D. Gliadin-primed CD4+ CD45RBlowCD25− T cells drive gluten-dependent small intestinal damage after adoptive transfer into lymphopenic mice. Gut 2009, 58, 1597–1605. [Google Scholar] [CrossRef] [PubMed]
- Freitag, T.L.; Loponen, J.; Messing, M.; Zevallos, V.; Andersson, L.C.; Sontag-Strohm, T.; Saavalainen, P.; Schuppan, D.; Salovaara, H.; Meri, S. Testing safety of germinated rye sourdough in a celiac disease model based on the adoptive transfer of prolamin-primed memory T cells into lymphopenic mice. Am. J. Physiol. Gastrointest. Liver. Physiol. 2014, 306, G526–G534. [Google Scholar] [CrossRef] [PubMed]
| Strains | Sandwich ELISA Assay (Residual Gluten) | Competitive ELISA Assay (Peptide Fragments) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 h | 16 h | 24 h | 36 h | 48 h | 6 h | 16 h | 24 h | 36 h | 48 h | ||
| Control | 1100 a ± 0.06 | 620 a ± 0.09 | 367 a ± 0.05 | 256 a ± 0.04 | 75 a ± 0.06 | 810 a ± 0.03 | 400 a ± 0.02 | 397 a ± 0.08 | 381 a ± 0.07 | 375 a ± 0.05 | |
| MC1 | Lp. plantarum DSM33370, DSM33363, DSM33364; Lc. paracasei DSM33373; Lv. brevis DSM33377; B. pumilus DSM33297, DSM33355, DSM33301 | 406 b ± 0.04 | 135 b ± 0.06 | 19 e ± 0.01 | 0 e | 0 e | 310 f ± 0.05 | 250 d ± 0.03 | 200 e ± 0.04 | 170 e ± 0.02 | 65 g ± 0.01 |
| MC2 | Lp. plantarum DSM33362, DSM33367, DSM33368; Lc. paracasei DSM33375; B. subtilis DSM33298; B. licheniformis DSM33354; B. megaterium DSM33300 | 346 c ± 0.07 | 121 b ± 0.03 | 15 e ± 0.01 | 0 e | 0 e | 332 f ± 0.05 | 226 ef ± 0.04 | 167 f ± 0.03 | 158 e ± 0.02 | 150 c ± 0.02 |
| MC3 | Lp. plantarum DSM33366, DSM33369; Ls. reuteri DSM33374; Lc. paracasei DSM33376; Ped. pentosaceus DSM33371; B. megaterium DSM33356; B. subtilis DSM33353 | 382 c ± 0.03 | 99 c ± 0.02 | 12 f ± 0.01 | 0 e | 0 e | 315 f ± 0.06 | 272 d ± 0.07 | 256 d ± 0.04 | 244 c ± 0.05 | 228 b ± 0.02 |
| MC4 | Lp. plantarum DSM33363, DSM33364; Lc. paracasei DSM33373; B. subtilis DSM33298; B. pumilus DSM33301 | 190 cd ± 0.05 | 0 g | 0 g | 0 e | 0 e | 399 e ± 0.08 | 233 e ± 0.07 | 112 g ± 0.05 | 0 j | 0 g |
| MC5 | Lv. brevis DSM33377; Ped. pentosaceus DSM33371; Lp. plantarum DSM33369; B. pumilus DSM33297; B. megaterium DSM33300 | 380 b ± 0.06 | 18 e ± 0.01 | 5 g ± 0.01 | 0 e | 0 e | 398 e ± 0.04 | 221 e ± 0.05 | 154 fg ± 0.03 | 46 i ± 0.02 | 0 g |
| MC6 | Lc. paracasei DSM33375; Lp. plantarum DSM33367, DSM33368; B. pumilus DSM33355; B. licheniformis DSM33354 | 350 c ± 0.06 | 15 e ± 0.02 | 2 g ± 0.01 | 0 e | 0 e | 404 e ± 0.06 | 245 de ± 0.05 | 100 g ± 0.08 | 79 h ± 0.04 | 0 g |
| MC7 | Lp. plantarum DSM33370, DSM33362, DSM33366; Ls. reuteri DSM33374; B. megaterium DSM33356; B. subtilis DSM33353 | 360 c ± 0.09 | 20 e ± 0.06 | 10 f ± 0.01 | 0 e | 0 e | 401 e ± 0.07 | 261 d ± 0.05 | 150 f ± 0.03 | 99 g ± 0.04 | 78 g ± 0.05 |
| MC8 | Lp. plantarum DSM33363, DSM33364; Lc. paracasei DSM33375; Ls. reuteri DSM33374; B. megaterium DSM33300; B. pumilus DSM33297 | 18 g ± 0.03 | 3 g ± 0.01 | 0 g | 0 e | 0 e | 323 f ± 0.08 | 228 e ± 0.06 | 218 e ± 0.05 | 157 e ± 0.06 | 0 g |
| MC9 | Lc. paracasei DSM33375; Lp. plantarum DSM33367; Ls. reuteri DSM33374; B. megaterium DSM33300; B. pumilus DSM33297; B. licheniformis DSM33354 | 60 f ± 0.04 | 12 f ± 0.01 | 0 g | 0 e | 0 e | 319 f ± 0.06 | 211 f ± 0.05 | 196 ef ± 0.03 | 195 de ± 0.07 | 152 c ± 0.02 |
| MC10 | Lp. plantarum DSM33363, DSM33364, DSM33370; Lv. brevis DSM33377; B. pumilus DSM33297; B. megaterium DSM33356 | 112 e ± 0.06 | 77 d ± 0.04 | 70 d ± 0.02 | 0 e | 0 e | 465 d ± 0.09 | 370 b ± 0.06 | 243 de ± 0.05 | 145 ef ± 0.04 | 97 f ± 0.03 |
| MC11 | Lp. plantarum DSM33368, DSM33362, DSM33367; Lc. paracasei DSM33375; B. megaterium DSM33300; B. subtilis DSM33353 | 221 d ± 0.05 | 89 c ± 0.07 | 69 d ± 0.06 | 50 d ± 0.04 | 43 d ± 0.03 | 512 c ± 0.06 | 367 b ± 0.08 | 340 b ± 0.09 | 300 b ± 0.06 | 123 de ± 0.05 |
| MC12 | Lp. plantarum DSM33366, DSM33369; Ls. reuteri DSM33374; Lc. paracasei DSM33376; Ped. pentosaceus DSM33371; B. pumilus DSM33297, DSM33355 | 145 e ± 0.06 | 110 c ± 0.05 | 89 c ± 0.03 | 75 c ± 0.02 | 63 b ± 0.03 | 601 b ± 0.09 | 312 c ± 0.06 | 289 c ± 0.07 | 288 b ± 0.05 | 143 cd ± 0.03 |
| MC13 | Lv. brevis DSM33377; Ped. pentosaceus DSM33371; Fr. sanfranciscensis DSM33379; B. megaterium DSM33300; B. pumilus DSM33297 | 163 de ± 0.06 | 122 b ± 0.04 | 82 c ± 0.02 | 45 d ± 0.03 | 0 e | 523 c ± 0.07 | 322 c ± 0.07 | 321 b ± 0.06 | 215 d ± 0.07 | 134 d ± 0.05 |
| MC14 | Lp. plantarum DSM33368; Lc. paracasei DSM33375; Fr. sanfranciscensis DSM33378; B. megaterium DSM33300; B. pumilus DSM33297; B. licheniformis DSM33354 | 234 d ± 0.08 | 135 b ± 0.07 | 120 b ± 0.07 | 108 b ± 0.05 | 56 c ± 0.03 | 587 b ± 0.09 | 333 c ± 0.09 | 256 d ± 0.08 | 211 d ± 0.08 | 167 c ± 0.07 |
| MC15 | Lp. plantarum DSM33362, DSM33366, DSM33370; Ls. reuteri DSM33374; Fr. sanfranciscensis DSM33378, DSM33379; B. licheniformis DSM33354; B. subtilis DSM33353 | 199 d ± 0.05 | 100 c ± 0.04 | 81 c ± 0.05 | 59 d ± 0.04 | 40 d ± 0.03 | 498 c ± 0.08 | 318 c ± 0.04 | 280 c ± 0.03 | 256 c ± 0.08 | 118 e ± 0.05 |
| MC16 | Lp. plantarum DSM33363, DSM33364; Lc. paracasei DSM33373; Ls. reuteri DSM33374; B. megaterium DSM33300; B. pumilus DSM33297, DSM33355 | 19 g ± 0.03 | 11 f ± 0.01 | 0 g | 0 e | 0 e | 280 g ± 0.06 | 200 f ± 0.05 | 50 h ± 0.03 | 10 j ± 0.01 | 0 g |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Angelis, M.; Siragusa, S.; Vacca, M.; Di Cagno, R.; Cristofori, F.; Schwarm, M.; Pelzer, S.; Flügel, M.; Speckmann, B.; Francavilla, R.; et al. Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions. Nutrients 2021, 13, 992. https://doi.org/10.3390/nu13030992
De Angelis M, Siragusa S, Vacca M, Di Cagno R, Cristofori F, Schwarm M, Pelzer S, Flügel M, Speckmann B, Francavilla R, et al. Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions. Nutrients. 2021; 13(3):992. https://doi.org/10.3390/nu13030992
Chicago/Turabian StyleDe Angelis, Maria, Sonya Siragusa, Mirco Vacca, Raffaella Di Cagno, Fernanda Cristofori, Michael Schwarm, Stefan Pelzer, Monika Flügel, Bodo Speckmann, Ruggiero Francavilla, and et al. 2021. "Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions" Nutrients 13, no. 3: 992. https://doi.org/10.3390/nu13030992
APA StyleDe Angelis, M., Siragusa, S., Vacca, M., Di Cagno, R., Cristofori, F., Schwarm, M., Pelzer, S., Flügel, M., Speckmann, B., Francavilla, R., & Gobbetti, M. (2021). Selection of Gut-Resistant Bacteria and Construction of Microbial Consortia for Improving Gluten Digestion under Simulated Gastrointestinal Conditions. Nutrients, 13(3), 992. https://doi.org/10.3390/nu13030992









